Published online May 6, 2023. doi: 10.12998/wjcc.v11.i13.2934
Peer-review started: September 16, 2022
First decision: October 11, 2022
Revised: October 25, 2023
Accepted: February 21, 2023
Article in press: February 21, 2023
Published online: May 6, 2023
Processing time: 220 Days and 22.8 Hours
Complement overactivation is a major driver of lupus nephritis (LN). Impaired interactions of C-reactive protein (CRP) with complement factor H (CFH) have been shown as a pathogenic mechanism that contributes to the overactivation of complement in LN. However, genetic variations of neither CRP nor CFH show consistent influences on the risk of LN.
To examine whether genetic variations of CRP and CFH in combination can improve the risk stratification in Chinese population.
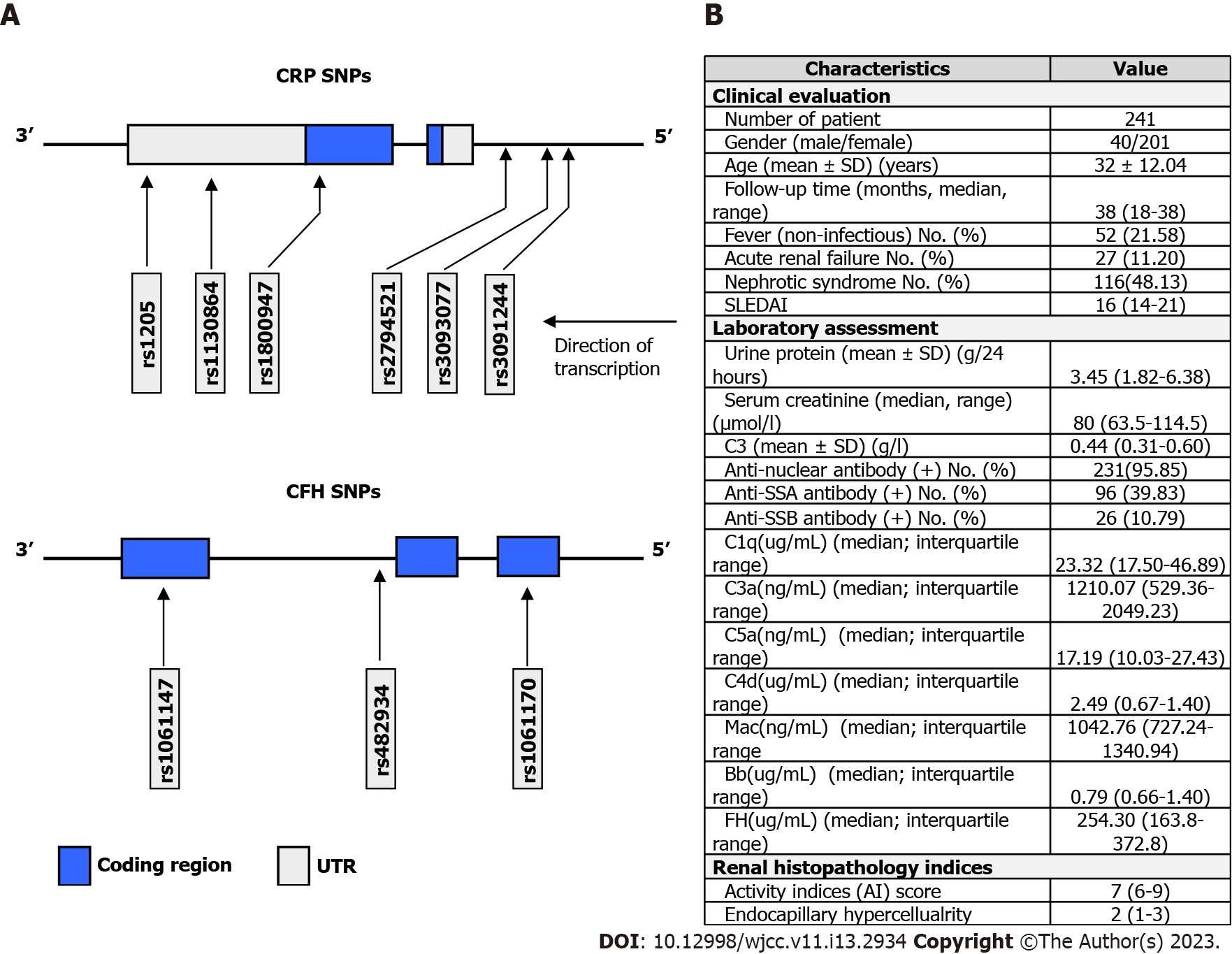
We genotyped six CRP single nucleotide polymorphisms (SNPs) (rs1205, rs3093062, rs2794521, rs1800947, rs3093077, and rs1130864) and three CFH SNPs (rs482934, rs1061170, and rs1061147) in 270 LN patients and 303 healthy subjects.
No linkage was found among CRP and CFH SNPs, indicating lack of genetic interactions between the two genes. Moreover, CRP and CFH SNPs, neither individually nor in combination, are associated with the risk or clinical manifestations of LN. Given the unambiguous pathogenic roles of the two genes.
These findings suggest that the biological effects of most genetic variations of CRP and CFH on their expressions or activities are not sufficient to influence the disease course of LN.
Core Tip: In spite of the unambiguous pathogenic roles of C-reactive protein (CRP)and complement factor H (CFH) in lupus nephritis (LN), our present study involving a Chinese population has failed to reveal any significant associations of their genetic variations with LN risk. These findings suggest that most genetic variations of CRP and CFH might possess limited biological effects on their expressions or activities and are thus not sufficient to influence the disease course of LN. Overall, we concluded that genetic variations of CRP and CFH could not be used to improve the risk stratification of LN in Chinese population.
- Citation: Li QY, Lv JM, Liu XL, Li HY, Yu F. Association of C-reactive protein and complement factor H gene polymorphisms with risk of lupus nephritis in Chinese population. World J Clin Cases 2023; 11(13): 2934-2944
- URL: https://www.wjgnet.com/2307-8960/full/v11/i13/2934.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i13.2934
As an autoimmune disease, the pathogenesis of systemic lupus erythematosus (SLE) involves clearance defect of apoptotic cells, uncontrolled activation of complement and massive production of autoantibodies[1,2].Previously studies have shown that 40% of SLE patients have clinical manifestations of renal dysfunction and that about 80% of SLE patients have different degrees of pathological renal damage, including lupus nephritis (LN), a common and severe complication of SLE, which is regarded as the main factor for the poor prognosis of patients[3-5].
reactive protein (CRP) is an acute phase reactant and a commonly used clinical marker of inflammation[6-10]. Besides, CRP could promote the elimination of damaged cells or pathogens by activating and regulating the complement system. Previous clinical studies suggest that CRP and SLE/LN have certain correlations. Firstly, CRP gene locates in the chromosome 1q23, an SLE linkage region[11,12]. Secondly, elevation of CRP levels is generally impaired in SLE patients especially those with kidney and skin involvement[13]. Thirdly, CRP autoantibodies can be detected in a considerable number of patients, which may relate to abnormal plasma levels in SLE[14-16]. Lastly, several animal studies on lupus-prone mouse strain (NZB/NZW) revealed that human CRP could help to produce less proteinuria and prolonged survival[17-19]. In the past decades, studies have been made to clarify the associations of CRP Single Nucleotide Polymorphisms (SNPs) with SLE susceptibility, while conclusions remained inconsistent[20-24]. Moreover, no CRP SNPs studies have been performed in the Chinese population.
In addition to CRP, complement factor H (CFH), a factor that negatively regulates alternative complement pathway, is another candidate associated with SLE. Firstly, CFH deficiency has been proven to accelerate the development of LN[25], and serum CFH level was observed to be associated with clinical and pathological activities of SLE patients with LN[26]. Secondly, several families suffering from SLE were reported to possess CFH deficiency or mutations[27,28]. Genetic variations of CFH can sometimes affect its bioactivities, in which several exotic SNPs were found to be related to various human diseases[29]. Among CFH SNPs, rs1061170 corresponds to a CFH variant Tyr402His, which exhibits impaired CFH-CRP binding efficiency. Because CRP could inhibit the complement overactivation by recruiting CFH, so Tyr402His theoretically results in dysregulated complement activation[30].
Despite that LN is a complement-related disease, and that both CRP and CFH are involved in complement regulation, it remains unclear whether CRP and CFH SNPs directly impact the pathogenesis of LN. In this scenario, we carried out the present study, in which six CRP SNPs and three CFH SNPs were genotyped in 270 LN patients and 303 healthy controls of a Chinese cohort. Association analysis was subsequently performed for these SNPs and LN risk from the perspectives of allele, genotype, combined SNPs and haplotype. As far as we know, this is the first study to consider SNPs of CRP and CFH together when evaluating their relationship with LN risk in the Chinese population. Our data show that SNPs of both genes have no significant association with LN risk. Given the unambiguous pathogenic role of the two genes, these findings suggest that the biological effects of genetic variations of CRP and CFH on their expression or activities are not sufficient to influence the disease course of LN in the Chinese population.
Renal histopathological data of 270 patients with renal biopsy-proven LN, diagnosed between January 2000 and July 2017 in Peking University First Hospital, were reviewed and reclassified according to the International Society of Nephrology and Renal Pathology Society (ISN/RPS) 2003 classification[31]. 303 age and gender matched healthy controls were collected. The work was approved by the Ethics Committee of Peking University First Hospital [Approval No. 2017(1333)].
Blood samples of the 270 SLE patients and 303 healthy controls of Chinese Han individuals were collected with the approval of the Ethics Committee of Peking University First Hospital. Human genomic DNA was extracted using Qiagen Blood DNA Kit (QIAGEN China, Shanghai) according to the manufacturer's instructions. Subsequently, the CRP SNPs (rs1205, rs3093077, rs3091244, rs1130864, rs1800947, rs2794521), and CFH SNPs (rs1061170, rs482934 and rs1061147) was genotyped by SNaPshot (ABI PRISM® SNaPshot™ Multiplex Kit, ABI) with specific primers.
For clinical evaluation, the disease activities of all patients were assessed by the SLE Disease Activity Index[32,33]. Briefly, the following items were collected and analyzed: Sex, fever, malar rash, photosensitivity, oral ulcer, alopecia, arthritis, serositis, neurologic disorder, anemia, leukocytopenia, thrombocytopenia, hematuria, and leukocyturia. For laboratory assessment, the following items were collected as we previously reported[34]: Complete blood count, plasma lactate dehydrogenase, liver enzymes, peripheral blood smear, urine analysis, serum creatinine, serum antinuclear antibodies, anti-double-stranded DNA antibodies, anti-extractable nuclear antigen antibodies, anti-cardiolipin antibodies and C3. For renal histopathology, all renal biopsy specimens were examined by light microscopy, direct immunofluorescence, and electron microscopy techniques as our previous reports[35]. All samples were double-blind reviewed by two experienced pathologists based on the 2003 ISN/RPS recommendation on LN classification[36]. The pathologists classified and scored the biopsies separately, especially for the activity indices, chronicity indices. Differences in scoring between pathologists were resolved by re-reviewing the biopsies and thus reaching a consensus. Renal histopathological data of 270 LN patients was classified according to the ISN/RPS 2003 classification, which was an improved version of World Health Organization (WHO)[37].
Hardy-Weinberg equilibrium testing was performed for all healthy controls using a chi-squared test. Distributions of genotype, allele and haplotype were compared between control and case groups using Pearson’s chi-squared test or Fisher’s exact test. For comparison of clinical, laboratory and pathological features of patients, student’s t-test and one way analysis of variance (ANOVA) were used. A P value less than 0.05 was considered to be significant. Pairwise Linkage Disequilibrium (LD) and haplotype analysis were both conducted using the SHEs is platform[38]. SNP pairs with D’ value great than 0.8 and r2 value great than 0.33 were considered to be in significant LD. Meta-analysis was performed by using Stata 15 software. Relative risks of SLE/LN were estimated according to ORs with 95% CIs. The inconsistency index I2 was calculated to quantify the heterogeneity: If I² < 50%, suggesting that the degree of heterogeneity was low, and the meta-analysis was performed using the fixed effect model; otherwise, the random effects model was used. P > 0.05 means no statistical significance.
Six CRP SNPs (rs1205, rs3093077, rs3091244, rs1130864, rs1800947 and rs2794521) were genotyped in 270 LN patients and 303 healthy controls (Figure 1). Chi-squared test showed that genotype frequency distributions of CRP SNPs in all healthy controls were all in Hardy-Weinberg equilibrium (Supplementary Table 1). In subsequent comparisons of case and control groups (Table 1), none of the alleles or genotypes was observed to be significantly associated with the SLE risk (P > 0.05). In further analysis, we examined the effects of CRP SNP combinations, and again failed to observe any significant difference in genotype distributions of LN patients and healthy controls (Supplementary Table 2).
| Genotype | LN | Normal | P value | Allele | LN | Normal | P value |
| rs1205 (C>T) | |||||||
| CC | 53 (19.63) | 49 (16.17) | 0.469 | C | 245 (45.37) | 254 (41.91) | 0.239 |
| CT | 139 (51.48) | 156 (51.49) | T | 295 (54.63) | 352 (58.09) | ||
| TT | 78 (28.89) | 98 (32.34) | |||||
| rs3093077 (A>C) | |||||||
| AA | 173 (64.07) | 208 (68.65) | 0.436 | A | 436 (80.74) | 502 (82.84) | 0.358 |
| AC | 90 (33.33) | 86 (28.38) | C | 104 (19.26) | 104 (17.16) | ||
| CC | 7 (2.59) | 9 (2.97) | |||||
| rs3091244 (C>T>A) | |||||||
| CC | 153 (56.67) | 180 (59.41) | 0.9351 | C | 410 (75.93) | 467 (77.06) | 0.712 |
| CT | 23 (8.52) | 28 (9.24) | T | 29 (5.37) | 36 (5.94) | ||
| CA | 81 (30.00) | 79 (26.07) | A | 101 (18.70) | 103 (17.00) | ||
| TT | 1 (0.37) | 1 (0.33) | |||||
| TA | 4 (1.48) | 6 (1.98) | |||||
| AA | 8 (2.96) | 9 (2.97) | |||||
| rs1130864 (G>A) | |||||||
| GG | 241 (89.26) | 270 (89.11) | 0.7971 | G | 509 (94.26) | 572 (94.39) | 0.924 |
| GA | 27 (10.00) | 32 (10.56) | A | 31 (5.74) | 34 (5.61) | ||
| AA | 2 (0.74) | 1 (0.33) | |||||
| rs1800947 (C>G) | |||||||
| CC | 245 (90.74) | 273 (90.10) | 1.0001 | C | 515 (95.37) | 575 (94.88) | 0.703 |
| CG | 25 (9.26) | 29 (9.57) | G | 25 (4.63) | 31 (5.12) | ||
| GG | 0 (0.00) | 1 (0.33) | |||||
| rs2794521 (T>C) | |||||||
| TT | 187 (69.26) | 222 (73.27) | 0.177 | T | 446 (82.59) | 520 (85.81) | 0.135 |
| TC | 72 (26.67) | 76 (25.08) | C | 94 (17.41) | 86 (14.19) | ||
| CC | 11 (4.07) | 5 (1.65) |
Subsequently, pairwise LD analysis was conducted for CRP SNPs in healthy controls. Of all SNP pairs, rs3091244/rs3093077 and rs3091244/rs1205 pairs were found to be in significant LD (Table 2). Considering that SNP haplotype may provide more informative details, CRP haplotypes were thus included for further investigation. Given the acceptable number, we included all 6 CRP SNPs in the haplotype analysis. Finally, 6 CRP haplotypes were observed at frequencies greater than 3.0% in both healthy controls and LN patients. However, no significant differences were found in the distribution frequencies of those haplotypes between the two groups (Table 3).
| r2\D' | rs3093077 | rs1205 | rs1130864 | rs1800947 | rs3091244 | rs2794521 |
| rs3093077 | 1.000 | 0.990 | 0.985 | 0.979 | 0.748 | |
| rs1205 | 0.278 | 0.999 | 0.999 | 0.984 | 1.000 | |
| rs1130864 | 0.012 | 0.082 | 0.986 | 1.000 | 0.974 | |
| rs1800947 | 0.011 | 0.039 | 0.003 | 0.993 | 0.986 | |
| rs3091244 | 0.675 | 0.360 | 0.212 | 0.014 | 0.996 | |
| rs2794521 | 0.019 | 0.229 | 0.009 | 0.009 | 0.044 |
| CRP haplotype | SLE-freq (%, n = 270) | Normal-freq (%, n = 303) | χ2 | Overall P value | |
| H1 | G G G G T A | 17.8 | 16.7 | 3.293 | 0.655 |
| H2 | T A G C G A | 4.3 | 5.1 | ||
| H3 | T A G G G A | 50.2 | 52.8 | ||
| H4 | T G A G A A | 5.4 | 5.6 | ||
| H5 | T G G G G A | 4.1 | 5.1 | ||
| H6 | T G G G G G | 16.6 | 13.7 | ||
To further confirm whether these CRP SNPs are indeed unrelated to LN in the present population, we thus further checked the association of these SNPs with clinical, laboratory and pathological features of all patients (Supplementary Tables 3-6). In line with the conclusions above, most indexes exhibited no significant differences between genotypes of these SNPs. Notably, WHO classification for all LN patients was performed and association between pathological subclass and SNPs were further analyzed, whereas no significant differences were observed. However, several items showed differences, which might imply potential relevance of these SNPs with LN to some extent, suggesting that conclusion should cautiously draw.
Similarly, we subsequently genotyped 3 CFH SNPs, namely rs1061170, rs482934 and rs1061147, in 270 LN patients and 303 healthy controls (Figure 1). After Hardy-Weinberg equilibrium was checked for all genotype frequencies of controls (Supplementary Table 7), association of CFH polymorphism with LN risk were examined. Of note, no significant enrichment or depletion of allele and genotype distribution has been observed in LN patients (Table 4). In further exploration, pairwise LD for those CFH SNPs was examined as before. Dramatically, all three SNP pairs were found to be in strong LD (Table 5). Based on this, haplotype analysis was conducted, in which two CFH haplotypes were observed at frequencies greater than 3.0% in both healthy controls and LN patients. However, no significant associations were found between those two haplotypes and LN risk (Table 6).
| Genotype | LN | Normal | P value | Allele | LN | Normal | P value |
| rs1061170 (T>C) | |||||||
| TT | 235 (87.04) | 258 (85.15) | 0.2201 | T | 503 (93.15) | 561 (92.57) | 0.707 |
| TC | 33 (12.22) | 45 (14.85) | C | 37 (6.85) | 45 (7.43) | ||
| CC | 2 (0.74) | 0 (0.00) | |||||
| rs482934 (A>C) | |||||||
| AA | 236 (87.41) | 258 (85.15) | 0.1891 | A | 504 (0.93) | 561 (0.93) | 0.617 |
| AC | 32 (11.85) | 45 (14.85) | C | 36 (0.07) | 45 (0.07) | ||
| CC | 2 (0.74) | 0 (0.00) | |||||
| rs1061147 (C>A) | |||||||
| CC | 235 (87.04) | 257 (84.82) | 0.1941 | C | 503 (0.93) | 560 (0.92) | 0.63 |
| CA | 33 (12.22) | 46 (15.18) | A | 37 (0.07) | 46 (0.08) | ||
| AA | 2 (0.74) | 0 (0.00) |
| r2\D' | rs1061147 | rs482934 | rs1061170 |
| rs1061147 | 0.976 | 1.000 | |
| rs482934 | 0.930 | 0.976 | |
| rs1061170 | 0.977 | 0.952 | |
| CRP Haplotype | LN-freq (%, n = 270) | Normal-freq (% n = 303) | χ2 | Overall P value | |
| H1 | A C G | 6.3 | 7.3 | 0.426 | 0.514 |
| H2 | C A A | 93.3 | 92.2 | ||
In further analysis, we checked the association of rs1061170 with clinical, laboratory and pathological features of all LN patients (Supplementary Table 8). Similar to CRP SNPs, except for a few items, most indexes exhibited no significant differences between genotypes of these SNPs.
Given the key roles of complement overactivation in SLE pathogenesis and the capacity of CRP to inhibit this process via interaction with CFH, we next asked whether any potential associations could be found in SNP combinations of CRP and CFH. Specifically, CFH SNP rs1061170, which corresponds to a variant Tyr402His with impaired capacity to bind CRP[39,40], was combined with 6 CRP SNPs and evaluated individually. However, we failed to observe any significant associations in all SNP combinations included (Table 7). Besides, cross pairs of the CRP and CFH SNPs were further included for pairwise LD evaluation, in which no significant LD was observed (Supplementary Table 9).
| CRP-CFH genotype combination | LN | Normal | P value |
| rs1205 + rs1061170 | |||
| CCTT | 46 (17.10) | 45 (14.85) | 0.105 |
| CCTC | 7 (2.60) | 4 (1.32) | |
| CTTT | 124 (46.10) | 125 (41.25) | |
| CTTC | 14 (5.20) | 31 (10.23) | |
| TTTT | 65 (24.16) | 88 (29.04) | |
| TTTC | 13 (4.83) | 10 (3.30) | |
| rs3093077 + rs1061170 | |||
| AATT | 150 (56.39) | 180 (59.80) | 0.322 |
| AATC | 23 (8.65) | 28 (9.30) | |
| ACTT | 81 (30.45) | 71 (23.59) | |
| ACTC | 8 (3.01) | 15 (4.98) | |
| CCTT | 4 (1.50) | 7 (2.33) | |
| rs3091244 + rs1061170 | |||
| CCTT | 133 (50.57) | 155 (52.19) | 0.756 |
| CCTC | 21 (7.98) | 25 (8.42) | |
| CTTT | 21 (7.98) | 25 (8.42) | |
| CATT | 71 (27.00) | 64 (21.55) | |
| CATC | 9 (3.42) | 15 (5.05) | |
| TATT | 4 (1.52) | 6 (2.02) | |
| AATT | 4 (1.52) | 7 (2.36) | |
| rs1130864 + rs1061170 | |||
| GGTT | 208 (78.49) | 228 (76.25) | 0.772 |
| GGTC | 32 (12.08) | 42 (14.05) | |
| GATT | 25 (9.43) | 29 (9.70) | |
| rs1800947 + rs1061170 | |||
| CCTT | 214 (80.75) | 232 (77.85) | 0.655 |
| CCTC | 30 (11.32) | 41 (13.76) | |
| CGTT | 21 (7.92) | 25 (8.39) | |
| rs2794521 + rs1061170 | |||
| TTTT | 160 (59.93) | 187 (61.92) | 0.431 |
| TTTC | 26 (9.74) | 35 (11.59) | |
| TCTT | 66 (24.72) | 67 (22.19) | |
| TCTC | 6 (2.25) | 9 (2.98) | |
| CCTT | 9 (3.37) | 4 (1.32) |
In the past decades, studies have been focused on revealing the associations of CRP/CFH genetic variations with SLE/LN[41]. However, those studies were mainly based on the European or American populations, and often gained inconsistent conclusions. Moreover, although CRP/CFH interaction theoretically plays a role in LN pathogenesis, they have not been considered together when evaluating the association of their genetic variations with the LN risk.
In this study, we enrolled 6 CRP SNPs and 3 CFH SNPs of a Chinese cohort, and studied their relationship with LN risk, which has not yet been systematically reported. Our study revealed that there were no significant associations between these SNPs and LN susceptibility in the Chinese population. All patients in this study were selected from the same center and their diagnosis were all confirmed by renal biopsy. Moreover, the complete clinical, laboratory and pathological indexes were also included to test the results. Therefore, although no statistical associations were observed in our study, valid and useful information could still be revealed.
In addition, these negative results are generally consistent with the conclusions of previous researches to a large extent. Among the 6 CRP SNPs, rs1800947[20,24-26] and rs2794521[23,25] were included in several studies, which were repeatedly reported to be unrelated to SLE in various populations, consistent with our results. For rs1205[20,21,24-26], rs3091244[20,21,24,25] and rs1130864[20-22], conclusions remained inconsistent among these studies, which might rationalize the existence of our negative results to some extent. For the CFH SNPs, Zhao et al[29]evaluated an Asian group involving 200 Chinese SLE cases and found no significant association between rs1061147 and SLE (without LN). Tan et al[41] enrolled 334 LN patients, 269 SLE patients without LN and 350 healthy controls from China, but failed to observe any significant differences in allele and genotype frequencies of rs1061170 among groups. Both conclusions were consistent with our present findings.
Given that interaction of CRP and CFH would theoretically help to regulate complement and therefore play roles in SLE/LN pathogenesis, we combined 6 CRP SNPs individually with CFH SNP rs1061170, which corresponds to a CFH variant with impaired capacity in CRP binding. To our knowledge, this is the first study to combine these two genes when performing a correlation analysis with LN risk. However, we still failed to observe any significant associations from this perspective.
Overall, our results suggest that CRP and CFH genetic variation and interaction do not affect the occurrence of LN at the gene level in a Chinese population. In future studies, multiple-center sampling is needed to expand the study scale, whereas SLE patients without LN from other rheumatism departments should also be included. Moreover, more SNPs should be examined for these two genes, while other molecules along the pathogenesis pathway of CRP and CFH should be involved for a joint analysis.
In spite of the unambiguous pathogenic roles of CRP and CFH in LN, our present study involving a Chinese population has failed to reveal any significant associations of their genetic variations with LN risk. These findings suggest that most genetic variations of CRP and CFH might possess limited biological effects on their expressions or activities, and are thus not sufficient to influence the disease course of LN. Overall, we concluded that genetic variations of CRP and CFH could not be used to improve the risk stratification of LN in Chinese population.
Both C-reactive protein (CRP) and complement factor H (CFH) play roles in pathogenesis of lupus nephritis (LN).
It still keeps unclear whether genetic variations of CRP and CFH are involved in risk of LN.
To examine whether genetic variations of CRP and CFH are associated with the susceptibility to LN in the Chinese population.
A case control study was conducted, in which six CRP Single Nucleotide Polymorphisms (SNPs) and three CFH SNPs were genotyped and analysed in 270 LN patients and 303 healthy subjects.
CRP and CFH SNPs, neither individually nor in combination, are associated with the risk or clinical manifestations of LN. Moreover, no linkage was found among CRP and CFH SNPs, indicating lack of genetic interactions between the two genes.
Biological effects of most genetic variations of CRP and CFH on their expressions or activities are not sufficient to influence the disease course of LN.
Future studies involving multiple-center sampling are needed to expand the study scale. Moreover, more SNPs should be examined for these two genes, while other molecules along the pathogenesis pathway of CRP and CFH should be involved for a joint analysis.
| 1. | Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358:929-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1314] [Cited by in RCA: 1345] [Article Influence: 74.7] [Reference Citation Analysis (1)] |
| 2. | Almaani S, Meara A, Rovin BH. Update on Lupus Nephritis. Clin J Am Soc Nephrol. 2017;12:825-835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 474] [Cited by in RCA: 634] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 3. | Johanneson B, Lima G, von Salomé J, Alarcón-Segovia D, Alarcón-Riquelme ME; Collaborative Group on the Genetics of SLE, The BIOMED II Collaboration on the Genetics of SLE and Sjögrens syndrome. A major susceptibility locus for systemic lupus erythemathosus maps to chromosome 1q31. Am J Hum Genet. 2002;71:1060-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 85] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | Tsao BP, Cantor RM, Grossman JM, Kim SK, Strong N, Lau CS, Chen CJ, Shen N, Ginzler EM, Goldstein R, Kalunian KC, Arnett FC, Wallace DJ, Hahn BH. Linkage and interaction of loci on 1q23 and 16q12 may contribute to susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2002;46:2928-2936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Tsao BP. Update on human systemic lupus erythematosus genetics. Curr Opin Rheumatol. 2004;16:513-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 111] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 6. | Caprio V, Badimon L, Di Napoli M, Fang WH, Ferris GR, Guo B, Iemma RS, Liu D, Zeinolabediny Y, Slevin M. pCRP-mCRP Dissociation Mechanisms as Potential Targets for the Development of Small-Molecule Anti-Inflammatory Chemotherapeutics. Front Immunol. 2018;9:1089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 7. | Jimenez RV, Wright TT, Jones NR, Wu J, Gibson AW, Szalai AJ. C-Reactive Protein Impairs Dendritic Cell Development, Maturation, and Function: Implications for Peripheral Tolerance. Front Immunol. 2018;9:372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 8. | Molins B, Romero-Vázquez S, Fuentes-Prior P, Adan A, Dick AD. C-Reactive Protein as a Therapeutic Target in Age-Related Macular Degeneration. Front Immunol. 2018;9:808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 9. | Romero-Vázquez S, Adán A, Figueras-Roca M, Llorenç V, Slevin M, Vilahur G, Badimon L, Dick AD, Molins B. Activation of C-reactive protein proinflammatory phenotype in the blood retinal barrier in vitro: implications for age-related macular degeneration. Aging (Albany NY). 2020;12:13905-13923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Zeinolabediny Y, Kumar S, Slevin M. Monomeric C-Reactive Protein - A Feature of Inflammatory Disease Associated With Cardiovascular Pathophysiological Complications? In Vivo. 2021;35:693-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 11. | Moser KL, Neas BR, Salmon JE, Yu H, Gray-McGuire C, Asundi N, Bruner GR, Fox J, Kelly J, Henshall S, Bacino D, Dietz M, Hogue R, Koelsch G, Nightingale L, Shaver T, Abdou NI, Albert DA, Carson C, Petri M, Treadwell EL, James JA, Harley JB. Genome scan of human systemic lupus erythematosus: evidence for linkage on chromosome 1q in African-American pedigrees. Proc Natl Acad Sci U S A. 1998;95:14869-14874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 348] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 12. | Cantor RM, Yuan J, Napier S, Kono N, Grossman JM, Hahn BH, Tsao BP. Systemic lupus erythematosus genome scan: support for linkage at 1q23, 2q33, 16q12-13, and 17q21-23 and novel evidence at 3p24, 10q23-24, 13q32, and 18q22-23. Arthritis Rheum. 2004;50:3203-3210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Pereira Da Silva JA, Elkon KB, Hughes GR, Dyck RF, Pepys MB. C-reactive protein levels in systemic lupus erythematosus: a classification criterion? Arthritis Rheum. 1980;23:770-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 53] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Li QY, Li HY, Fu G, Yu F, Wu Y, Zhao MH. Autoantibodies against C-Reactive Protein Influence Complement Activation and Clinical Course in Lupus Nephritis. J Am Soc Nephrol. 2017;28:3044-3054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 15. | Bell SA, Faust H, Schmid A, Meurer M. Autoantibodies to C-reactive protein (CRP) and other acute-phase proteins in systemic autoimmune diseases. Clin Exp Immunol. 1998;113:327-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 67] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 16. | Sjöwall C, Eriksson P, Almer S, Skogh T. Autoantibodies to C-reactive protein is a common finding in SLE, but not in primary Sjögren's syndrome, rheumatoid arthritis or inflammatory bowel disease. J Autoimmun. 2002;19:155-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 17. | Rodriguez W, Mold C, Kataranovski M, Hutt J, Marnell LL, Du Clos TW. Reversal of ongoing proteinuria in autoimmune mice by treatment with C-reactive protein. Arthritis Rheum. 2005;52:642-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 18. | Szalai AJ, Weaver CT, McCrory MA, van Ginkel FW, Reiman RM, Kearney JF, Marion TN, Volanakis JE. Delayed lupus onset in (NZB x NZW)F1 mice expressing a human C-reactive protein transgene. Arthritis Rheum. 2003;48:1602-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Szalai AJ, Wu J, Lange EM, McCrory MA, Langefeld CD, Williams A, Zakharkin SO, George V, Allison DB, Cooper GS, Xie F, Fan Z, Edberg JC, Kimberly RP. Single-nucleotide polymorphisms in the C-reactive protein (CRP) gene promoter that affect transcription factor binding, alter transcriptional activity, and associate with differences in baseline serum CRP level. J Mol Med (Berl). 2005;83:440-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 133] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 20. | Russell AI, Cunninghame Graham DS, Shepherd C, Roberton CA, Whittaker J, Meeks J, Powell RJ, Isenberg DA, Walport MJ, Vyse TJ. Polymorphism at the C-reactive protein locus influences gene expression and predisposes to systemic lupus erythematosus. Hum Mol Genet. 2004;13:137-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 223] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 21. | Atisha-Fregoso Y, Lima G, Carrillo-Maravilla E, Posadas-Sánchez R, Pérez-Hernández N, Baños-Peláez M, Iturralde-Chávez A, Hernández-Díaz N, Jakez-Ocampo J, Rodríguez-Pérez JM, Vargas-Alarcón G, Llorente L, Romero-Díaz J. C-reactive protein (CRP) polymorphisms and haplotypes are associated with SLE susceptibility and activity but not with serum CRP levels in Mexican population. Clin Rheumatol. 2018;37:1817-1824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 22. | Delongui F, Lozovoy MAB, Iriyoda TMV, Costa NT, Stadtlober NP, Alfieri DF, Flauzino T, Dichi I, Simão ANC, Reiche EMV. C-reactive protein +1444CT (rs1130864) genetic polymorphism is associated with the susceptibility to systemic lupus erythematosus and C-reactive protein levels. Clin Rheumatol. 2017;36:1779-1788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Kim HA, Chun HY, Kim SH, Park HS, Suh CH. C-reactive protein gene polymorphisms in disease susceptibility and clinical manifestations of Korean systemic lupus erythematosus. J Rheumatol. 2009;36:2238-2243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 24. | Shih PB, Manzi S, Shaw P, Kenney M, Kao AH, Bontempo F, Barmada MM, Kammerer C, Kamboh MI. Genetic variation in C-reactive protein (CRP) gene may be associated with risk of systemic lupus erythematosus and CRP concentrations. J Rheumatol. 2008;35:2171-2178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Wang FM, Yu F, Tan Y, Song D, Zhao MH. Serum complement factor H is associated with clinical and pathological activities of patients with lupus nephritis. Rheumatology (Oxford). 2012;51:2269-2277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 26. | Fijen CA, Kuijper EJ, Te Bulte M, van de Heuvel MM, Holdrinet AC, Sim RB, Daha MR, Dankert J. Heterozygous and homozygous factor H deficiency states in a Dutch family. Clin Exp Immunol. 1996;105:511-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 27. | Sánchez-Corral P, Bellavia D, Amico L, Brai M, Rodríguez de Córdoba S. Molecular basis for factor H and FHL-1 deficiency in an Italian family. Immunogenetics. 2000;51:366-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 28. | Rodríguez de Córdoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sánchez-Corral P. The human complement factor H: functional roles, genetic variations and disease associations. Mol Immunol. 2004;41:355-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 423] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 29. | Zhao J, Wu H, Khosravi M, Cui H, Qian X, Kelly JA, Kaufman KM, Langefeld CD, Williams AH, Comeau ME, Ziegler JT, Marion MC, Adler A, Glenn SB, Alarcón-Riquelme ME; BIOLUPUS Network; GENLES Network, Pons-Estel BA, Harley JB, Bae SC, Bang SY, Cho SK, Jacob CO, Vyse TJ, Niewold TB, Gaffney PM, Moser KL, Kimberly RP, Edberg JC, Brown EE, Alarcon GS, Petri MA, Ramsey-Goldman R, Vilá LM, Reveille JD, James JA, Gilkeson GS, Kamen DL, Freedman BI, Anaya JM, Merrill JT, Criswell LA, Scofield RH, Stevens AM, Guthridge JM, Chang DM, Song YW, Park JA, Lee EY, Boackle SA, Grossman JM, Hahn BH, Goodship TH, Cantor RM, Yu CY, Shen N, Tsao BP. Association of genetic variants in complement factor H and factor H-related genes with systemic lupus erythematosus susceptibility. PLoS Genet. 2011;7:e1002079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 166] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 30. | Li Q, Song D, Wang F, Tan Y, Yu F, Zhao M. Clinicopathological characteristics and outcomes of Chinese patients with scanty immune deposits lupus nephritis: a large cohort study from a single center. ScientificWorldJournal. 2014;212597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 31. | Liang MH, Socher SA, Larson MG, Schur PH. Reliability and validity of six systems for the clinical assessment of disease activity in systemic lupus erythematosus. Arthritis Rheum. 1989;32:1107-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 594] [Cited by in RCA: 602] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 32. | Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35:630-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3258] [Cited by in RCA: 3560] [Article Influence: 104.7] [Reference Citation Analysis (0)] |
| 33. | Huang J, Han SS, Qin DD, Wu LH, Song Y, Yu F, Wang SX, Liu G, Zhao MH. Renal Interstitial Arteriosclerotic Lesions in Lupus Nephritis Patients: A Cohort Study from China. PLoS One. 2015;10:e0141547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Song D, Wu LH, Wang FM, Yang XW, Zhu D, Chen M, Yu F, Liu G, Zhao MH. The spectrum of renal thrombotic microangiopathy in lupus nephritis. Arthritis Res Ther. 2013;15:R12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 169] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 35. | Weening JJ, D'Agati VD, Schwartz MM, Seshan SV, Alpers CE, Appel GB, Balow JE, Bruijn JA, Cook T, Ferrario F, Fogo AB, Ginzler EM, Hebert L, Hill G, Hill P, Jennette JC, Kong NC, Lesavre P, Lockshin M, Looi LM, Makino H, Moura LA, Nagata M; International Society of Nephrology Working Group on the Classification of Lupus Nephritis; Renal Pathology Society Working Group on the Classification of Lupus Nephritis. The classification of glomerulonephritis in systemic lupus erythematosus revisited. Kidney Int. 2004;65:521-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 976] [Cited by in RCA: 1133] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 36. | Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1414] [Cited by in RCA: 1795] [Article Influence: 85.5] [Reference Citation Analysis (0)] |
| 37. | Schaumberg DA, Christen WG, Kozlowski P, Miller DT, Ridker PM, Zee RY. A prospective assessment of the Y402H variant in complement factor H, genetic variants in C-reactive protein, and risk of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:2336-2340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 38. | Molins B, Fuentes-Prior P, Adán A, Antón R, Arostegui JI, Yagüe J, Dick AD. Complement factor H binding of monomeric C-reactive protein downregulates proinflammatory activity and is impaired with at risk polymorphic CFH variants. Sci Rep. 2016;6:22889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 39. | Rhodes B, Fürnrohr BG, Vyse TJ. C-reactive protein in rheumatology: biology and genetics. Nat Rev Rheumatol. 2011;7:282-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 153] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 40. | Rhodes B, Wong A, Navarra SV, Villamin C, Vyse TJ. Genetic determinants of basal C-reactive protein expression in Filipino systemic lupus erythematosus families. Genes Immun. 2008;9:153-160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 41. | Tan M, Hao JB, Chu H, Wang FM, Song D, Zhu L, Yu F, Li YZ, Song Y, Zhao MH. Genetic variants in FH are associated with renal histopathologic subtypes of lupus nephritis: a large cohort study from China. Lupus. 2017;26:1309-1317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Reda R, Italy; Tanaka H, Japan S-Editor: Liu GL L-Editor: A P-Editor: Liu GL