©The Author(s) 2025.
World J Clin Oncol. Dec 24, 2025; 16(12): 112161
Published online Dec 24, 2025. doi: 10.5306/wjco.v16.i12.112161
Published online Dec 24, 2025. doi: 10.5306/wjco.v16.i12.112161
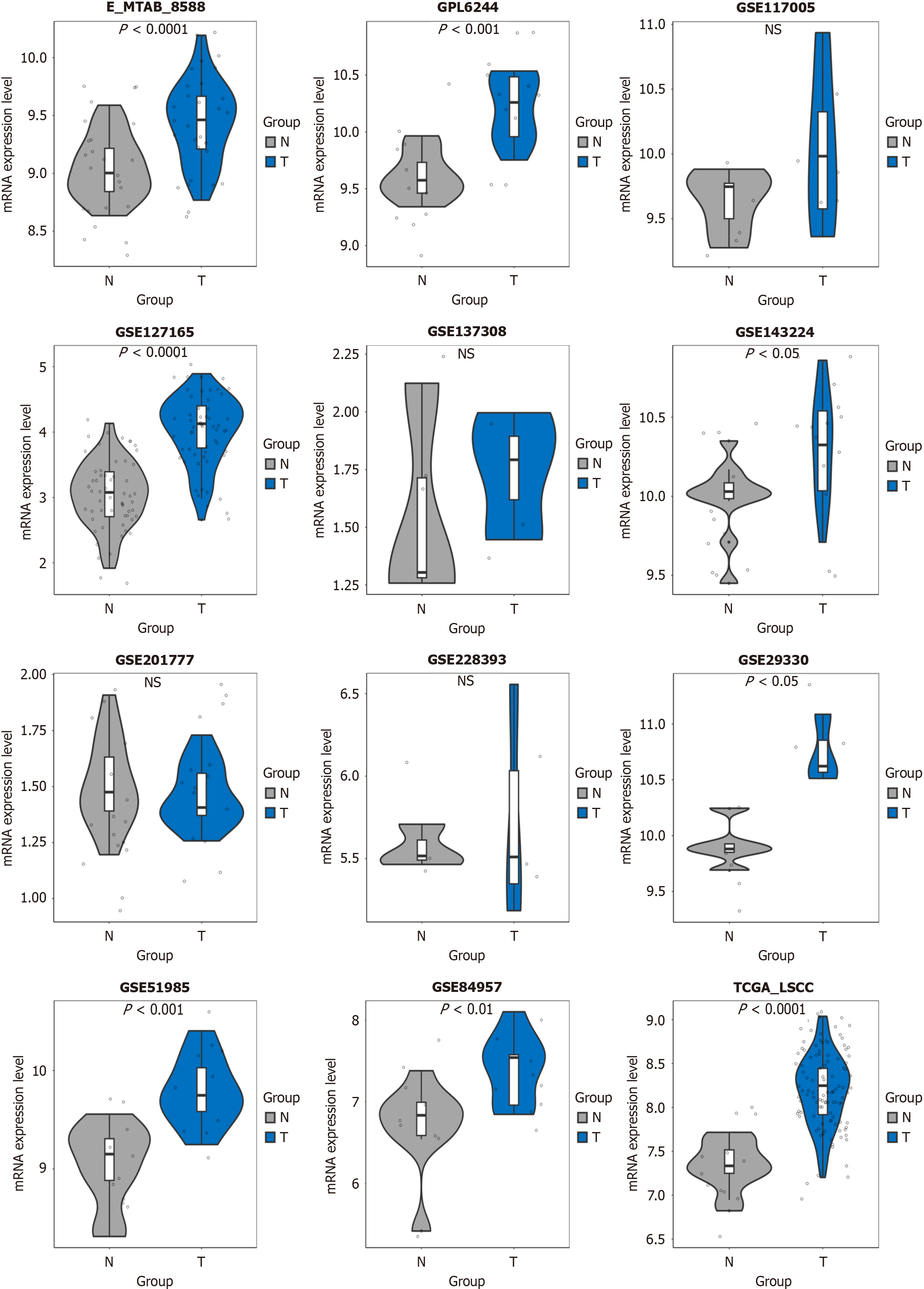
Figure 1 Chaperonin-containing TCP1 subunit 3 mRNA levels in laryngeal squamous cell carcinoma are consistently higher compared to non-cancerous tissue.
NS: Not significant.
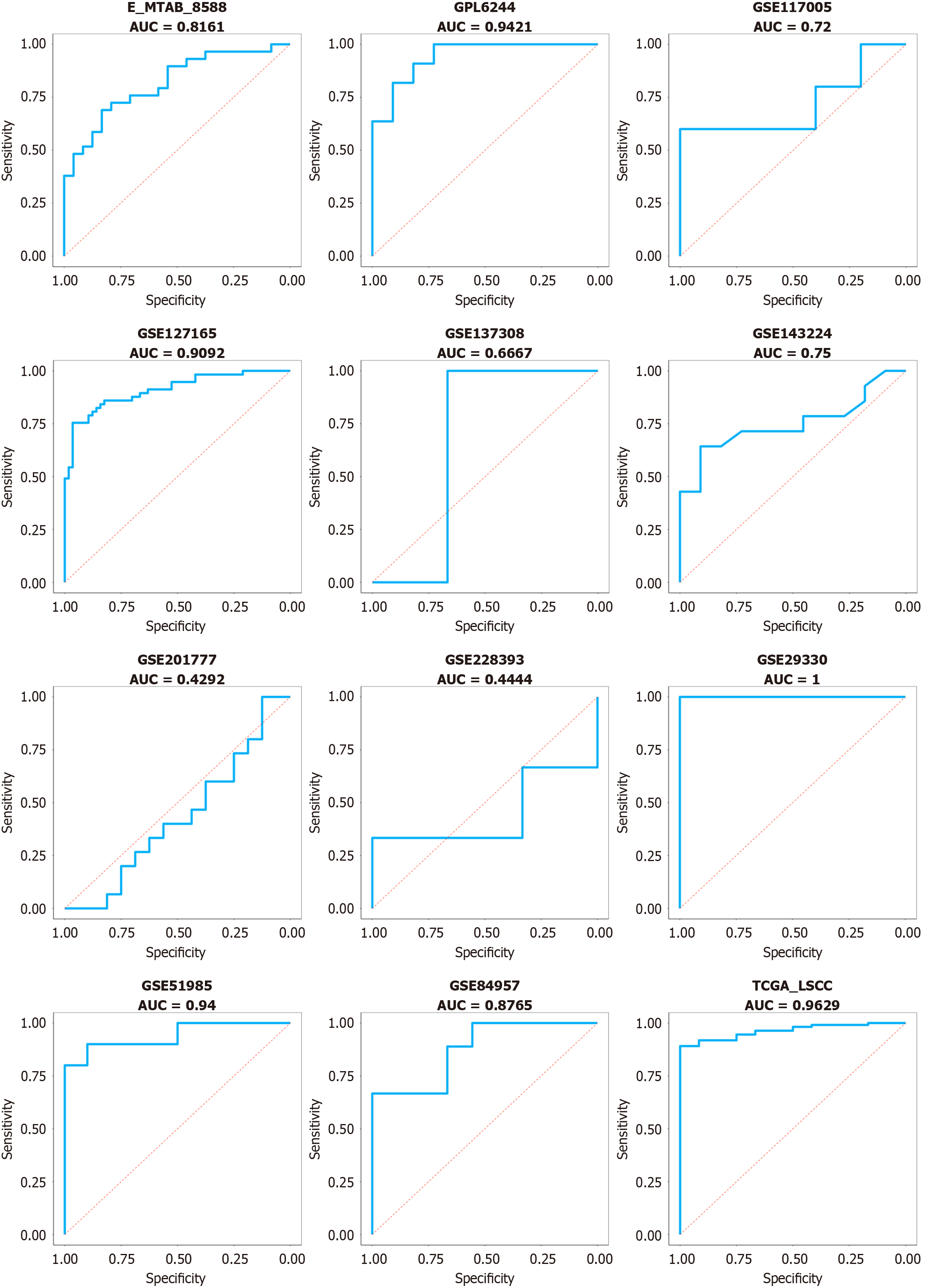
Figure 2 Chaperonin-containing TCP1 subunit 3 mRNA has a marvelous performance in discriminating between laryngeal squamous cell carcinoma and non-cancerous tissue.
AUC: Area under the curve.
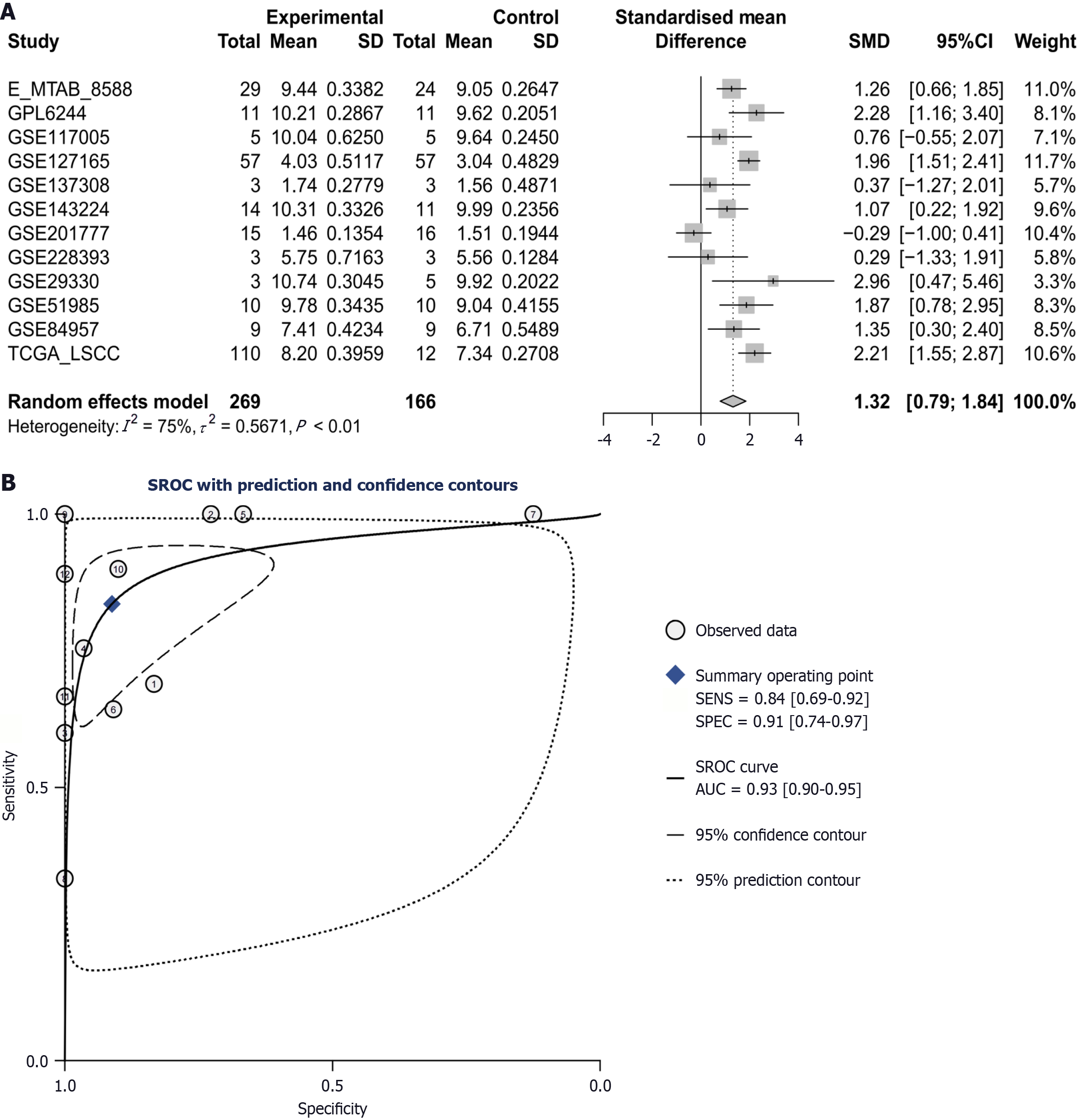
Figure 3 Chaperonin-containing TCP1 subunit 3 mRNA was significantly increased and has a strong discriminatory performance between laryngeal squamous cell carcinoma and non-cancerous tissue.
A: The expression level of chaperonin-containing TCP1 subunit 3 mRNA in the Laryngeal squamous cell carcinoma group was significantly higher than that in the control group overall (the standardized mean difference = 1.32, 95% confidence interval: 0.79-1.84), but there was significant heterogeneity among the studies (I2 = 75%); B: The summary receiver operating characteristic curve indicates that the discriminative performance of chaperonin-containing TCP1 subunit 3 mRNA is strong (area under the curve = 0.93), with sensitivity and specificity of 0.84 and 0.91, respectively. SMD: Standardized mean difference; CI: Confidence interval; SROC: Summary receiver operating characteristic; SENS: Sensitivity; SPEC: Specificity; AUC: Area under the curve.
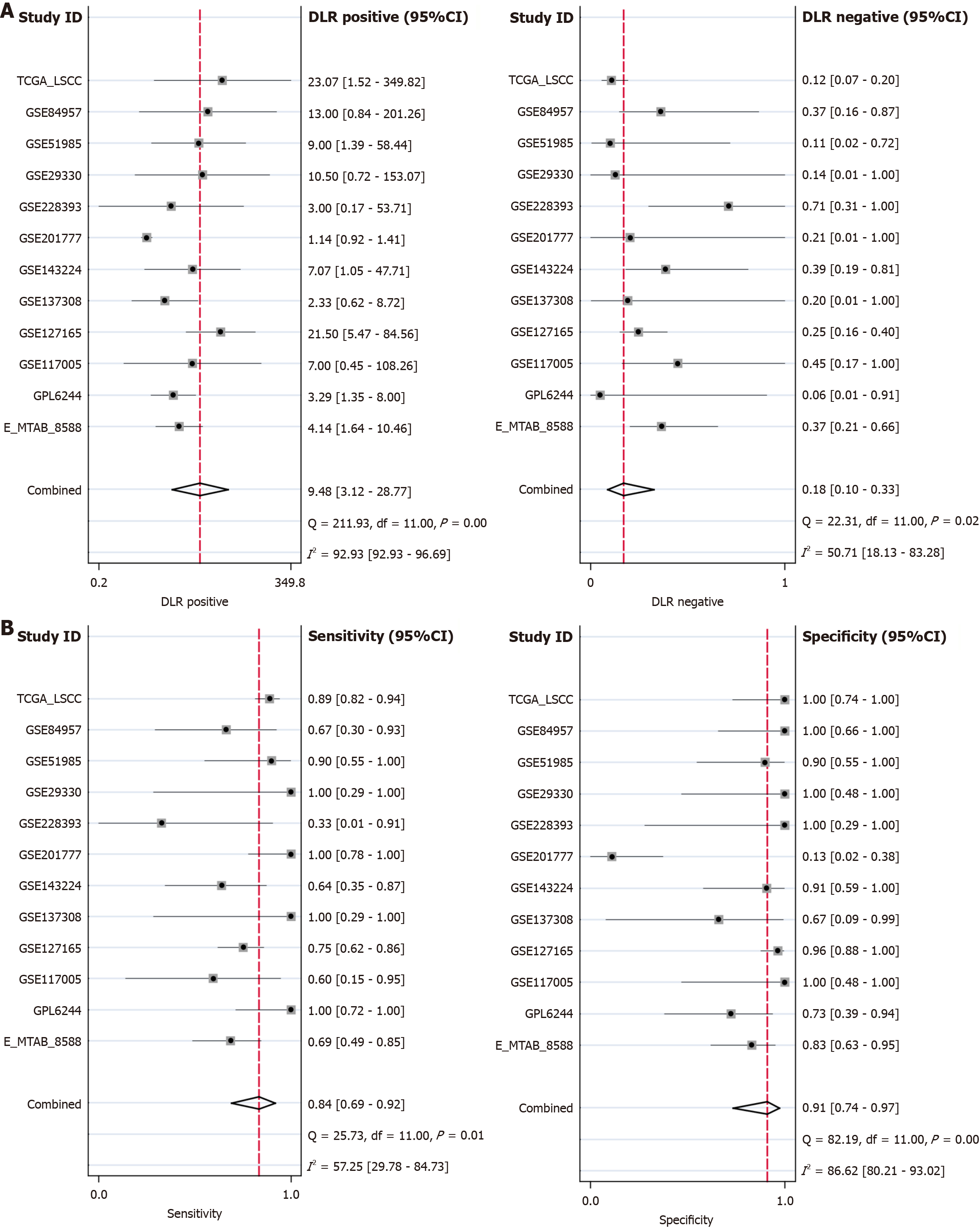
Figure 4 Chaperonin-containing TCP1 subunit 3 mRNA has a strong discriminatory performance in laryngeal squamous cell carcinoma.
A: The positive diagnostic likelihood ratio is 9.48, and the negative diagnostic likelihood ratio is 0.18; B: The overall sensitivity of the test is 0.84, and the specificity is 0.91.
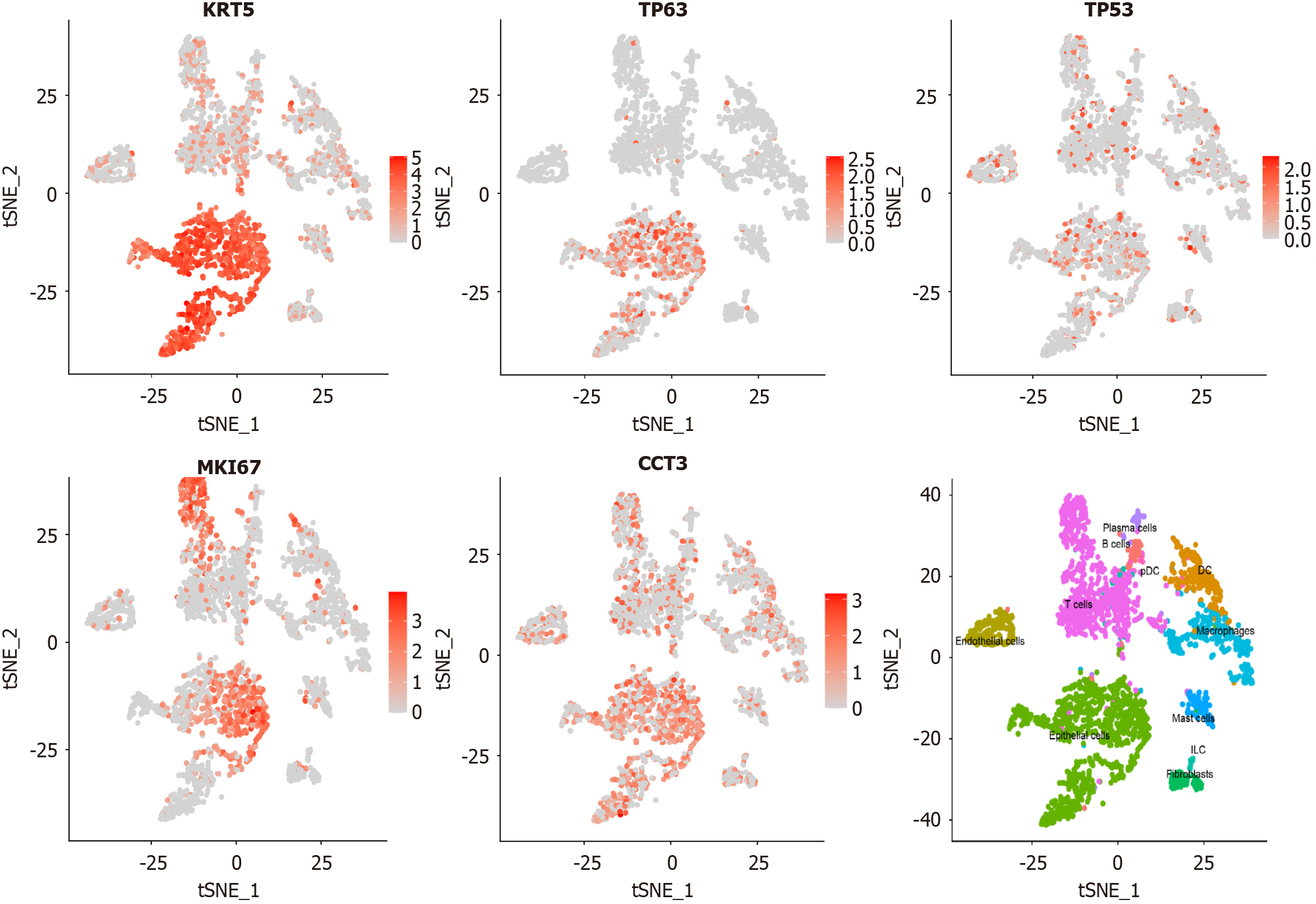
Figure 5 Single cell expression analysis of chaperonin-containing TCP1 subunit 3 in laryngeal squamous cell carcinoma.
These plots illustrate the expression patterns of Chaperonin-containing TCP1 subunit 3 and laryngeal squamous cell carcinoma-related marker genes, as well as the distribution and clustering of different cell types. tSNE: T-distributed stochastic neighbor embedding.
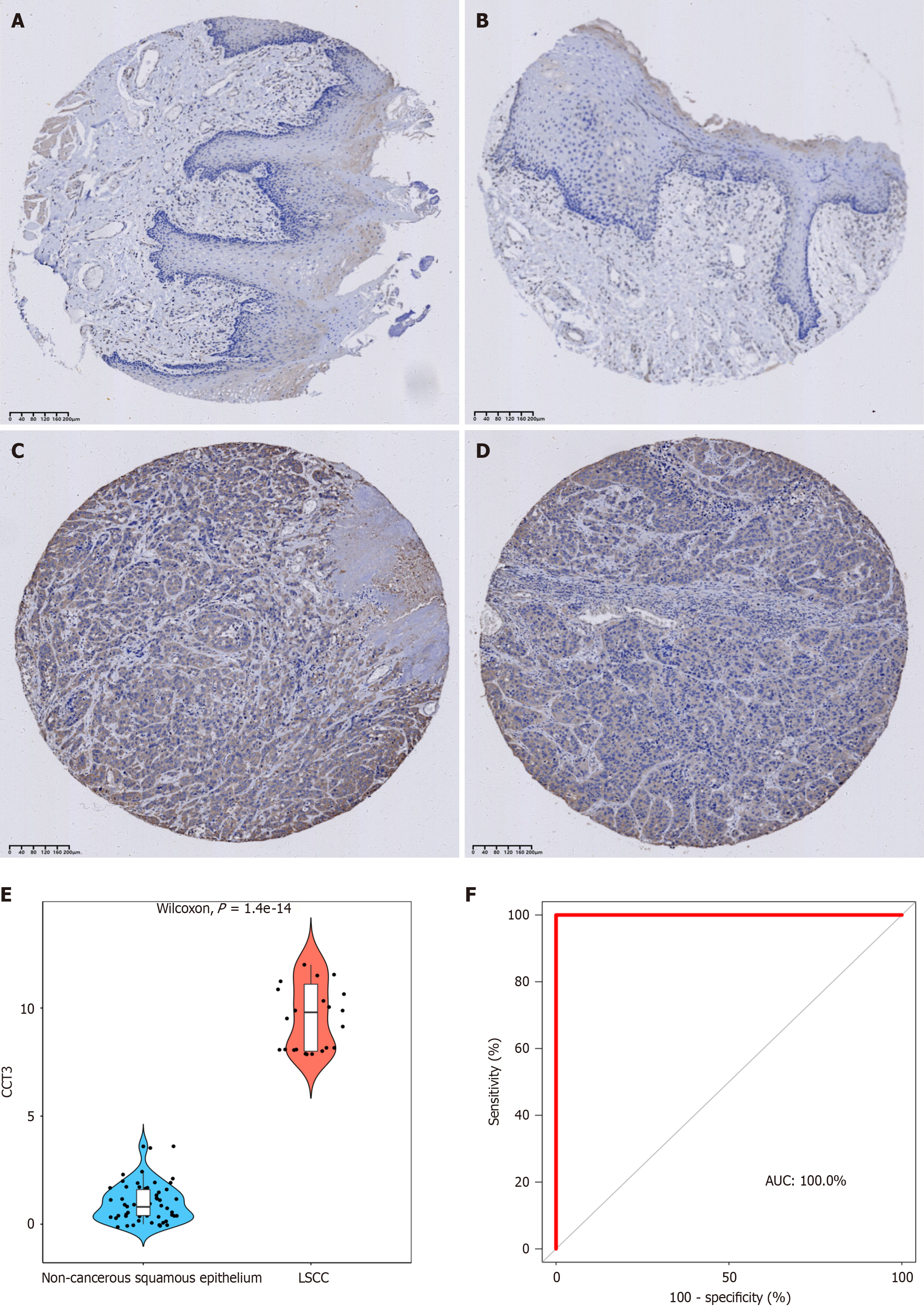
Figure 6 Immunohistochemical verification of chaperonin-containing TCP1 subunit 3 protein in laryngeal squamous cell carcinoma tissue.
A and B: Chaperonin-containing TCP1 subunit 3 (CCT3) protein show negative intensity in non-cancerous squamous epithelium; C and D: CCT3 protein show positive intensity in the cytoplasm of laryngeal squamous cell carcinoma cell; E: The expression of CCT3 protein is significantly higher in laryngeal squamous cell carcinoma than in non-cancerous squamous epithelium; F: CCT3 protein has perfect diagnostic performance, accurately distinguishing between laryngeal squamous cell carcinoma and non-cancerous squamous epithelium. CCT3: Chaperonin-containing TCP1 subunit 3; LSCC: Laryngeal squamous cell carcinoma; AUC: Area under the curve.
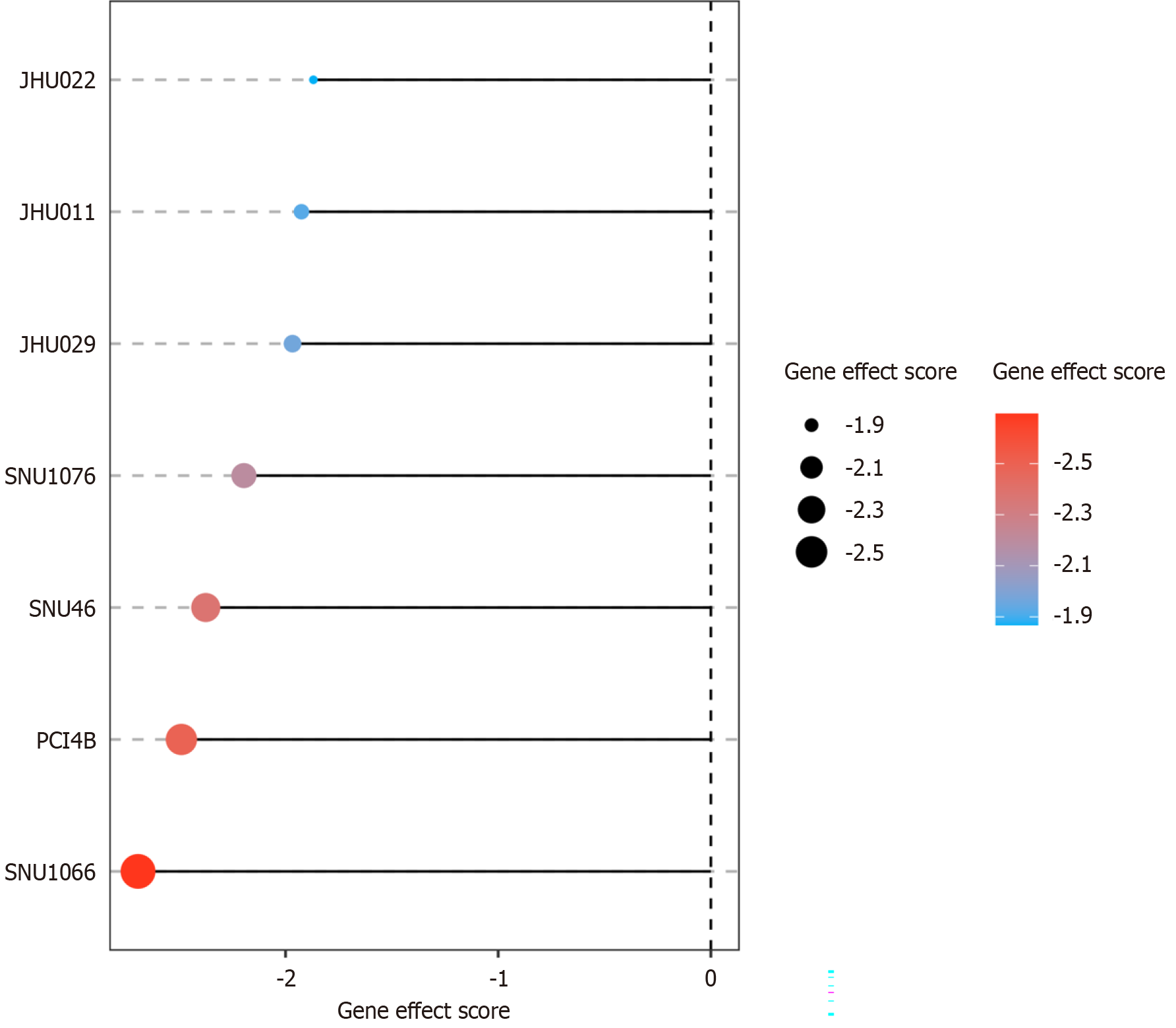
Figure 7 Clustered regularly interspaced short palindromic repeats screen is used to identify the function of chaperonin-containing TCP1 subunit 3 by knocking them out and observing the effect on laryngeal squamous cell carcinoma.
Laryngeal squamous cell carcinoma cell lines show a highly negative effect score, indicating that chaperonin-containing TCP1 subunit 3 knocking out may induce strongly inhibitory effect on laryngeal squamous cell carcinoma.
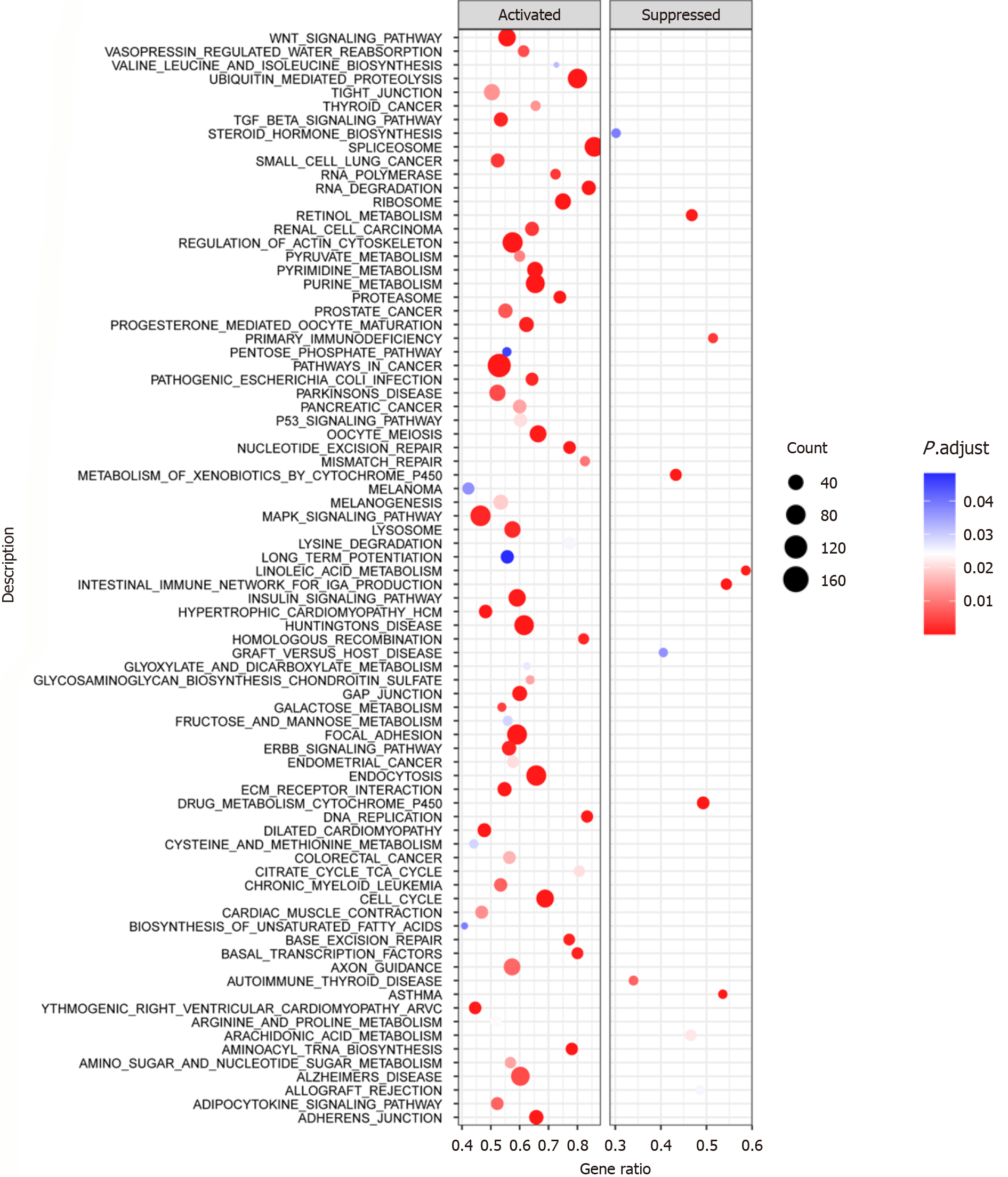
Figure 8 Gene set enrichment analysis of chaperonin-containing TCP1 subunit 3 in laryngeal squamous cell carcinoma.
This gene set enrichment analysis plot for chaperonin-containing TCP1 subunit 3 provides a visual representation of which biological pathways are significantly enriched in the activated or suppressed conditions, helping to understand the functional impact of chaperonin-containing TCP1 subunit 3 on cellular processes.
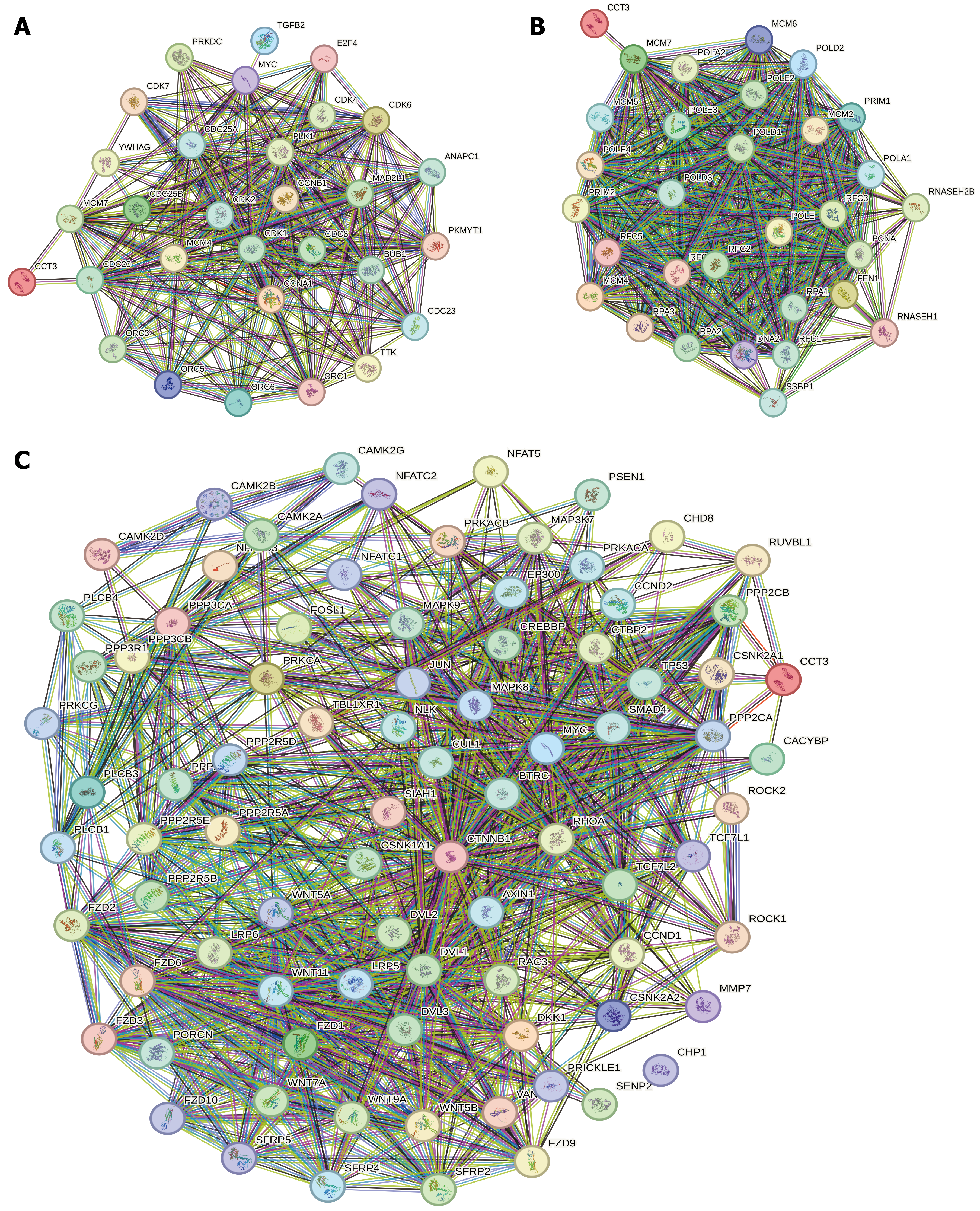
Figure 9 Analysis of chaperonin-containing TCP1 subunit 3 protein interaction network.
A: Kyoto Encyclopedia of Genes and Genomes (KEGG) cell cycle; B: KEGG DNA replication; C: KEGG Wnt signaling pathway.
- Citation: Mo BY, Wen JY, Chen GQ, Ling JW, He H, Qin ZL, Tian FY, Li Q, Li B, Li JD, He RQ, Qin DY, Li ZY, Chen G, Mo CH, Chen C, Yin SH, Yang L. Expression patterns and clinical implications of chaperonin subunit 3 mRNA and protein in laryngeal squamous cell carcinoma. World J Clin Oncol 2025; 16(12): 112161
- URL: https://www.wjgnet.com/2218-4333/full/v16/i12/112161.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i12.112161