Published online Jan 24, 2026. doi: 10.5306/wjco.v17.i1.116090
Revised: November 26, 2025
Accepted: December 29, 2025
Published online: January 24, 2026
Processing time: 79 Days and 7.4 Hours
Pancreatic cancer remains a highly lethal malignancy, primarily due to late-stage diagnosis. Current screening paradigms, which focus exclusively on high-risk individuals, leave the vast “low-risk” population unscreened. This conventional binary risk stratification, based predominantly on family history and known genetic syndromes, fails to incorporate emerging risk dimensions such as poly
Core Tip: This commentary argues that the conventional “low-risk” label for pancreatic cancer, based solely on family history and known genetic syndromes, is obsolete. We propose a paradigm shift towards a dynamic, multidimensional risk model. This framework continuously integrates polygenic risk scores, liquid biopsy biomarkers (e.g., circulating tumor DNA), lifestyle factors, and artificial intelligence to enable real-time, personalized risk stratification. This approach can reclassify a significant portion of the “low-risk” population, paving the way for precision screening and potentially transforming early detection strategies for this lethal malignancy.
- Citation: Wang RG. Beyond sensitivity and specificity: Redefining the era connotation of “low-risk” in pancreatic cancer screening. World J Clin Oncol 2026; 17(1): 116090
- URL: https://www.wjgnet.com/2218-4333/full/v17/i1/116090.htm
- DOI: https://dx.doi.org/10.5306/wjco.v17.i1.116090
The most prevalent histological type of pancreatic cancer (PC) is pancreatic ductal adenocarcinoma (PDAC), which constitutes 80%-90% of all pancreatic malignancies[1]. The origin of this neoplasm is the epithelial cells that line the pancreatic ducts, leading to a high degree of invasiveness and a poor prognosis for patients[2,3]. PDAC is projected to become the second leading cause of cancer-related deaths by 2030, with a 5-year survival rate of only 12%[4,5]. This dismal outlook is largely a consequence of late diagnosis, highlighting the critical need for effective early detection strategies. However, current screening paradigms exclusively target high-risk individuals, leaving the vast “low-risk” general population without surveillance options[6,7], explicitly recommend against screening asymptomatic, non-high-risk individuals. This recommendation hinges on the low incidence of PDAC in the general population and the suboptimal positive predictive value of current screening tools.
The conventional definition of “low-risk” is predominantly based on the absence of known hereditary syndromes (e.g., breast cancer susceptibility gene 2, cyclin-dependent kinase inhibitor 2A, serine/threonine kinase 11 mutations) or a strong family history of PC[7,8]. However, this definition is increasingly outdated. Landmark studies, such as the cancer of pancreas screening trials, have demonstrated that screening high-risk individuals with magnetic resonance imaging and endoscopic ultrasonography can detect early-stage PDAC and its precursors, improving survival[9,10]. Meanwhile, advancements in molecular profiling have unveiled novel genetic risk variants beyond the classic syndromes[11-13]. For instance, a recent genome-wide association study identified five new PDAC susceptibility loci, each increasing risk by 15%-25%[12]. Concurrently, blood-based biomarkers, including circulating tumor DNA (ctDNA) and refined carbohy
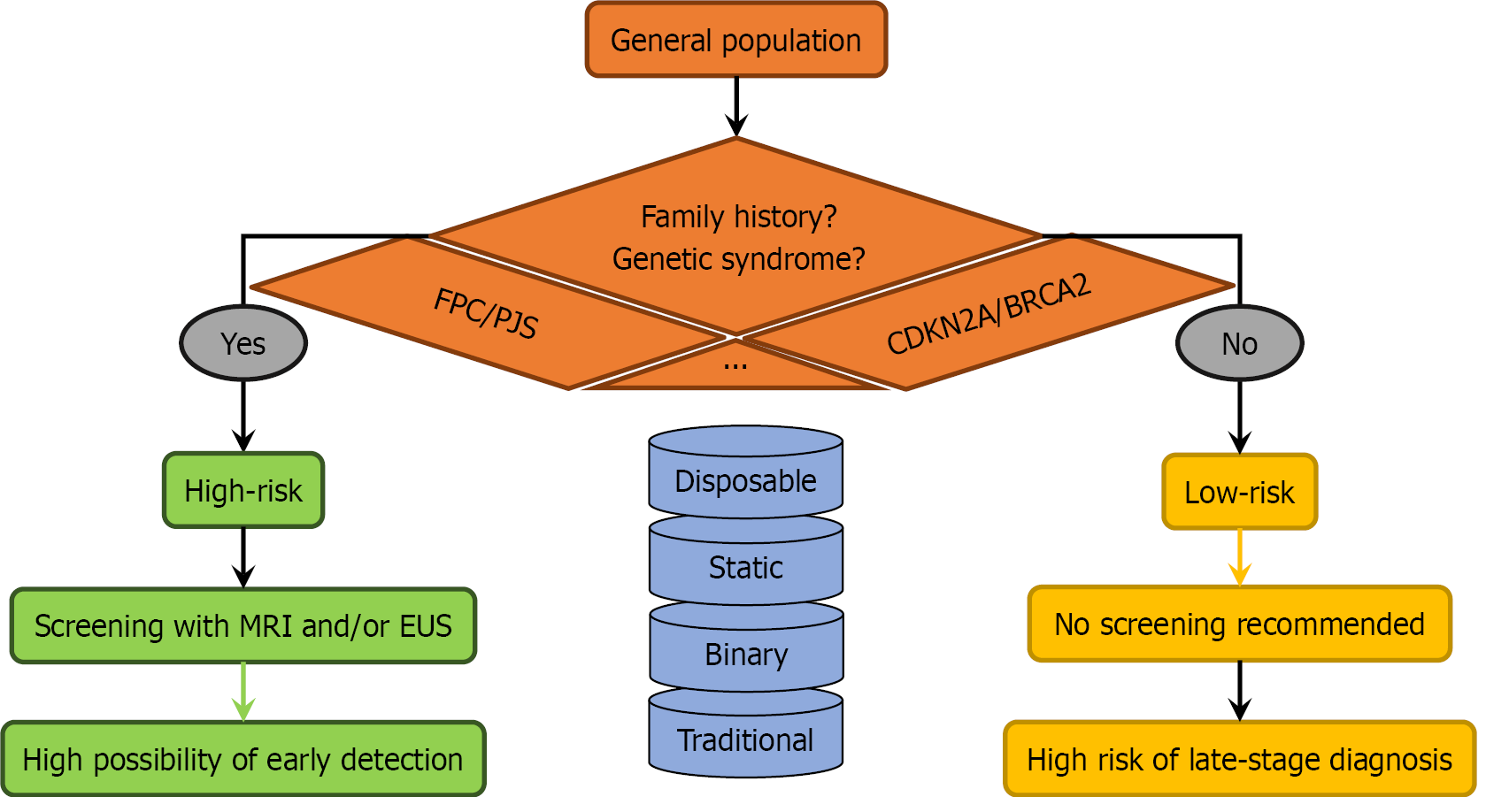
The prevailing model for PC screening is not only binary but fundamentally static, relying on a one-time assessment of a limited set of criteria (Figure 1). Individuals are categorized as “high-risk” if they meet specific criteria, such as having two or more first-degree relatives with PC, carrying a pathogenic mutation in a gene like cyclin-dependent kinase inhibitor 2A or breast cancer susceptibility gene 2, or having Peutz-Jeghers syndrome[6,7,9]. While this approach effectively targets a narrow subset of the population, it overlooks up to 90% of PC cases that occur in individuals classified as “low-risk” due to the absence of these specific criteria[12].
This approach is flawed for several reasons. First, a significant proportion of PDAC cases occur in individuals without a recognized family history or known genetic syndrome. Relying solely on these factors misses a substantial at-risk population. Second, the model fails to account for the cumulative impact of lower-penetrance genetic variants. The polygenic nature of PDAC risk means that an individual with several common risk variants, each conferring a modest increase in risk, may have a lifetime risk comparable to someone with a single high-penetrance mutation. Third, non-genetic factors, such as new-onset diabetes (NOD), chronic pancreatitis, and obesity, are not adequately integrated into the initial risk assessment[7,16]. NOD, in particular, is a well-established early indicator of PDAC, yet it is often not acted upon until symptoms develop.
The limitations of current imaging modalities underscore the need for complementary, non-invasive tools. Liquid biopsy has emerged as a promising frontier, offering a multifaceted view of the disease through several complementary components: (1) ctDNA: The detection of kirsten rat sarcoma viral oncogene homolog and tumor protein 53 mutations in ctDNA boasts high specificity for PDAC. Emerging technologies now allow for the identification of these mutations in plasma and even duodenal fluids with improving and clinically informative sensitivity for early-stage disease[11,17]. Moreover, the presence of post-operative ctDNA is a powerful predictor of recurrence and poor survival[18,19]; (2) Novel protein and glycomic biomarkers: Multi-marker panels that combine CA19-9 with other analytes, such as mucin 1 glycoprotein or specific glycomic profiles, are under active investigation[14,20]. For instance, the proximity-assisted click-mediated antibody-neutralization assay, an artificial intelligence (AI)-driven model combined with CA19-9, reportedly achieved high sensitivity for stage I PDAC[21], highlighting the power of integrated diagnostic approaches; and (3) Multimodal blood tests: Prospective studies are evaluating dual-screening strategies that combine clinical risk models with multi-analyte blood tests (e.g., including circulating microRNAs, extracellular vesicles) to identify high-risk individuals within general population cohorts[22]. The true potential of these biomarkers lies not in single measurements but in dynamic monitoring. Serial assessment of ctDNA variant allele frequency or CA19-9 kinetics can provide unparalleled insights into tumor evolution, thereby serving as the cornerstone for a continuously updated, dynamic risk model.
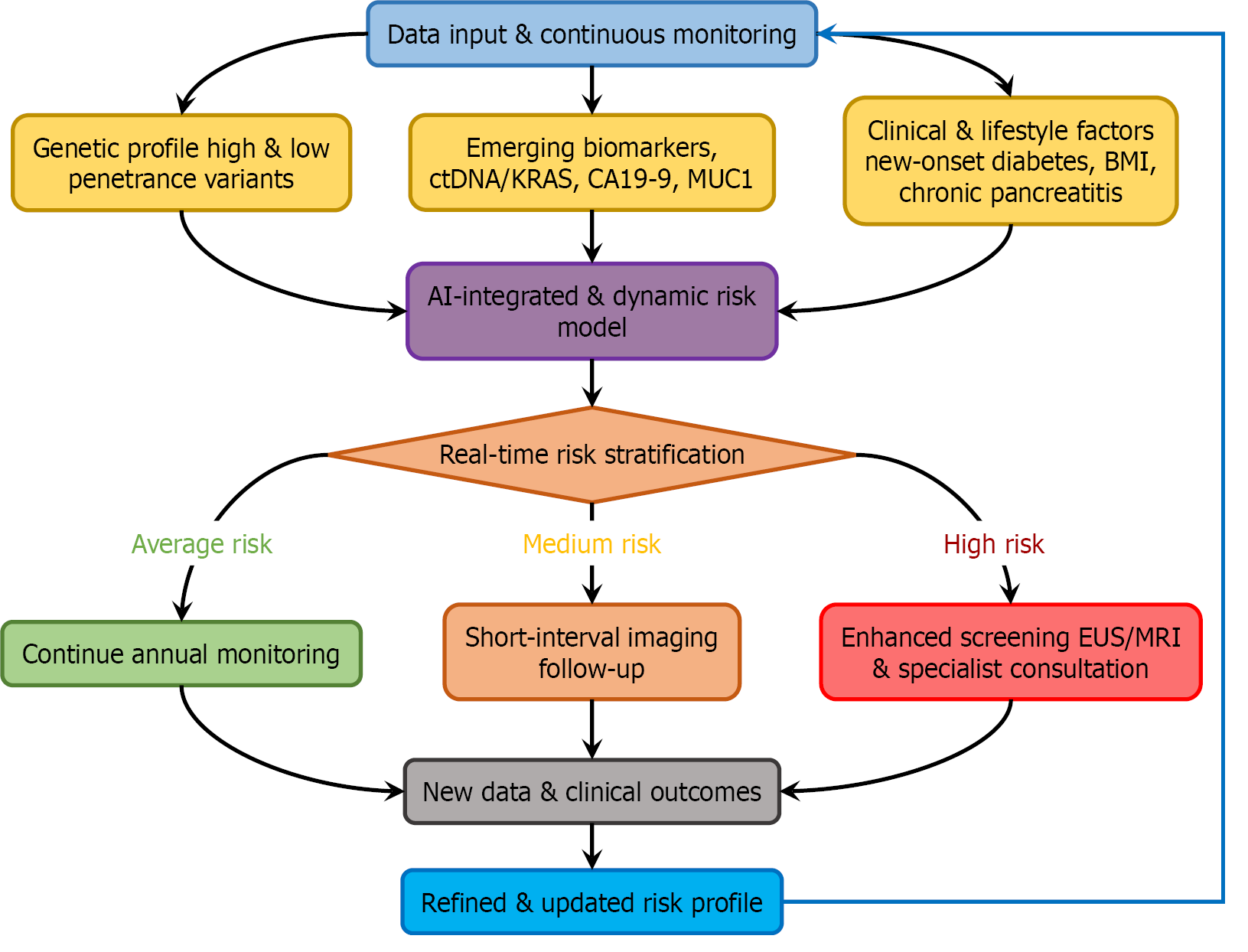
A modern risk stratification model must be multidimensional, quantifiable, and inherently dynamic, capable of evolving with new patient data (Figure 2). This involves several key components: (1) Polygenic risk scores (PRS): A PRS aggregates the effects of numerous common genetic variants into a single metric. An individual with a high PRS but no family history could be reclassified into a higher-risk category[23]; (2) Lifestyle and clinical factors: Factors like long-term smoking, high body mass index, and especially NOD after age 50 should be formally incorporated into risk algorithms[7]. The model should assign quantitative weights to these factors; and (3) AI and deep learning: AI serves as the essential computational engine for this model, capable of integrating these complex, non-linear data streams to generate predictive, dynamic risk scores[24-26]. For example, deep learning models applied to electronic health records have demonstrated high accuracy in predicting future cancer risk[27]. Such tools are ideally suited for the continuous re-evaluation required by our proposed framework.
Implementing this model necessitates a shift in clinical practice. It begins with an initial risk assessment (e.g., PRS + baseline clinical data around age 50). This risk score is continuously updated with longitudinal data, such as the onset of NOD, a significant weight change, or a rise in CA19-9. When an individual’s dynamic risk score crosses a predefined threshold, it triggers eligibility for imaging surveillance (e.g., endoscopic ultrasonography/magnetic resonance imaging). This transforms screening from a one-time qualification into a continuous, evidence-based monitoring process. The following graph illustrates how these emerging biomarkers can be integrated with traditional and novel risk factors to create a dynamic risk model (Figures 1 and 2).
Despite the promise of dynamic risk stratification, several challenges must be addressed before widespread clinical implementation. First, the ethical and psychological implications of reclassifying “low-risk” individuals based on genetic or biomarker data require careful consideration. Labeling patients as “intermediate” or “high-risk” may induce anxiety and lead to over-surveillance, underscoring the need for robust patient education and shared decision-making frameworks[28]. Second, the logistical and economic hurdles associated with genetic testing, repeated biomarker assays, and AI-based analytics remain substantial. However, cost-effectiveness analyses suggest that targeting screening to a redefined high-risk group could prove economically viable by focusing resources more efficiently[29]. Prospective validation in diverse populations is critical. Ongoing initiatives like the PC Early Detection (PRECEDE) Consortium and the PC Cohort Consortium (PanScan) are addressing this by developing multi-ethnic cohorts and adjusted risk algorithms[30]. Looking ahead, the convergence of continuous biomarker monitoring, AI-driven risk integration, and patient-specific screening intervals holds the potential to transform PC from a lethal disease into a detectable one. Future research should prioritize the standardization of assays, the refinement of AI algorithms through federated learning across institutions, and the execution of large-scale, risk-adaptive randomized trials to demonstrate the ultimate impact of dynamic risk stratification on PC mortality.
The definition of “low-risk” for PC is no longer fit for purpose in the modern era. Clinging to a binary, static model based solely on family history and a limited set of genetic syndromes inevitably misses opportunities for early detection in a significant portion of the population. The tools for a revolution are at hand: An expanding catalog of genetic risk variants, sophisticated liquid biopsy biomarkers, and powerful AI-based integration tools. By embracing a dynamic, multidimensional risk stratification model that continuously updates an individual’s risk based on genetics, biomarkers, and lifestyle, we can transition from a reactive to a proactive screening paradigm. This approach holds the promise of redefining “low-risk”, ultimately allowing us to extend the benefits of early detection to a broader population and finally change the trajectory of this devastating disease.
| 1. | Taherian M, Wang H, Wang H. Pancreatic Ductal Adenocarcinoma: Molecular Pathology and Predictive Biomarkers. Cells. 2022;11:3068. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 2. | Cai J, Chen H, Lu M, Zhang Y, Lu B, You L, Zhang T, Dai M, Zhao Y. Advances in the epidemiology of pancreatic cancer: Trends, risk factors, screening, and prognosis. Cancer Lett. 2021;520:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 318] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 3. | Jamal MH, Khan MN. Developments in pancreatic cancer emerging therapies, diagnostic methods, and epidemiology. Pathol Res Pract. 2025;271:156012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 4. | Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913-2921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5379] [Cited by in RCA: 5344] [Article Influence: 445.3] [Reference Citation Analysis (0)] |
| 5. | Dou Z, Lu C, Shen X, Gu C, Shen Y, Xu W, Qin S, Zhu J, Xu C, Li J. Development of a radiomics-3D deep learning fusion model for prognostic prediction in pancreatic cancer. BMC Cancer. 2025;25:1612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Conroy T, Pfeiffer P, Vilgrain V, Lamarca A, Seufferlein T, O'Reilly EM, Hackert T, Golan T, Prager G, Haustermans K, Vogel A, Ducreux M; ESMO Guidelines Committee. Pancreatic cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34:987-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 339] [Article Influence: 113.0] [Reference Citation Analysis (0)] |
| 7. | Pancreatic Disease Collaborative Group of Chinese Society of Digestive Endoscopology. [Chinese consensus for pancreatic cancer screening and surveillance in high-risk individuals (2021, Nanjing)]. Zhonghua Xiaohua Neijing Zazhi. 2022;39:85-95. [DOI] [Full Text] |
| 8. | Stoop TF, Javed AA, Oba A, Koerkamp BG, Seufferlein T, Wilmink JW, Besselink MG. Pancreatic cancer. Lancet. 2025;405:1182-1202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 120] [Article Influence: 120.0] [Reference Citation Analysis (0)] |
| 9. | Goggins M, Overbeek KA, Brand R, Syngal S, Del Chiaro M, Bartsch DK, Bassi C, Carrato A, Farrell J, Fishman EK, Fockens P, Gress TM, van Hooft JE, Hruban RH, Kastrinos F, Klein A, Lennon AM, Lucas A, Park W, Rustgi A, Simeone D, Stoffel E, Vasen HFA, Cahen DL, Canto MI, Bruno M; International Cancer of the Pancreas Screening (CAPS) consortium. Management of patients with increased risk for familial pancreatic cancer: updated recommendations from the International Cancer of the Pancreas Screening (CAPS) Consortium. Gut. 2020;69:7-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 441] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 10. | Dbouk M, Katona BW, Brand RE, Chak A, Syngal S, Farrell JJ, Kastrinos F, Stoffel EM, Blackford AL, Rustgi AK, Dudley B, Lee LS, Chhoda A, Kwon R, Ginsberg GG, Klein AP, Kamel I, Hruban RH, He J, Shin EJ, Lennon AM, Canto MI, Goggins M. The Multicenter Cancer of Pancreas Screening Study: Impact on Stage and Survival. J Clin Oncol. 2022;40:3257-3266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 162] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 11. | Abdel-Razeq R, Mansour A, Barbar M, Abu Shanap M, Sharaf B, Tamimi F, Mansour R, Muhanna A, Al-Othman Y, Hammad H, Shakhatreh M, Mahafdah S, Bani Hani H, Abdel-Razeq H. Enhancing Early Detection of Pancreatic Cancer in Genetically Predisposed Individuals: Integrating Advanced Imaging Modalities with Emerging Biomarkers and Liquid Biopsy. Biologics. 2025;19:511-523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 12. | Klein AP, Wolpin BM, Risch HA, Stolzenberg-Solomon RZ, Mocci E, Zhang M, Canzian F, Childs EJ, Hoskins JW, Jermusyk A, Zhong J, Chen F, Albanes D, Andreotti G, Arslan AA, Babic A, Bamlet WR, Beane-Freeman L, Berndt SI, Blackford A, Borges M, Borgida A, Bracci PM, Brais L, Brennan P, Brenner H, Bueno-de-Mesquita B, Buring J, Campa D, Capurso G, Cavestro GM, Chaffee KG, Chung CC, Cleary S, Cotterchio M, Dijk F, Duell EJ, Foretova L, Fuchs C, Funel N, Gallinger S, M Gaziano JM, Gazouli M, Giles GG, Giovannucci E, Goggins M, Goodman GE, Goodman PJ, Hackert T, Haiman C, Hartge P, Hasan M, Hegyi P, Helzlsouer KJ, Herman J, Holcatova I, Holly EA, Hoover R, Hung RJ, Jacobs EJ, Jamroziak K, Janout V, Kaaks R, Khaw KT, Klein EA, Kogevinas M, Kooperberg C, Kulke MH, Kupcinskas J, Kurtz RJ, Laheru D, Landi S, Lawlor RT, Lee IM, LeMarchand L, Lu L, Malats N, Mambrini A, Mannisto S, Milne RL, Mohelníková-Duchoňová B, Neale RE, Neoptolemos JP, Oberg AL, Olson SH, Orlow I, Pasquali C, Patel AV, Peters U, Pezzilli R, Porta M, Real FX, Rothman N, Scelo G, Sesso HD, Severi G, Shu XO, Silverman D, Smith JP, Soucek P, Sund M, Talar-Wojnarowska R, Tavano F, Thornquist MD, Tobias GS, Van Den Eeden SK, Vashist Y, Visvanathan K, Vodicka P, Wactawski-Wende J, Wang Z, Wentzensen N, White E, Yu H, Yu K, Zeleniuch-Jacquotte A, Zheng W, Kraft P, Li D, Chanock S, Obazee O, Petersen GM, Amundadottir LT. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun. 2018;9:556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 202] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 13. | Zhang YD, Hurson AN, Zhang H, Choudhury PP, Easton DF, Milne RL, Simard J, Hall P, Michailidou K, Dennis J, Schmidt MK, Chang-Claude J, Gharahkhani P, Whiteman D, Campbell PT, Hoffmeister M, Jenkins M, Peters U, Hsu L, Gruber SB, Casey G, Schmit SL, O'Mara TA, Spurdle AB, Thompson DJ, Tomlinson I, De Vivo I, Landi MT, Law MH, Iles MM, Demenais F, Kumar R, MacGregor S, Bishop DT, Ward SV, Bondy ML, Houlston R, Wiencke JK, Melin B, Barnholtz-Sloan J, Kinnersley B, Wrensch MR, Amos CI, Hung RJ, Brennan P, McKay J, Caporaso NE, Berndt SI, Birmann BM, Camp NJ, Kraft P, Rothman N, Slager SL, Berchuck A, Pharoah PDP, Sellers TA, Gayther SA, Pearce CL, Goode EL, Schildkraut JM, Moysich KB, Amundadottir LT, Jacobs EJ, Klein AP, Petersen GM, Risch HA, Stolzenberg-Solomon RZ, Wolpin BM, Li D, Eeles RA, Haiman CA, Kote-Jarai Z, Schumacher FR, Al Olama AA, Purdue MP, Scelo G, Dalgaard MD, Greene MH, Grotmol T, Kanetsky PA, McGlynn KA, Nathanson KL, Turnbull C, Wiklund F; Breast Cancer Association Consortium (BCAC); Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON); Colon Cancer Family Registry (CCFR); Transdisciplinary Studies of Genetic Variation in Colorectal Cancer (CORECT); Endometrial Cancer Association Consortium (ECAC); Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO); Melanoma Genetics Consortium (GenoMEL); Glioma International Case-Control Study (GICC); International Lung Cancer Consortium (ILCCO); Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Consortium; International Consortium of Investigators Working on Non-Hodgkin’s Lymphoma Epidemiologic Studies (InterLymph); Ovarian Cancer Association Consortium (OCAC); Oral Cancer GWAS; Pancreatic Cancer Case-Control Consortium (PanC4); Pancreatic Cancer Cohort Consortium (PanScan); Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL); Renal Cancer GWAS; Testicular Cancer Consortium (TECAC), Chanock SJ, Chatterjee N, Garcia-Closas M. Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat Commun. 2020;11:3353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 14. | Maekawa A, Oba A, Mie T, Sawa Y, Baba H, Kobayashi K, Ono Y, Sato T, Ito H, Sasaki T, Ozaka M, Sasahira N, Takamatsu M, Ban D, Inoue Y, Takahashi Y. Exploring the role of CA 19-9 dynamics in pancreatic cancer multidisciplinary treatment: Proposal of the PANC classification. Surgery. 2025;188:109783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | Lin WC, Yu LY, Kuo YC, Chang CW, Wang HY, Shih SC, Chang CW, Lin HH, Chan YH, Lin YC, Hu KC. Role of endoscopic ultrasonography or magnetic resonance imaging for screening of pancreatic cancer in low-risk individuals. World J Clin Oncol. 2025;16:112030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Blackford AL, Canto MI, Dbouk M, Hruban RH, Katona BW, Chak A, Brand RE, Syngal S, Farrell J, Kastrinos F, Stoffel EM, Rustgi A, Klein AP, Kamel I, Fishman EK, He J, Burkhart R, Shin EJ, Lennon AM, Goggins M. Pancreatic Cancer Surveillance and Survival of High-Risk Individuals. JAMA Oncol. 2024;10:1087-1096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 79] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 17. | Yachida S, Yoshinaga S, Shiba S, Urabe M, Tanaka H, Takeda Y, Shimizu A, Sakamoto Y, Hijioka S, Haba S, Ashida R, Kushiyama Y, Asano K, Kobayashi M, Murawaki Y, Onishi K, Yamashita T, Kimura H, Totoki Y, Kamada H, Isomoto H, Hattori S, Morizane C, Ohkawa K, Kitano M, Hara K, Ikezawa K, Hanada K, Matsumoto K. KRAS Mutations in Duodenal Lavage Fluid after Secretin Stimulation for Detection of Pancreatic Cancer. Ann Surg. 2025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 18. | Eckhoff AM, Kanu E, Fletcher A, Bao M, Aushev VN, Spickard E, Nussbaum DP, Allen PJ. Initial Report: Personalized Circulating Tumor DNA and Survival in Patients with Resectable Pancreatic Cancer. Ann Surg Oncol. 2024;31:1444-1446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 19. | Lee B, Lipton L, Cohen J, Tie J, Javed AA, Li L, Goldstein D, Burge M, Cooray P, Nagrial A, Tebbutt NC, Thomson B, Nikfarjam M, Harris M, Haydon A, Lawrence B, Tai DWM, Simons K, Lennon AM, Wolfgang CL, Tomasetti C, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann Oncol. 2019;30:1472-1478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 171] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 20. | Engels MML, Berger CK, Mahoney DW, Hoogenboom SA, Sarwal D, Klatte DCF, De La Fuente J, Gandhi S, Taylor WR, Foote PH, Doering KA, Delgado AM, Burger KN, Abu Dayyeh BK, Bofill-Garcia A, Brahmbhatt B, Chandrasekhara V, Gleeson FC, Gomez V, Kumbhari V, Law RJ, Lukens FJ, Raimondo M, Rajan E, Storm AC, Vargas Valls EJ, van Hooft JE, Wallace MB, Kisiel JB, Majumder S. Multimodal Pancreatic Cancer Detection Using Methylated DNA Biomarkers in Pancreatic Juice and Plasma CA 19-9: A Prospective Multicenter Study. Clin Gastroenterol Hepatol. 2025;23:766-775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 21. | Montoya Mira JL, Quentel A, Patel RK, Keith D, Sousa M, Minnier J, Kingston BR, David L, Esener SC, Sears RC, Lopez CD, Sheppard BC, Demirci U, Wong MH, Fischer JM. Early detection of pancreatic cancer by a high-throughput protease-activated nanosensor assay. Sci Transl Med. 2025;17:eadq3110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 22. | Toledano-Fonseca M, Brozos-Vázquez E, García-Ortiz MV, Costa-Fraga N, Díaz-Lagares Á, Rodríguez-Ariza A, López-López R, Aranda E. Unveilling the potential of liquid biopsy in pancreatic cancer for molecular diagnosis and treatment guidance. Crit Rev Oncol Hematol. 2025;212:104807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 23. | Galeotti AA, Gentiluomo M, Rizzato C, Obazee O, Neoptolemos JP, Pasquali C, Nentwich M, Cavestro GM, Pezzilli R, Greenhalf W, Holleczek B, Schroeder C, Schöttker B, Ivanauskas A, Ginocchi L, Key TJ, Hegyi P, Archibugi L, Darvasi E, Basso D, Sperti C, Bijlsma MF, Palmieri O, Hlavac V, Talar-Wojnarowska R, Mohelnikova-Duchonova B, Hackert T, Vashist Y, Strouhal O, van Laarhoven H, Tavano F, Lovecek M, Dervenis C, Izbéki F, Padoan A, Małecka-Panas E, Maiello E, Vanella G, Capurso G, Izbicki JR, Theodoropoulos GE, Jamroziak K, Katzke V, Kaaks R, Mambrini A, Papanikolaou IS, Szmola R, Szentesi A, Kupcinskas J, Bursi S, Costello E, Boggi U, Milanetto AC, Landi S, Gazouli M, Vodickova L, Soucek P, Gioffreda D, Gemignani F, Brenner H, Strobel O, Büchler M, Vodicka P, Paiella S, Canzian F, Campa D. Polygenic and multifactorial scores for pancreatic ductal adenocarcinoma risk prediction. J Med Genet. 2021;58:369-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 24. | Farinella R, Felici A, Peduzzi G, Testoni SGG, Costello E, Aretini P, Blazquez-Encinas R, Oz E, Pastore A, Tacelli M, Otlu B, Campa D, Gentiluomo M. From classical approaches to artificial intelligence, old and new tools for PDAC risk stratification and prediction. Semin Cancer Biol. 2025;112:71-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 25. | Javed S, Qureshi TA, Gaddam S, Wang L, Azab L, Wachsman AM, Chen W, Asadpour V, Jeon CY, Wu B, Xie Y, Pandol SJ, Li D. Risk prediction of pancreatic cancer using AI analysis of pancreatic subregions in computed tomography images. Front Oncol. 2022;12:1007990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 26. | Huang B, Huang H, Zhang S, Zhang D, Shi Q, Liu J, Guo J. Artificial intelligence in pancreatic cancer. Theranostics. 2022;12:6931-6954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 27. | Mishra AK, Chong B, Arunachalam SP, Oberg AL, Majumder S. Machine Learning Models for Pancreatic Cancer Risk Prediction Using Electronic Health Record Data-A Systematic Review and Assessment. Am J Gastroenterol. 2024;119:1466-1482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 28. | Chavarri-Guerra Y, Slavin TP, Longoria-Lozano O, Weitzel JN. Genetic cancer predisposition syndromes among older adults. J Geriatr Oncol. 2020;11:1054-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Schwartz NRM, Matrisian LM, Shrader EE, Feng Z, Chari S, Roth JA. Potential Cost-Effectiveness of Risk-Based Pancreatic Cancer Screening in Patients With New-Onset Diabetes. J Natl Compr Canc Netw. 2021;20:451-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 30. | Gonda TA, Everett JN, Wallace M, Simeone DM; PRECEDE Consortium. Recommendations for a More Organized and Effective Approach to the Early Detection of Pancreatic Cancer From the PRECEDE (Pancreatic Cancer Early Detection) Consortium. Gastroenterology. 2021;161:1751-1757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/