Published online Dec 24, 2025. doi: 10.5306/wjco.v16.i12.112161
Revised: September 13, 2025
Accepted: November 20, 2025
Published online: December 24, 2025
Processing time: 157 Days and 2.8 Hours
Laryngeal squamous cell carcinoma (LSCC) is a prevalent head and neck malignancy with suboptimal survival rates due to late detection and therapeutic resistance.
To investigate chaperonin-containing TCP1 subunit 3 (CCT3) expression and its clinical implications, and its effects on LSCC cell growth.
Systematic data on CCT3 mRNA expression were collected from biomedical databases, and integrated further based on the standardized mean difference and the summary receiver operating characteristic curve. Single-cell RNA-seq data were mined to validate the expression level of CCT3 mRNA. In-house immunohistochemistry was performed to explore the CCT3 protein levels of clinical LSCC samples and their relationship with clinical pa
CCT3 mRNA was significantly overexpressed in 269 LSCC tissues cases across multiple independent datasets (standardized mean difference = 32, area under the curve = 0.93); At the translational level, the in-house immunohistochemical analysis further demonstrated the consistent upregulation of CCT3 protein in 88 cases of LSCC samples (58 non-LSCC samples vs 30 LSCC samples, P = 1.4e-14). Analysis of clinical parameters showed no significant differences among subgroup. Functional characterization with clustered regularly interspaced short palindromic repeats--mediated gene knockout revealed that depletion of CCT3 potently suppressed LSCC cell viability in vitro. Gene set enrichment analysis indicated that CCT3 was markedly associated with several key oncogenic pathways, including extracellular matrix receptor interaction and cell cycle regulation pathways.
CCT3 upregulation in LSCC may influence cellular growth by regulating related pathways, indicating its potential as a biomarker and therapeutic target for LSCC.
Core Tip: In this study, single-cell RNA-seq and functional clustered regularly interspaced short palindromic repeats screening demonstrated the overexpression of chaperonin-containing TCP1 subunit 3 in laryngeal squamous cell carcinoma, thereby showing its role in promoting cell viability. Gene set enrichment analysis indicated that chaperonin-containing TCP1 subunit 3 is involved in extracellular matrix receptor interaction and cell cycle pathways, highlighting its potential as a biomarker and therapeutic target for laryngeal squamous cell carcinoma.
- Citation: Mo BY, Wen JY, Chen GQ, Ling JW, He H, Qin ZL, Tian FY, Li Q, Li B, Li JD, He RQ, Qin DY, Li ZY, Chen G, Mo CH, Chen C, Yin SH, Yang L. Expression patterns and clinical implications of chaperonin subunit 3 mRNA and protein in laryngeal squamous cell carcinoma. World J Clin Oncol 2025; 16(12): 112161
- URL: https://www.wjgnet.com/2218-4333/full/v16/i12/112161.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i12.112161
Laryngeal squamous cell carcinoma (LSCC) represents one of the most prevalent malignancies within the head and neck region, accounting for approximately 20% of head and neck cancer diagnoses globally[1,2]. Despite advances in mul
Attention has recently turned to the role of molecular chaperones in oncogenesis, particularly the chaperonin-containing TCP1 (CCT) complex, which facilitates the folding of nascent polypeptides such as actin, tubulin, and cell cycle regulators[10]. Among the subunits of this complex, CCT subunit 3 (CCT3) has emerged as a critical player in tumorigenesis, with studies indicating that its overexpression in malignancies such as hepatocellular carcinoma[11], lung cancer[12], and cervical cancer[13] predicts worse survival rates. CCT3 supports cancer cell survival by stabilizing oncoproteins and modulating signaling pathways (e.g., phosphoinositide 3-kinase/protein kinase B)[14-16]. However, its involvement in LSCC remains underexplored, leading to a gap in the understanding of how proteostatic mechanisms influence LSCC biology.
This study was conducted to determine the clinical relevance of CCT3 in case of LSCC by investigating its expression patterns, its effects on cell growth, and its putative functional contributions to LSCC occurrence and aggressiveness.
This study was designed to comprehensively evaluate the role of CCT3 mRNA and protein in LSCC through a com
A systematic search and collection of high throughput data was conducted to identify datasets containing expression levels of CCT3 mRNA in LSCC and non-cancerous laryngeal squamous epithelium tissues. A total of 269 cases were included after screening. The inclusion criteria for the data were as follows: Human subjects, primary LSCC tissues, and tissues containing control groups. The exclusion criterion was the absence of CCT3 expression values. After removing the batch effect using the sva package in R (v4.2.1), the differential CCT3 mRNA expression levels obtained were compared across multiple public repositories, namely The Gene Expression Omnibus, The Cancer Genome Atlas (TCGA), the Sequence Read Archive, and ArrayExpress. The standardized mean difference (SMD) was calculated to compare the comprehensive expression levels of CCT3. Heterogeneity among the studies was assessed using the I² statistic.
Publication bias was assessed using Begg’s test and Egger’s test to ensure the reliability of the SMD results based on State (v12.0) software. To explore the potential sources of heterogeneity, a sensitivity analysis was conducted by excluding individual studies one at a time to evaluate their impact on the overall results and to identify key studies that might affect the outcomes.
A summary receiver operating characteristic curve was generated using State (v12.0) software to evaluate the comprehensive discriminatory performance of CCT3 mRNA. As a result, the area under the curve (AUC), sensitivity, specificity, positive diagnostic likelihood ratio, and negative diagnostic likelihood ratio were calculated accordingly.
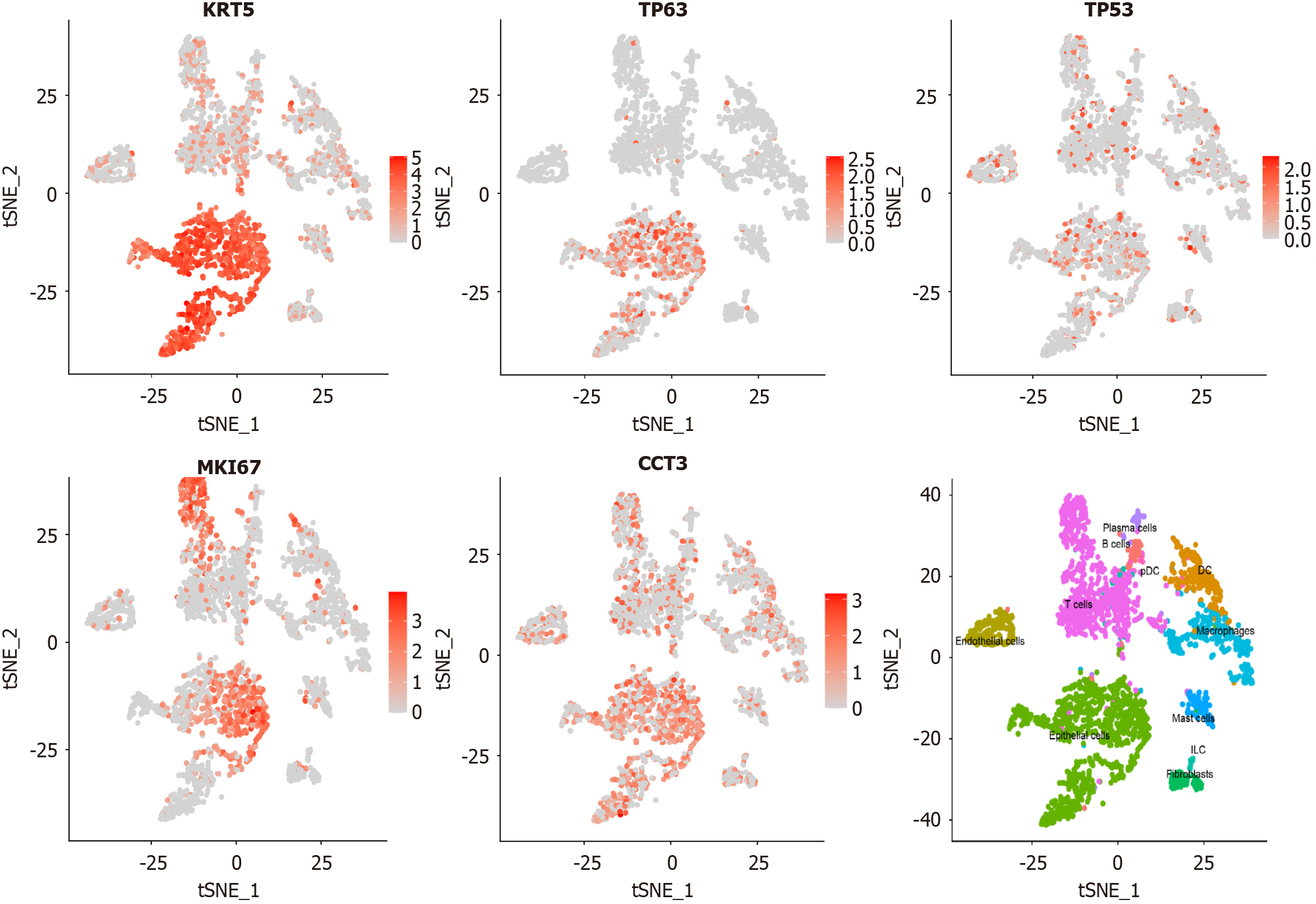
Single-cell RNA sequencing data from untreated LSCC tissues (GSM4546858[17]) were analyzed to investigate the expression patterns of CCT3 and LSCC-related marker genes. The cell selection criteria included cells with nCount_RNA > 1000, nFeature_RNA < 5000, percent.mt < 30%, and nFeature_RNA > 600. The seurat package (v5.0.0) was used to process the dataset. The filtered dataset was normalized using the LogNormalize method. Dimensionality reduction was performed via principal component analysis, t-distributed stochastic neighbor embedding, and uniform manifold approximation and projection. Based on CellTypist[18], the distribution and clustering of different cell types were visualized to elucidate the cellular heterogeneity and the role of CCT3 in various cell populations.
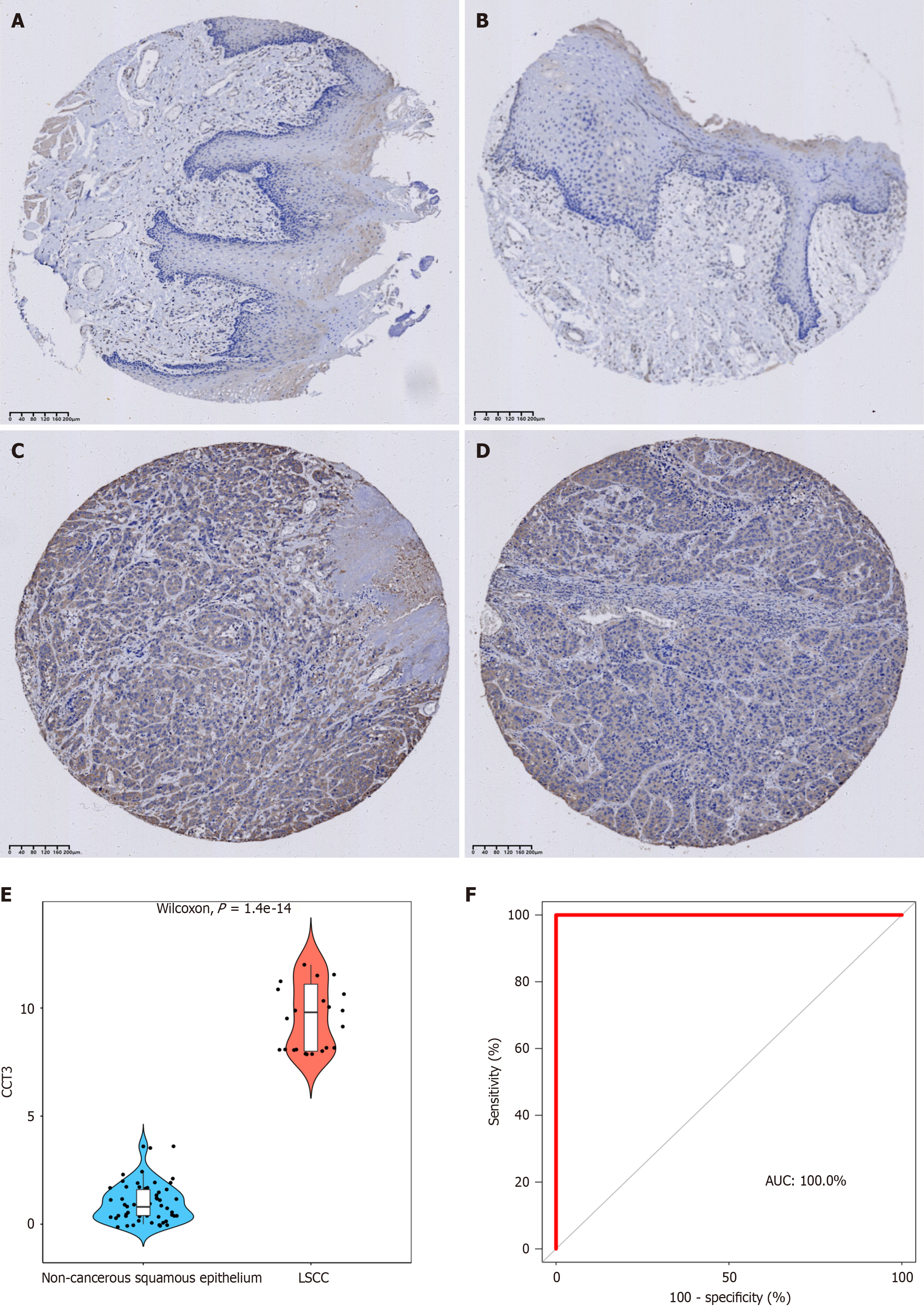
Immunohistochemical staining was performed on in-house LSCC and non-cancerous squamous epithelial tissue samples to validate the protein expression levels of CCT3. The clinical samples were purchased from Guilin Fanpu Biotech (Guangxi Zhuang Autonomous Region, China), a wholly owned subsidiary of Pantomics, Inc HNT961. Other path
SPSS 23.0 software was used to explore the relationship between the expression levels of CCT3 protein and clinical parameters in 30 patients from the immunohistochemistry (IHC) dataset, thereby elaborating on the clinical significance in greater depth.
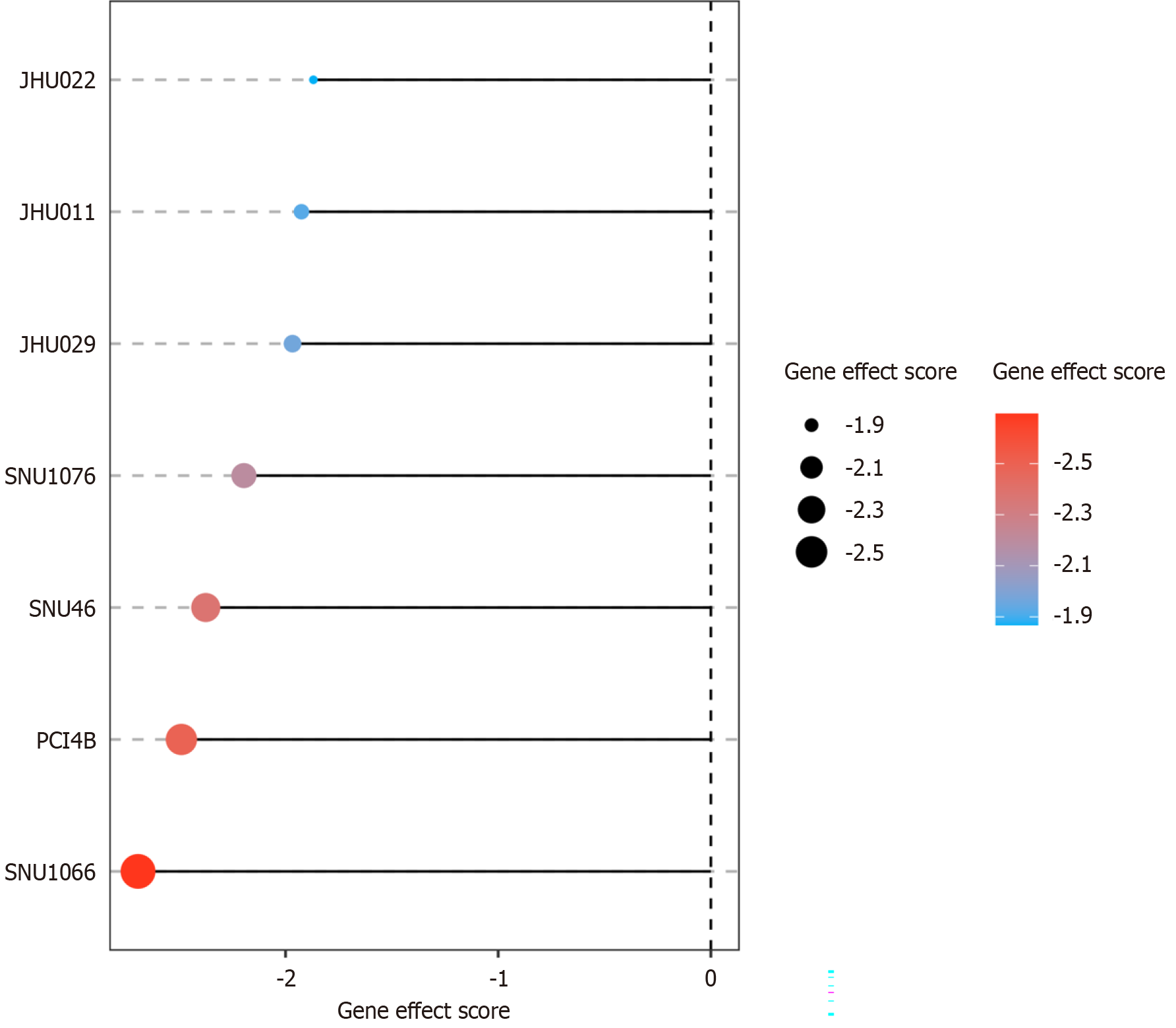
On DepMap[22,23], a clustered regularly interspaced short palindromic repeats(CRISPR)/CRISPR-associated protein 9 (CRISPR-Cas9) knockout screen analysis was conducted with seven LSCC cell lines, including JHU029, JHU022, JHU011, PCI4B, SNU1066, SNU46, and SNU1076. The effect of CCT3 knockout on cell growth was evaluated by measuring the negative effect scores: Which indicated the potential inhibitory role of CCT3 in LSCC cells. The result was plotted using the ggplot2 (v3.5.2).
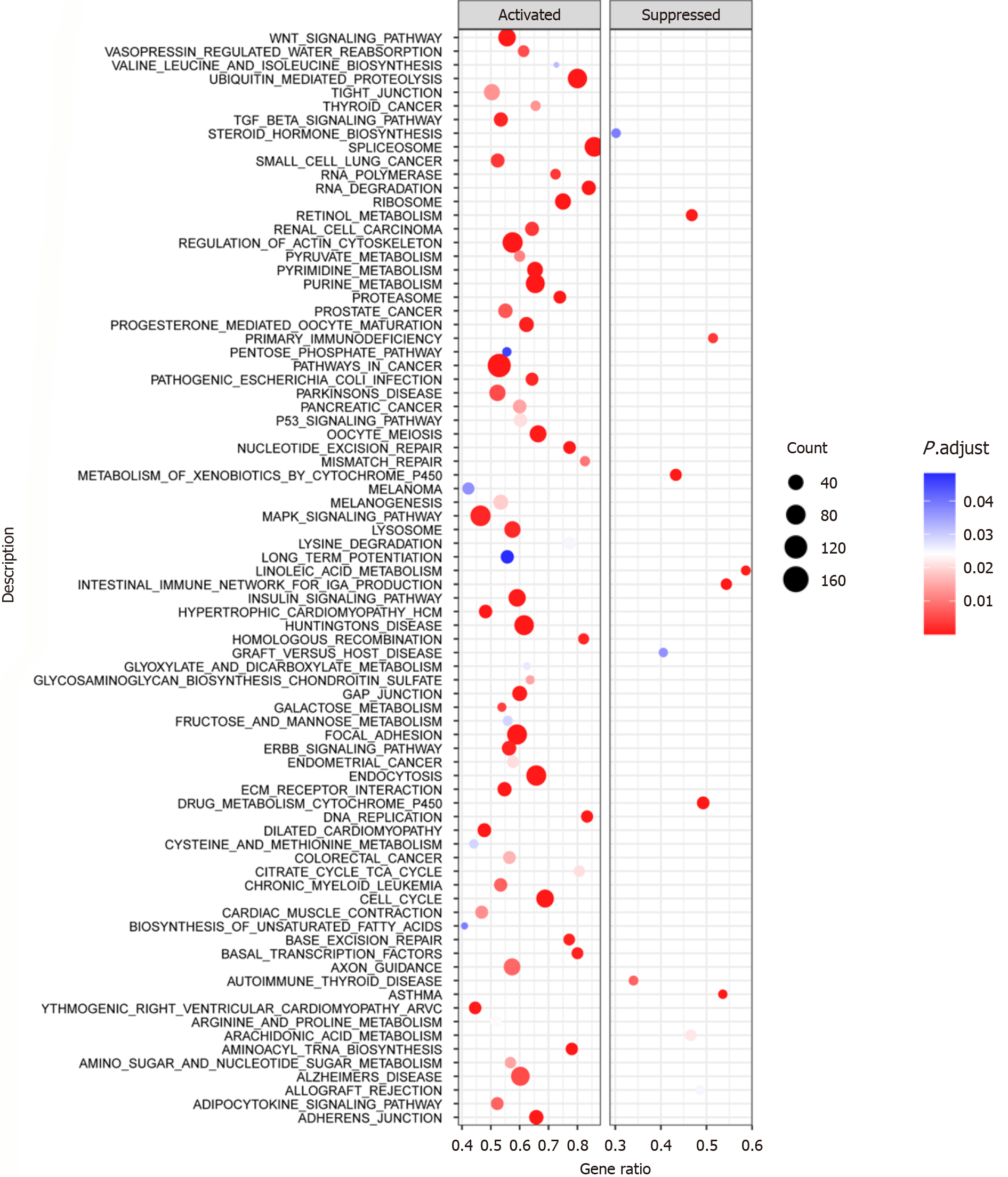
The clusterProfiler package[24-26] was used to perform a gene set enrichment analysis (GSEA) of genes sorted by log2FC from a differential analysis between the CCT3 high-expression group and CCT3 low-expression group, to screen out the signaling pathways enriched in CCT3 high expression. This analysis aimed to provide insights into the functional impact of CCT3 on cellular processes and its potential role in tumorigenesis.
To explore the downstream molecular mechanism by which CCT3 exerts its function, this study took CCT3 as the core node and constructed its related protein-protein interaction (PPI) network using the Search Tool for the Retrieval of Interacting Genes (STRING; Version 12.0) database, so as to observe the correlation and interaction between proteins.
Statistical analyses were performed using Stata (v12.0) and R (v4.2.1) software, with a P value < 0.05 considered statistically significant. SMD was calculated using R (v4.2.1) and the meta (v4.18-2). A random-effects model was applied for SMD calculation when significant heterogeneity was indicated by a P value < 0.05 and I² value > 50%; otherwise, a fixed-effects model was used.
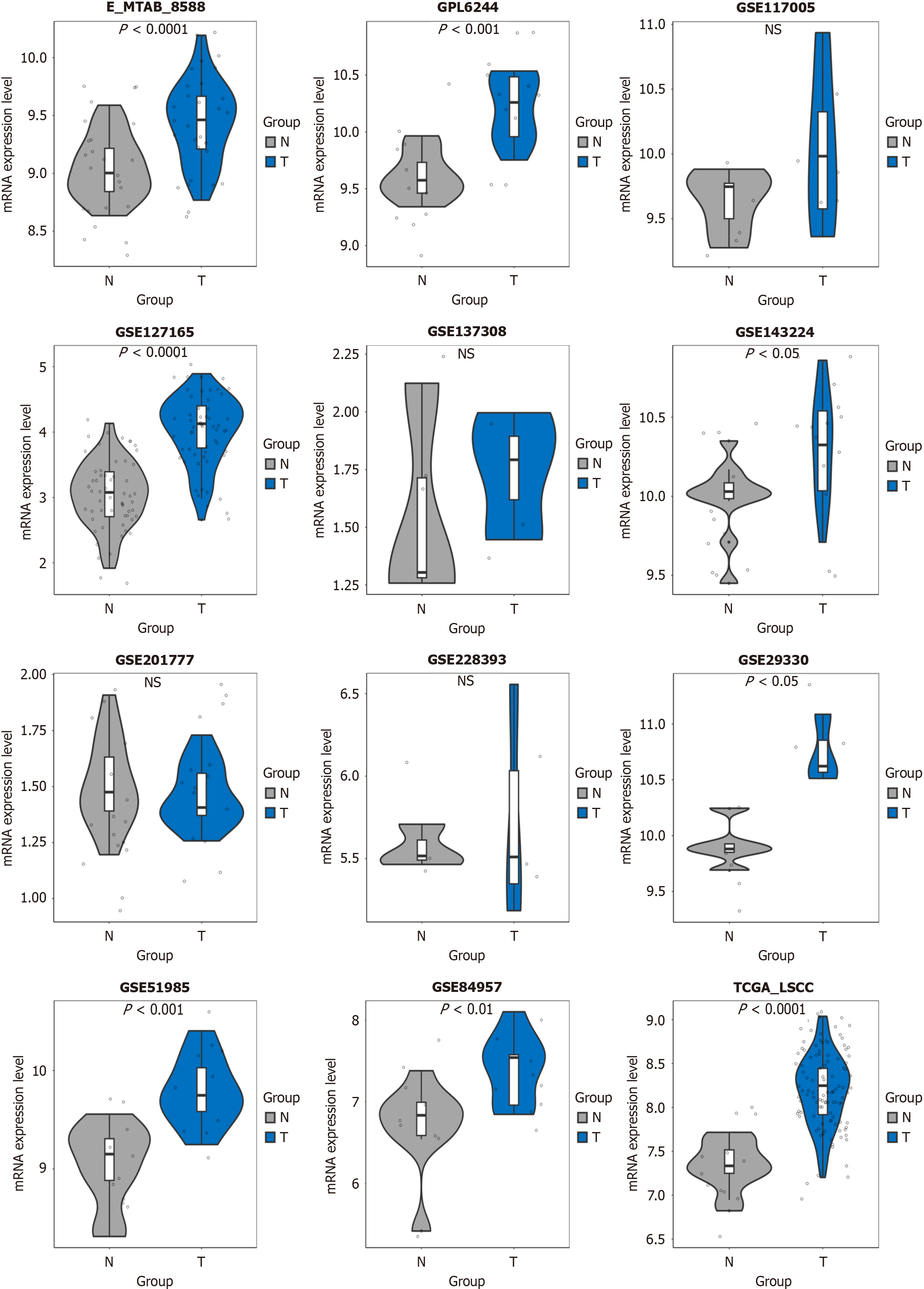
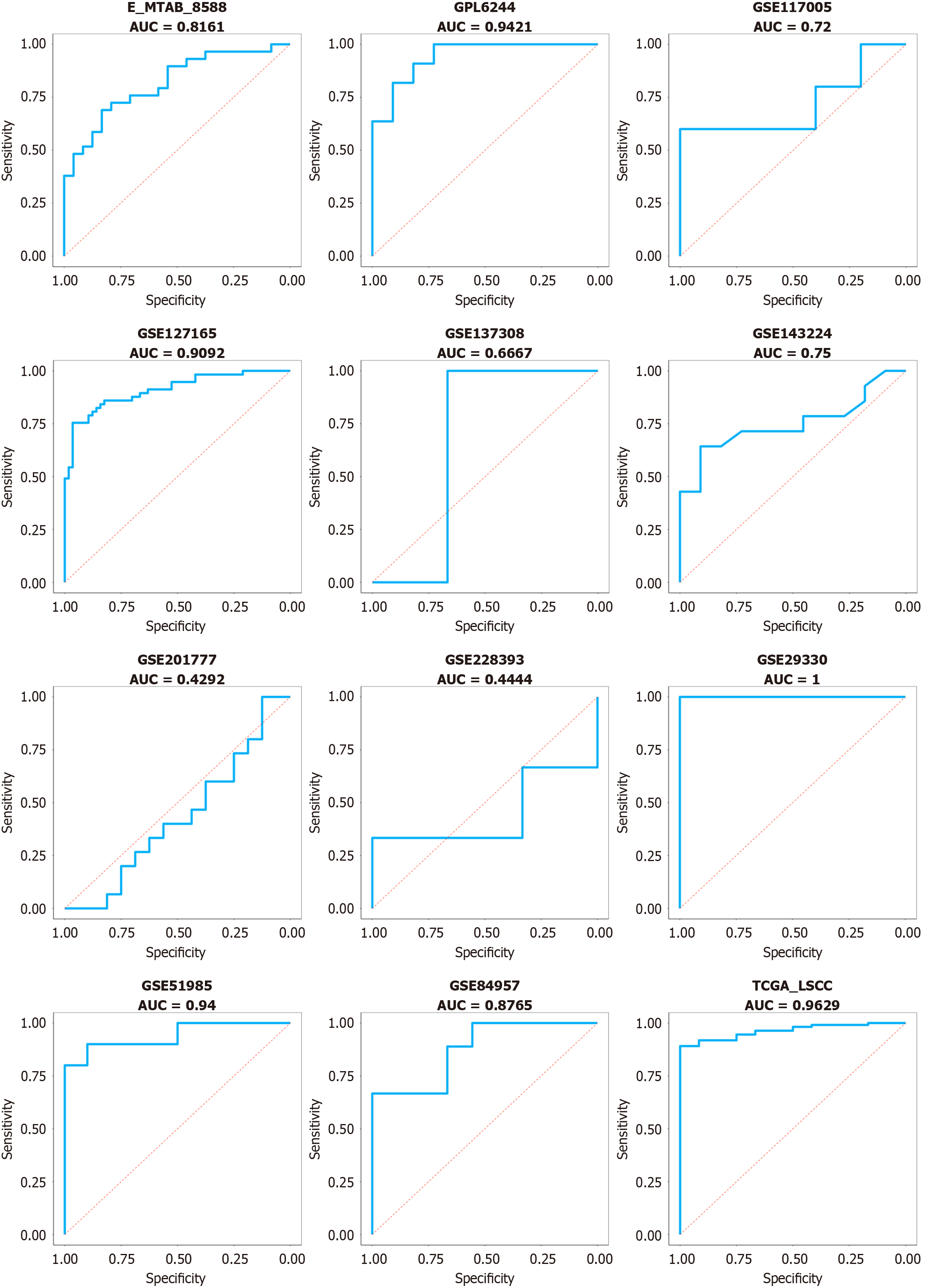
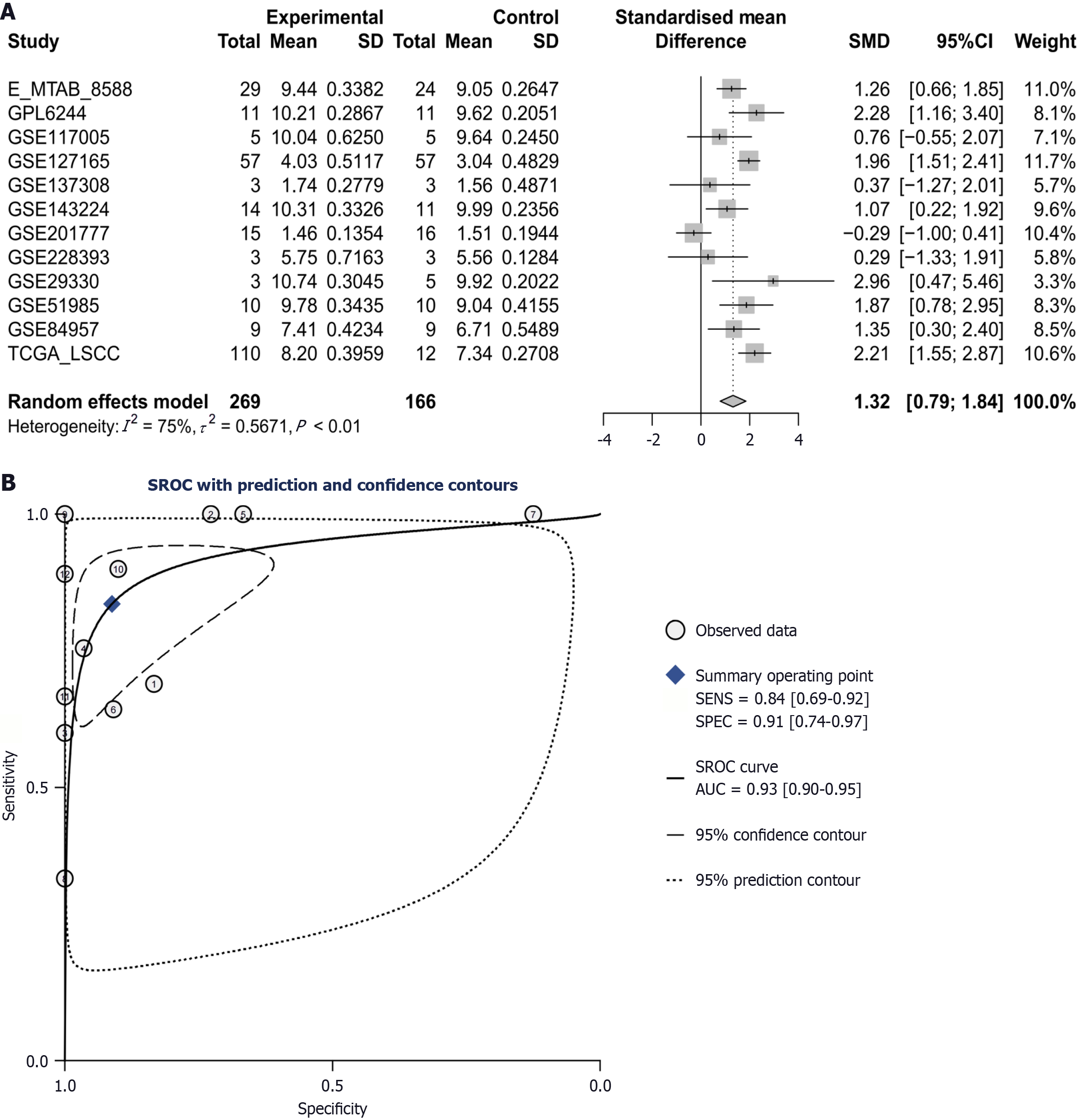
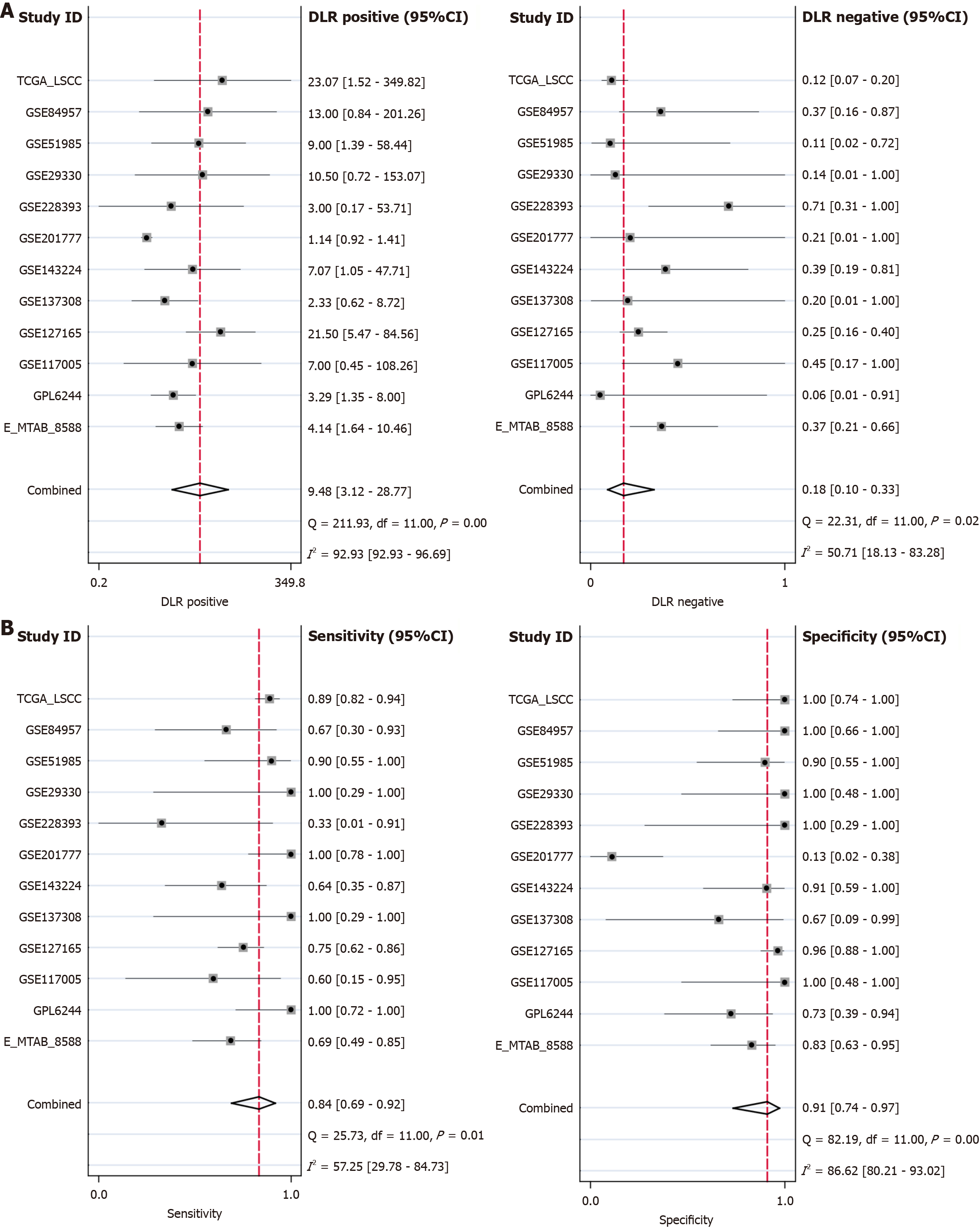
Following the above inclusion and exclusion processes, a total of 13 datasets were integrated into 12 platforms for inclusion (Figure 1). In most datasets (e.g., E-MTAB-8588, GPL6244, GSE127165, GSE143224, GSE29330, GSE51985, GSE84957, and TCGA-LSCC), the CCT3 mRNA levels in LSCC tissue were consistently higher than in non-cancerous tissue (P < 0.05; Figure 1). CCT3 mRNA exhibited a strong discriminative performance in GPL6244, GSE127165, GSE29330, GSE51985, and TCGA-LSCC (AUC > 0.90; Figure 2). After integration, the comprehensive mRNA level of CCT3 showed a notable upregulation in 269 cases of LSCC tissue compared to non-cancerous tissues (SMD = 1.32, 95%confidence interval: 0.79-1.84) (Figure 3A). Significant heterogeneity was observed among the studies (I2 = 75%, τ² = 0.5671, P < 0.01), so the random effects model was employed. The summary receiver operating characteristic curve analysis revealed a strong discriminative performance of CCT3 mRNA [AUC = 0.93, (0.90-0.95), with sensitivity and specificity values of 0.84 and 0.91, respectively (Figure 3B). The positive diagnostic likelihood ratio was 9.48, and the negative diagnostic likelihood ratio was 0.18, indicating a strong diagnostic potential for CCT3 (Figure 4).
Both Begg’s test and Egger’s test indicated the absence of publication bias (Supplementary Figure 1). The sensitivity analysis demonstrated that the overall results were robust, with no single study significantly affecting the overall estimates (Supplementary Figure 2).
Single-cell RNA sequencing analysis revealed distinct expression patterns of CCT3 and LSCC-related marker genes (P < 0.05; Figure 5). The plots illustrated the distribution and clustering of different cell types, highlighting the cellular heterogeneity in LSCC tissues and the potential role of CCT3 in specific cell populations.
The positive signals of the CCT3 protein were mainly localized in the cell membrane and the cytoplasm of non-cancerous epithelial cells and LSCC cells (LSCC samples = 30 vs non-LSCC samples = 58). A small number of positive signals appeared in the stroma, in accordance with the findings from scRNA-seq (Figure 5). Hence, the in-house immunohistochemical staining confirmed that CCT3 protein expression was significantly higher in LSCC tissues compared to non-cancerous squamous epithelium (median of non-LSCC samples = 9.8 vs median of LSCC samples = 0.8, P < 0.05, Figure 6).
Thirty patients from the IHC dataset were enrolled as subjects in this study. The results showed that there was no statistical significance between the protein levels and general clinical parameters such as age, gender, pathological grade, and tumor node metastasis stage (Table 1). This indicates that CCT3 was overexpressed in LSCC tissue, with no diff
| Clinical features | CCT3 protein expression | t or F (ANOVA test) | P value | ||
| Number | Mean | SD | |||
| Age | 0.538 | 0.5951 | |||
| > 55 | 12 | 9.500 | 1.698 | ||
| ≤ 55 | 18 | 9.822 | 1.548 | ||
| Sex | 0.448 | 0.6573 | |||
| Male | 28 | 9.729 | 1.608 | ||
| Female | 2 | 9.200 | 1.697 | ||
| Clinical stage | 0.537 | 0.7097 | |||
| Stage I | 12 | 9.533 | 1.533 | ||
| Stage II | 8 | 9.900 | 1.720 | ||
| Stage III | 4 | 8.900 | 1.800 | ||
| Stage I-II | 2 | 9.800 | 0.0283 | ||
| Stage II-III | 4 | 10.050 | 1.915 | ||
| TNM-N | |||||
| N1 | 24 | 9.683 | 1.565 | -0.678 | 0.9465 |
| N2 | 6 | 9.733 | 1.836 | ||
| TNM-T | 2.533 | 0.0982 | |||
| T1 | 2 | 9.800 | 1.979 | ||
| T2 | 26 | 9.508 | 1.519 | ||
| T3 | 2 | 12.000 | 0.000 | ||
The CRISPR-Cas9 knockout screen in the LSCC cell lines showed a highly negative effect score, indicating that CCT3 knockout induced a strongly inhibitory effect on LSCC cell growth (gene effect score < -1) (Figure 7).
With a statistical threshold of normalized enrichment score ≥ 1.39 and false discovery rate ≤ 0.048, GSEA identified several biological pathways enriched in the CCT3 overexpression group of LSCC tissues. The results provide insights into the potential functional impact of CCT3 on cellular processes, such as ECM receptor interaction, the cell cycle, pathways in cancer, DNA replication, the Wnt signaling pathway, basal transcription factors, biosynthesis of unsaturated fatty acids, epidermal growth factor receptor family, transforming growth factor-beta, adipocytokine, and mitogen-activated protein kinase signaling pathway (Figure 8).
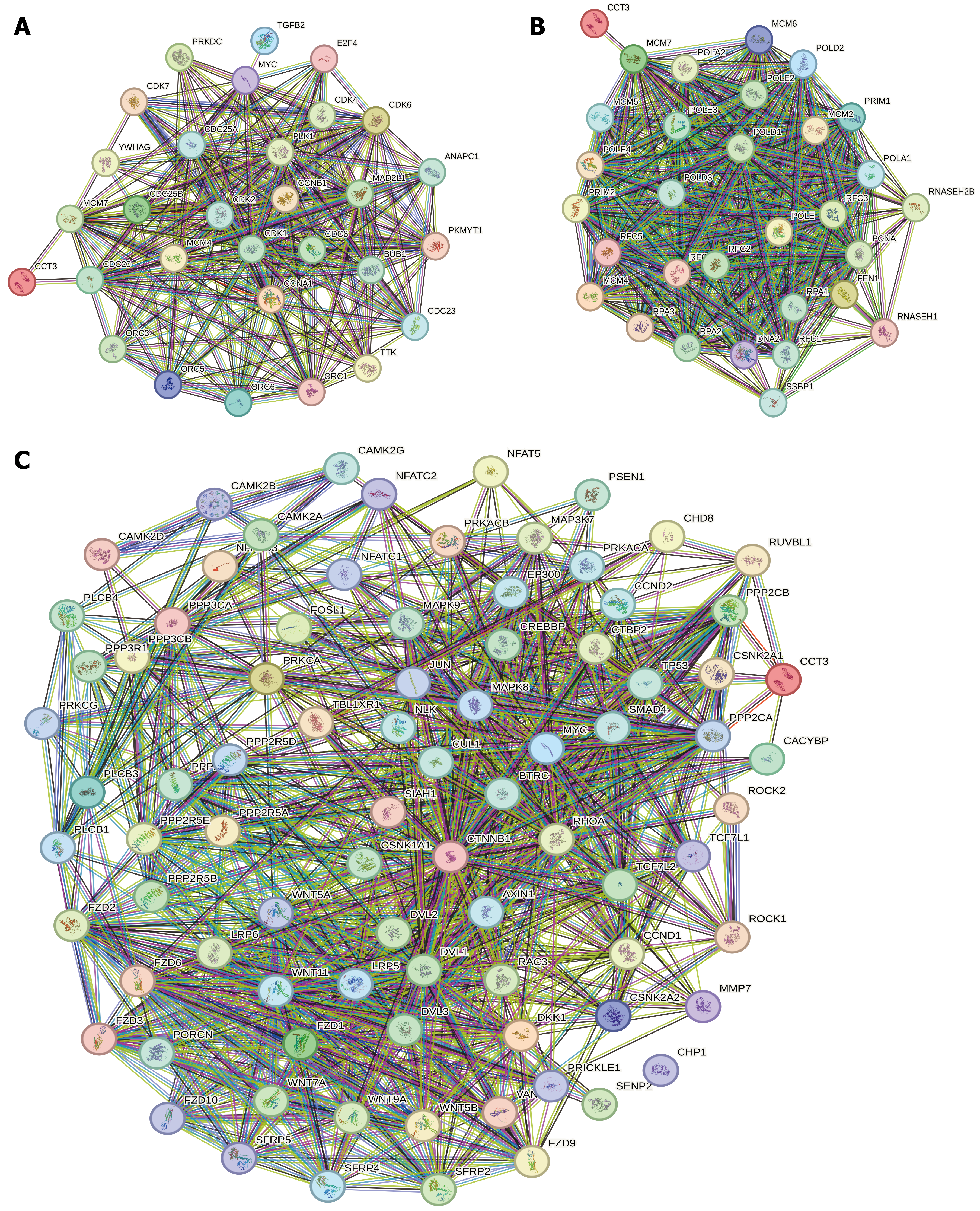
The STRING database was used to perform PPI network analysis of the CCT3 gene and its pathway-related genes. The core objective was to leverage the database’s interaction prediction and visualization functionalities to identify the key interacting genes of CCT3. Ultimately, six genes-CDC20, MCM7, PPP2CA, PPP2CB, CSNK2A1, RUVBL1, and CACYBP were screened out, and these genes exhibited stable associations with CCT3 (Figure 9).
This study investigated into the multifaceted role of CCT3 in LSCC - a malignancy that remains a formidable challenge despite therapeutic advancements. The findings highlight the discriminatory and functional significance of CCT3 in LSCC progression, thereby underscoring its potential as a biomarker and therapeutic target. However, existing studies have not yet fully explored the mechanism underlying the role of CCT3 in LSCC.
The consistent overexpression of CCT3 mRNA in LSCC tissues across various datasets suggested a fundamental role in tumor biology. The robust discriminatory performance of CCT3 mRNA, as evidenced by the high AUC value and favorable sensitivity and specificity metrics, indicated its potential as a reliable biomarker for distinguishing LSCC tissues from non-cancerous tissues. This was further corroborated by in-house immunohistochemical validation, which confirmed elevated CCT3 protein expression in LSCC tissues. Meanwhile, the results of clinical parameter analysis further confirmed this point. Despite the small sample size and certain limitations of this study, the protein’s discriminatory performance showed promising accuracy, indicating that CCT3 may hold potential as a useful tool in clinical settings for the early detection and diagnosis of LSCC.
The functional analysis via CRISPR-Cas9 knockout screening revealed a critical role for CCT3 in sustaining LSCC cell viability and proliferation. The significant negative effect scores observed in the LSCC cell lines upon CCT3 knockout implied that this chaperone subunit might be essential for tumor cell survival. This finding aligned with the broader understanding of molecular chaperones in cancer, where they often stabilize oncoproteins and modulate the signaling pathways crucial for tumor progression[27-30]. Targeting CCT3 could potentially disrupt these processes, offering a novel therapeutic strategy for cancers[31,32], especially LSCC.
The single-cell RNA sequencing analysis provided a granular view of CCT3’s expression patterns within the heterogeneous cellular landscape of LSCC tissue. The distinct clustering and distribution of CCT3 highlighted its potential involvement in specific cell populations that contribute to tumor aggressiveness. This cellular heterogeneity is a hallmark of many cancers and underscores the complexity of targeting LSCC. Understanding the specific cell types and pathways influenced by CCT3 could pave the way for more precise therapeutic interventions.
GSEA further elucidated the biological pathways enriched in LSCC samples with high CCT3 expression. These CCT3-participating pathways, including cell cycle regulation[33], DNA replication[34], the WNT signaling pathway[35], and various signaling cascades, are integral to cancer development and progression. The enrichment of these pathways suggested that CCT3 might orchestrate a complex interplay of cellular processes that drive LSCC. This mechanistic insight is crucial for developing targeted therapies that can disrupt these pathways and inhibit tumor growth. Fur
Overall, this study integrated multiple lines of evidence to establish CCT3 as a key player in LSCC. The findings highlighted the potential of CCT3 as a discriminatory biomarker and therapeutic target, offering new avenues for improving the management of this challenging malignancy. Future research should focus on validating these findings in larger cohorts and exploring the detailed molecular mechanisms by which CCT3 influences LSCC biology. Additionally, the development of CCT3-targeted therapies and their evaluation in preclinical and clinical settings could significantly advance the treatment landscape for LSCC.
In summary, this study verified the overexpression of CCT3 at both the mRNA and protein levels in LSCC tissues. Additionally, the AUC value indicated high diagnostic performance, suggesting CCT3’s potential as a diagnostic biomarker for LSCC. After CRISPR-Cas9 knockout screening, the viability of LSCC cells was impaired, which confirmed that CCT3 is functionally involved in the growth of LSCC cells. GSEA revealed that CCT3 may promote the progression of LSCC by regulating multiple oncogenic pathways, such as extracellular matrix receptor interaction and cell cycle regulation, further demonstrating its potential as a therapeutic target.
The authors of this paper gratefully acknowledge the Clinical Pathology and Computational Pathology techniques provided by Chen G Lab at the First Affiliated Hospital of Guangxi Medical University. Additionally, we express our sincere appreciation for the public high throughput databases involved in this study.
| 1. | Qi H. Role and research progress of hematological markers in laryngeal squamous cell carcinoma. Diagn Pathol. 2023;18:50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 2. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 12410] [Article Influence: 6205.0] [Reference Citation Analysis (6)] |
| 3. | Ding W, Cai W, Wang H. P53 and pRB induction improves response to radiation therapy in HPV-positive laryngeal squamous cell carcinoma. Clinics (Sao Paulo). 2024;79:100415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 4. | Wang Q, Tan L, Lv Y, Yu T, Chang Y, Liu J, Sun Y. MiR-125a-5p regulates the radiosensitivity of laryngeal squamous cell carcinoma via HK2 targeting through the DDR pathway. Front Oncol. 2024;14:1438722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 5. | Peng J, Luo G, Yu Y, Ning K, Liu X. Retrospective assessment of neoadjuvant camrelizumab combined with induction chemotherapy: efficacy in laryngeal preservation for advanced hypopharyngeal and laryngeal squamous cell carcinoma. Cancer Immunol Immunother. 2024;73:54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 6. | Xu L, Li W, Liu D, Cao J, Ge J, Liu X, Wang Y, Teng Y, Liu P, Guo X, He C, Liu M, Tian L. ANXA3-Rich Exosomes Derived from Tumor-Associated Macrophages Regulate Ferroptosis and Lymphatic Metastasis of Laryngeal Squamous Cell Carcinoma. Cancer Immunol Res. 2024;12:614-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 7. | Prasad A, Carey RM, Panara K, Rajasekaran K, Cannady SB, Newman JG, Brant JA, Brody RM. Nodal metastasis in surgically treated laryngeal squamous cell carcinoma. Head Neck. 2023;45:2303-2312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 8. | Huang Q, Chen J, Huang Y, Xiong Y, Zhou J, Zhang Y, Lu M, Hu W, Zheng F, Zheng C. The prognostic role of coagulation markers in the progression and metastasis of laryngeal squamous cell carcinoma. BMC Cancer. 2023;23:901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 9. | Yang WJ, Zhao HP, Yu Y, Wang JH, Guo L, Liu JY, Pu J, Lv J. Updates on global epidemiology, risk and prognostic factors of gastric cancer. World J Gastroenterol. 2023;29:2452-2468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 343] [Cited by in RCA: 309] [Article Influence: 103.0] [Reference Citation Analysis (14)] |
| 10. | Han W, Jin M, Liu C, Zhao Q, Wang S, Wang Y, Yin Y, Peng C, Wang Y, Cong Y. Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT. Sci Adv. 2023;9:eade1207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 11. | Han C, Chen J, Huang J, Zhu R, Zeng J, Yu H, He Z. Single-cell transcriptome analysis reveals the metabolic changes and the prognostic value of malignant hepatocyte subpopulations and predict new therapeutic agents for hepatocellular carcinoma. Front Oncol. 2023;13:1104262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 12. | Huang J, Hu B, Yang Y, Liu H, Fan X, Zhou J, Chen L. Integrated analyzes identify CCT3 as a modulator to shape immunosuppressive tumor microenvironment in lung adenocarcinoma. BMC Cancer. 2023;23:241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 13. | Li M, Zeng J, Chang Y, Lv L, Ye G. CCT3 as a Diagnostic and Prognostic Biomarker in Cervical Cancer. Crit Rev Eukaryot Gene Expr. 2023;33:17-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 14. | Dong Y, Lu S, Wang Z, Liu L. CCTs as new biomarkers for the prognosis of head and neck squamous cancer. Open Med (Wars). 2020;15:672-688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 15. | Chen S, Tian Y, Ju A, Li B, Fu Y, Luo Y. Suppression of CCT3 Inhibits Tumor Progression by Impairing ATP Production and Cytoplasmic Translation in Lung Adenocarcinoma. Int J Mol Sci. 2022;23:3983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 16. | Zhu H, Liu Q, Meng Q, Zhang L, Ju S, Lang J, Zhu D, Chen Y, Aishan N, Ouyang X, Zhang S, Jin L, Xiao L, Wang L, Li L, Ji F. CCT3/ACTN4/TFRC axis protects hepatocellular carcinoma cells from ferroptosis by inhibiting iron endocytosis. J Exp Clin Cancer Res. 2024;43:245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 17. | Song L, Zhang S, Yu S, Ma F, Wang B, Zhang C, Sun J, Mao X, Wei L. Cellular heterogeneity landscape in laryngeal squamous cell carcinoma. Int J Cancer. 2020;147:2879-2890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 18. | Domínguez Conde C, Xu C, Jarvis LB, Rainbow DB, Wells SB, Gomes T, Howlett SK, Suchanek O, Polanski K, King HW, Mamanova L, Huang N, Szabo PA, Richardson L, Bolt L, Fasouli ES, Mahbubani KT, Prete M, Tuck L, Richoz N, Tuong ZK, Campos L, Mousa HS, Needham EJ, Pritchard S, Li T, Elmentaite R, Park J, Rahmani E, Chen D, Menon DK, Bayraktar OA, James LK, Meyer KB, Yosef N, Clatworthy MR, Sims PA, Farber DL, Saeb-Parsy K, Jones JL, Teichmann SA. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science. 2022;376:eabl5197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 641] [Article Influence: 160.3] [Reference Citation Analysis (0)] |
| 19. | Li H, Huang HQ, Huang ZG, He RQ, Fang YY, Song R, Luo JY, Zeng DT, Qin K, Wei DM, Chen G. Potential regulatory mechanism and clinical significance of synaptotagmin binding cytoplasmic RNA interacting protein in colorectal cancer. World J Clin Oncol. 2024;15:1412-1427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 20. | Li JD, He RQ, Dang YW, Huang ZG, Xiong DD, Zhang L, Du XF, Chen G. Unveiling expression patterns, mechanisms, and therapeutic opportunities of transmembrane protein 106C: From pan-cancers to hepatocellular carcinoma. World J Gastrointest Oncol. 2025;17:92437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 21. | Qin K, Xiong DD, Qin Z, Li MJ, Li Q, Huang ZG, Tang YX, Li JD, Zhan YT, He RQ, Luo J, Wang HQ, Zhang SQ, Chen G, Wei DM, Dang YW. Overexpression and clinicopathological significance of zinc finger protein 71 in hepatocellular carcinoma. World J Hepatol. 2025;17:101914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Arafeh R, Shibue T, Dempster JM, Hahn WC, Vazquez F. The present and future of the Cancer Dependency Map. Nat Rev Cancer. 2025;25:59-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 115] [Reference Citation Analysis (1)] |
| 23. | Corsello SM, Nagari RT, Spangler RD, Rossen J, Kocak M, Bryan JG, Humeidi R, Peck D, Wu X, Tang AA, Wang VM, Bender SA, Lemire E, Narayan R, Montgomery P, Ben-David U, Garvie CW, Chen Y, Rees MG, Lyons NJ, McFarland JM, Wong BT, Wang L, Dumont N, O'Hearn PJ, Stefan E, Doench JG, Harrington CN, Greulich H, Meyerson M, Vazquez F, Subramanian A, Roth JA, Bittker JA, Boehm JS, Mader CC, Tsherniak A, Golub TR. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat Cancer. 2020;1:235-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 581] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 24. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 24784] [Article Influence: 1770.3] [Reference Citation Analysis (2)] |
| 25. | Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 5856] [Article Influence: 1171.2] [Reference Citation Analysis (0)] |
| 26. | Xu S, Hu E, Cai Y, Xie Z, Luo X, Zhan L, Tang W, Wang Q, Liu B, Wang R, Xie W, Wu T, Xie L, Yu G. Using clusterProfiler to characterize multiomics data. Nat Protoc. 2024;19:3292-3320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 557] [Article Influence: 278.5] [Reference Citation Analysis (0)] |
| 27. | Liu W, Lu Y, Yan X, Lu Q, Sun Y, Wan X, Li Y, Zhao J, Li Y, Jiang G. Current understanding on the role of CCT3 in cancer research. Front Oncol. 2022;12:961733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 28. | Wang K, He J, Tu C, Xu H, Zhang X, Lv Y, Song C. Upregulation of CCT3 predicts poor prognosis and promotes cell proliferation via inhibition of ferroptosis and activation of AKT signaling in lung adenocarcinoma. BMC Mol Cell Biol. 2022;23:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 29. | Wang J, Guan X, Shang N, Wu D, Liu Z, Guan Z, Zhang Z, Jin Z, Wei X, Liu X, Song M, Zhu W, Dai G. Dysfunction of CCT3-associated network signals for the critical state during progression of hepatocellular carcinoma. Biochim Biophys Acta Mol Basis Dis. 2024;1870:167054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 30. | Macario AJL, Conway de Macario E. Chaperonins in cancer: Expression, function, and migration in extracellular vesicles. Semin Cancer Biol. 2022;86:26-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 31. | Temiz E, Koyuncu İ, Sahin E. CCT3 suppression prompts apoptotic machinery through oxidative stress and energy deprivation in breast and prostate cancers. Free Radic Biol Med. 2021;165:88-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 32. | Liu W, Zhang X, Chen C, Li Y, Yang C, Han Z, Jiang G, Liu Y. Suppression of CCT3 inhibits melanoma cell proliferation by downregulating CDK1 expression. J Cancer. 2022;13:1958-1971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 33. | Shi H, Zhang Y, Wang Y, Fang P, Liu Y, Li W. Restraint of chaperonin containing T-complex protein-1 subunit 3 has antitumor roles in non-small cell lung cancer via affection of YAP1. Toxicol Appl Pharmacol. 2022;439:115926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 34. | Hou JY, Wu HY, He RQ, Lin P, Dang YW, Chen G. Clinical and prognostic value of chaperonin containing T-complex 1 subunit 3 in hepatocellular carcinoma: A Study based on microarray and RNA-sequencing with 4272 cases. Pathol Res Pract. 2019;215:177-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Qian T, Cui L, Liu Y, Cheng Z, Quan L, Zeng T, Huang W, Dai Y, Chen J, Liu L, Chen J, Pang Y, Wu G, Ye X, Shi J, Fu L, Si C. High expression of chaperonin-containing TCP1 subunit 3 may induce dismal prognosis in multiple myeloma. Pharmacogenomics J. 2020;20:563-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Jeong SM, Bui QT, Kwak M, Lee JY, Lee PC. Targeting Cdc20 for cancer therapy. Biochim Biophys Acta Rev Cancer. 2022;1877:188824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 37. | Zhang XY, Tang LZ, Ren BG, Yu YP, Nelson J, Michalopoulos G, Luo JH. Interaction of MCM7 and RACK1 for activation of MCM7 and cell growth. Am J Pathol. 2013;182:796-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 38. | Brewer A, Sathe G, Pflug BE, Clarke RG, Macartney TJ, Sapkota GP. Mapping the substrate landscape of protein phosphatase 2A catalytic subunit PPP2CA. iScience. 2024;27:109302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 39. | Sun S, Wang Y, Wang J, Bi J. Wnt pathway-related three-mRNA clinical outcome signature in bladder urothelial carcinoma: computational biology and experimental analyses. J Transl Med. 2021;19:409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 40. | Zhang X, Cui D, Sun W, Yang G, Wang W, Mi C. Regulation of the βcatenin/LEF1 pathway by the siRNA knockdown of RUVBL1 expression inhibits breast cancer cell proliferation, migration and invasion. Oncol Rep. 2025;53:22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 41. | Zhang L, Yang J, Huang Y, You T, Huang Q, Shen X, Xue X, Feng S. Comprehensive landscape of gastric cancer-targeted therapy and identification of CSNK2A1 as a potential target. Heliyon. 2024;10:e36205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 42. | Mo B, Luo B, Wu Y. Pan-analysis reveals CACYBP to be a novel prognostic and predictive marker for multiple cancers. Am J Transl Res. 2024;16:12-26. [PubMed] [DOI] [Full Text] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/