Published online Oct 24, 2025. doi: 10.5306/wjco.v16.i10.107877
Revised: May 29, 2025
Accepted: September 22, 2025
Published online: October 24, 2025
Processing time: 208 Days and 8.9 Hours
Gastrointestinal malignancies, particularly pancreatobiliary and gastroesophageal cancers, are associated with poor prognosis due to their frequent late-stage dia
Core Tip: Gastrointestinal malignancies, more specifically pancreatobiliary and gastroesophageal cancers, harbor a bad prognosis mainly because tumors are frequently diagnosed in an advanced stage. Most of the tumors of the digestive tract associate anorexia-cachexia syndrome as well as malnutrition. Inflammation is a key feature of neoplasic proliferation and gut microbiome may have an interplay in inflammation and outcomes of these patients. In this article, we want to address this association and the impact of nutritional strategies in patients with gastrointestinal malignancies.
- Citation: Pacheco-Barcia V, Mariño-Mendez A, Hernandez-Jimenez E, Jimenez-Fonseca P, Muñoz Martín AJ, Custodio-Cabello S, Palka-Kotlowska M, Gonzalez-Diaz I, Cabezon-Gutierrez L. Gut microbiome and nutritional strategies in gastrointestinal cancers: Clinical implications and therapeutic perspectives. World J Clin Oncol 2025; 16(10): 107877
- URL: https://www.wjgnet.com/2218-4333/full/v16/i10/107877.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i10.107877
Cancer is considered as a main leading cause of death worldwide[1]. The International Agency for Research on Cancer reported an estimated 20 million new cancer cases and 9.7 million cancer deaths in 2022[2]. The hallmarks of cancer were early described to include fourteen biological capabilities which have essential roles in contributing to tumor complexity[3]. Among these are deregulating cellular metabolism, genome instability, tumor-promoting inflammation, and polymorphic microbiomes, which underscore the growing importance of the gut microbiome and inflammation in tumorigenesis.
In the late 19th century, German pathologist Rudolf Virchow proposed that cancer could arise as a consequence of chronic inflammation triggered by persistent exposure to harmful toxic agents, including infections[4]. Around the same time, pioneering research by Robert Koch and Louis Pasteur led to the identification of bacteria within tumor tissues, suggesting a potential link between bacterial infections and cancer development[4]. The term “human microbiome” refers to the diverse community of microorganisms residing within the human body, along with their collective genetic material[5,6].
There is a growing recognition that the complex ecosystems formed by resident bacteria and fungi - the microbiomes - play a crucial role in both health and disease[7]. This understanding has been significantly advanced by the development of next-generation sequencing and bioinformatics, enabling a detailed characterization of microbial populations. In the context of cancer, mounting evidence suggests that inter-individual variability in microbiome composition can profoundly influence cancer phenotypes[8,9]. Currently, approximately 20% of malignancies worldwide are linked to infections[10], accounting for an estimated 1.2 million cases annually[11]. Research into the microbiome, particularly the role of bacteria in cancer pathogenesis, is evolving rapidly, with more than about 100 trillion bacterial cells representing about 10000 species across the human population identified within the human body to date[12,13].
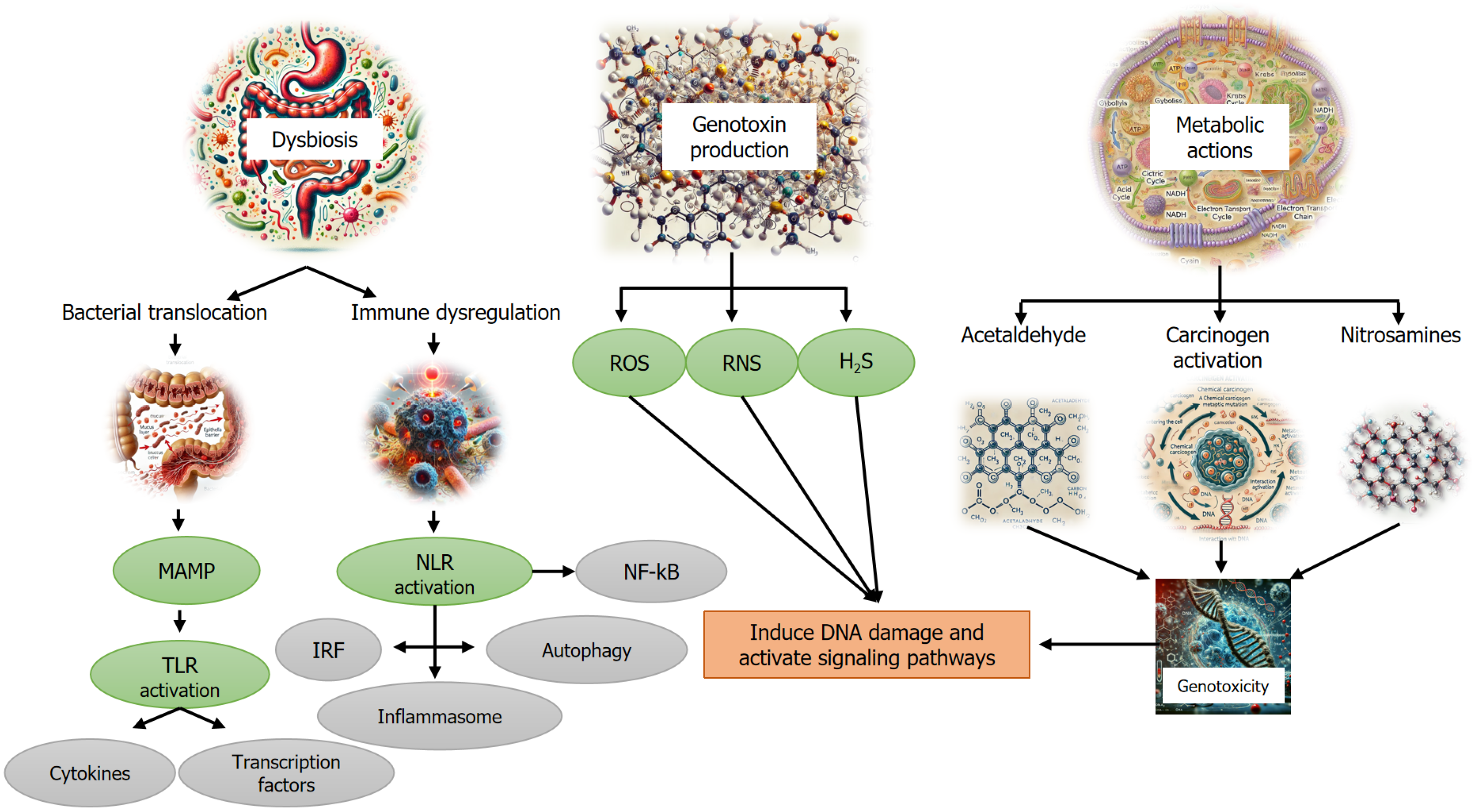
The microbiome has been implicated in tumor development and progression through various mechanisms, including DNA damage, activation of oncogenic pathways and epithelial cell proliferation[14,15], production of carcinogenic metabolites[16], induction of chronic inflammation[17,18], and suppression of antitumor immunity[14,16]. These insights have expanded the understanding of the intricate interactions between the tumor microenvironment and systemic microbial-immune networks, suggesting a broader and more complex influence than previously recognized[19].
Notably, microbial dysbiosis, an imbalance in microbial composition that disrupts host homeostasis, has been shown to significantly affect therapeutic responses to anticancer treatments[20,21]. This is largely attributed to the microbiome’s capacity to metabolize drugs, modulate inflammation, and influence immune responses within the tumor microenvironment, all of which play a crucial role in treatment efficacy and drug toxicity[22]. The relationship between the microbiome and anticancer therapy responses has been described as bidirectional, with both factors exerting significant effects on each other[20,23]. This has led to the emergence of “pharmacomicrobiomics”, a field dedicated to studying the interaction between drugs and microbial communities[20]. In this context, the role of probiotics and antibiotics, whether used alone or in combination with anticancer agents, is being explored as a strategy to manipulate the microbiome and potentially improve cancer prevention and treatment outcomes[20,24].
Increasing evidence supports the role of bacterial infections in the pathogenesis and progression of various gastrointestinal (GI) malignancies[25,26]. Additionally, substantial data indicate that the microbiome influences tumor responses to anticancer therapies, including conventional chemotherapy and targeted treatments[20,27]. Consequently, the bacterial microbiome represents a promising therapeutic target for the prevention and treatment of GI cancers. Moreover, dietary patterns play a significant role in shaping gut microbiome composition, serving as a key factor in cancer development[28].
This article explores the current understanding of the intricate relationship between the bacterial microbiome and inflammation in GI malignancies. Particular emphasis is placed on its potential as a therapeutic target, with a specific focus on nutritional interventions as a modifiable factor in disease management.
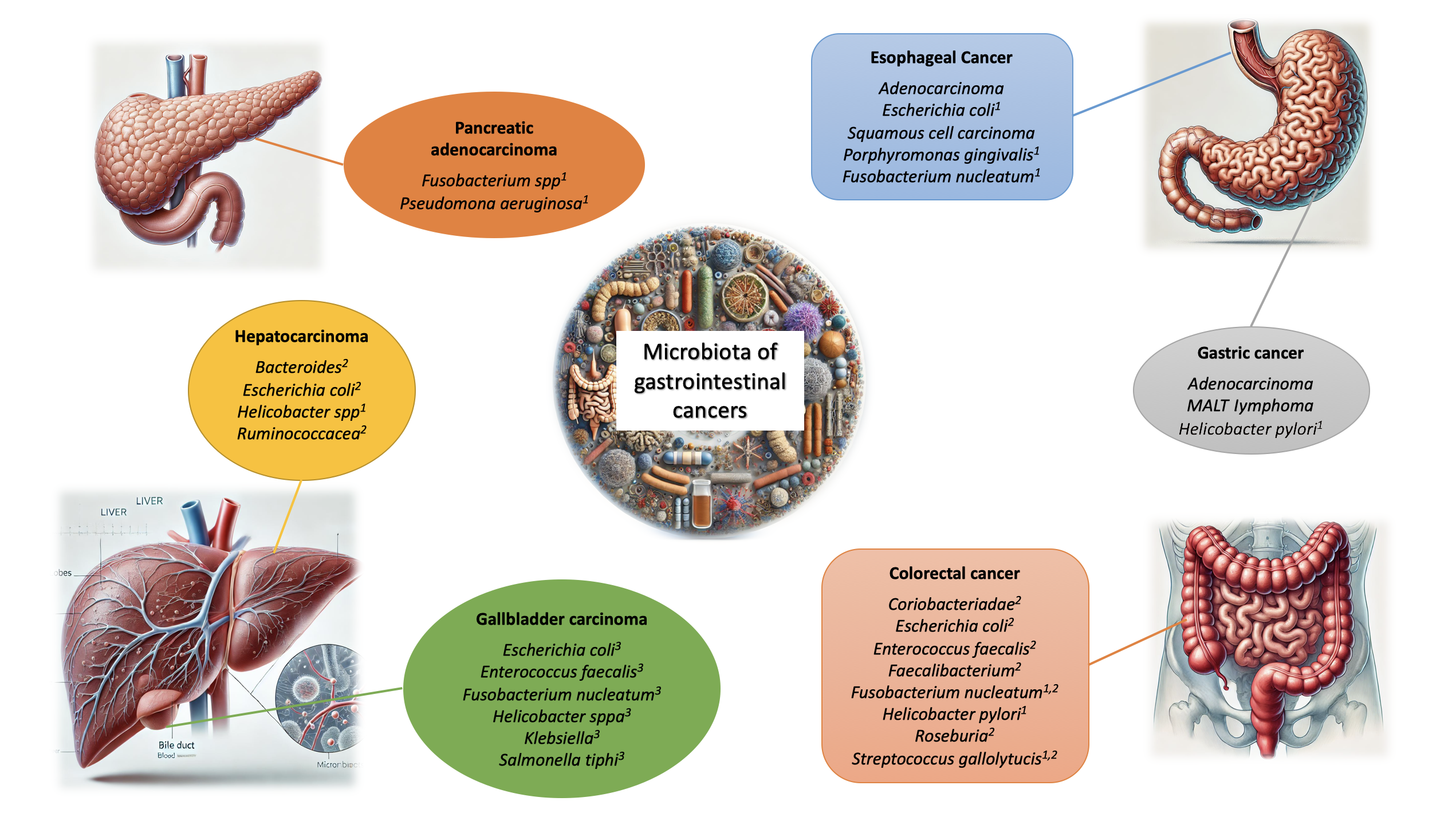
The majority of microorganisms comprising the human microbiome are commensal. The highest microbial diversity is observed in the GI tract, particularly in the cecum and proximal colon. This diversity is shaped by multiple factors, including mode of delivery at birth, infant feeding practices, dietary patterns, lifestyle factors, and host genetics[29]. However, substantial evidence has definitively shown that gut microbiome plays a role in the development of various medical conditions, including inflammatory, hepatic, pancreatic, and respiratory diseases, as well as neurological and dermatological disorders, and cancer[30]. Figure 1 summarizes the most common bacterial species associated with cancers affecting GI tract.
Cancer and the immune system are closely interconnected. Similarly, a robust interaction between gut microbiome and both innate and adaptive immunity has been well-documented. The interplay between bacteria, viruses, parasites, fungi, and mucosal immune cells is regulated by a complex network of cytokines[31].
Germ-free mouse models have demonstrated the critical role of gut microbiome in the development and regulation of various organs and systems, including the immune and endocrine systems, blood, liver, and lungs[32]. Within the intestine, the microbiome maintains epithelial homeostasis and supports the formation of gut-associated lymphoid tissue. Additionally, it promotes the production of epithelial cytokines, which in turn regulate the activity of T and B lymphocytes, macrophages, and polymorphonuclear cells[33,34]. Cytokines like interleukin (IL)-1β, tumor necrosis factor (TNF) α, IL-2, IL-6, IL-15, IL-21, and IL-23 can trigger an inflammatory response, whereas IL-10 and TGF-β exhibit anti-inflammatory properties. The balance between these pro-inflammatory and anti-inflammatory cytokines determines the gut’s inflammatory or homeostatic state[35].
Helicobater pylori (H. pylori) colonization induces chronic inflammation by overexpressing pro-inflammatory cytokines like IL-1β, IL-8, IL-17, and TNF-α, which increases the risk of gastric cancer (GC)[36]. In patients with gastric adenocarcinoma, higher abundances of Lactobacillus, Bifidobacterium, Lactococcus, and Streptococcus have been observed[37]. Lactic acid bacteria (LAB) contribute to DNA damage by producing reactive oxygen species (ROS) and reducing nitrates to nitrites, which drive mutagenesis, proto-oncogene overexpression, angiogenesis, and inhibit apoptosis[37]. LAB also plays a key role in epithelial-mesenchymal transition[38]. Escherichia coli (E. coli) produces colibactin, a compound involved in colorectal carcinoma development[39]. Additionally, Fusobacterium nucleatum (F. nucleatum), E. coli NC101, and Bacteroides fragilis promote colorectal cancer (CRC) by activating the Wnt-β-catenin signaling pathway[40]. Increased abundances of genera like Bacteroides fragilis, Campylobacter, Enterococcus, F. nucleatum, Streptococcus, Prevotella, and Peptostreptococcus, and decreased abundances of Bifidobacterium, Clostridium spp., Lactobacillus, Ruminococcus, and Roseburia have been noted in CRC patients[41].
Pathogen-associated molecular patterns are recognized by pattern recognition receptors, such as toll-like receptors (TLRs) on macrophages and dendritic cells. TLRs initiate immune responses by activating signaling cascades, including the nuclear factor kappa B (NF-κB), mitogen-activated protein kinase (MAPK), and interferon regulatory factor pathways[42]. For instance, TLR3 and TLR4 engage interferon regulatory factor to induce interferon-beta, while others signal through the myeloid differentiation primary response 88 (MyD88)-interleukin-1 receptor-associated kinase axis, triggering MAPK like p38 and c-Jun N-terminal kinase (JNK) that culminate in the transcription of pro-inflammatory genes[42]. Although TLR-mediated signaling is vital for maintaining epithelial integrity and immune homeostasis, aberrant activation can contribute to tumorigenesis[43-47]. Overexpression of TLRs has been observed in various cancers, and studies in MyD88-deficient mice demonstrate reduced tumor development[48], underscoring the oncogenic potential of dysregulated TLR signaling. Additionally, JNK activation downstream of TLRs may intersect with Ras pathways, promoting apoptosis resistance and increased matrix metalloproteinase expression-hallmarks of cancer progression[17,47,49,50].
The connection between TLR signaling, JNK activation, and Ras-driven oncogenesis in CRC is largely based on in vitro evidence using human colon epithelial cell lines. Several studies have shown that activation of TLR4 by bacterial ligands can stimulate the JNK cascade and downstream Ras signaling, resulting in increased expression of pro-survival and proliferative genes[51,52]. However, the relevance of this pathway in vivo remains speculative. Animal models have yielded inconsistent results, with some indicating tumor-promoting effects of TLR overactivation, while others report context-dependent roles for JNK that may be either oncogenic or tumor suppressive depending on isoform specificity and the inflammatory milieu[53,54]. Additionally, the crosstalk between TLR/JNK/Ras pathways and host immune responses, microbiota composition, and epithelial integrity is complex and not yet fully characterized in preclinical systems. Thus, while in vitro data support mechanistic hypotheses, further in vivo studies are required to clarify the functional significance of this signaling axis in colorectal tumorigenesis.
CD8+ cytotoxic T-lymphocytes (CTLs) are crucial for anti-cancer immunity, as they kill malignant cells upon recognizing antigenic peptides[35]. Tumor-specific CTL responses are supported by tumor-associated antigens and the presence of tumor-associated antigen-specific CD8+ T-cells in regressing tumors. In CRC, tumor-infiltrating lymphocytes are primarily CD4+ T cells, producing pro-inflammatory cytokines like induce interferon-γ and IL-17, whereas subsets producing IL-4 may promote oncogenesis[55]. Additionally, natural killer cells at tumor sites in long-surviving patients trigger tumor apoptosis and inhibit proliferation[56].
Cancer cells induce an immunosuppressive microenvironment by producing factors like TGF-β and recruiting regulatory immune cells, such as T-regs, which are inversely correlated with tumor prognosis[57]. T-regs suppress tumor-specific T-lymphocytes by producing IL-10 and TGF-β, consuming IL-2, or expressing CTL-associated protein 4 (CTLA-4). They also inhibit pro-inflammatory CD4+ T-helper (Th) cells and stimulate B cells to produce immunoglobulins. Th17 cells, involved in carcinogenesis by promoting cell proliferation and inhibiting apoptosis, produce cytokines like IL-17 and IL-23, contributing to tumor growth[58-62]. Th17 cells also promote the production of Th1-related cytokines, such as C-X-C motif chemokine ligand (CXCL) 9 and CXCL10, at the tumor site. T-cell exhaustion, a phenomenon where persistent tumor antigen stimulation impairs T-cell function, is a primary mechanism of immune escape, allowing tumors to continue growing despite initial immune responses[63-65]. Figure 2 summarizes how the microbiome promotes carcinogenesis through various mechanisms.
In a healthy state, there is a harmonious balance between the gut microbiome and the immune system at the gut interface[66]. Disruption of this balance leads to a pathological condition called “gut dysbiosis”, which fosters the overgrowth of pathogenic bacteria in the intestinal lumen. Dysbiosis can arise from an inflammatory state: In genetically predisposed individuals, factors such as dietary compounds, toxins, and antibiotics can trigger low-grade inflammation, promoting dysbiosis. For instance, high-calorie and high-fat diets common in Western societies have been linked to worsened gut inflammation in patients with inflammatory bowel disease (IBD)[67]. The following sections will develop and analyze the relationship between various treatments for GI tumors and the nutritional strategies that may influence the microbiome and inflammation.
While growing evidence implicates the gut microbiome in the initiation and progression of cancer through mechanisms such as chronic inflammation, genotoxin production, and immune modulation, this pathogenic potential also opens the door to therapeutic opportunities. Among the most accessible and modifiable factors influencing the gut microbiota is diet. The next section explores how targeted nutritional interventions - particularly those involving dietary fiber and microbiota-accessible carbohydrates - can reshape the gut microbial landscape to potentially prevent carcinogenesis and enhance treatment responses.
Probiotics, defined as live microorganisms that, when consumed in sufficient amounts, confer health advantages to the host, have gained attention for their potential role in CRC prevention and treatment. Their beneficial effects are primarily attributed to three mechanisms: Suppression of pathogenic bacterial colonization, modulation of gut immune responses, and reinforcement of intestinal barrier function[68]. The antimicrobial properties of probiotics against pathogenic bacteria have been well-documented. For example, Bacillus species were shown to prevent intestinal colonization by Staphylococcus aureus by disrupting its quorum sensing communication system[69]. Similarly, Clostridium butyricum suppressed biofilm formation by enterotoxigenic Bacteroides fragilis by regulating key virulence and efflux pump-related genes[70]. Certain Lactobacillus strains have also exhibited protective effects by preventing bacterial adhesion. Lactobacillus fermentum 88 and Lactobacillus plantarum 9 demonstrated strong adherence to human enteric cell line HT29 while simultaneously inhibiting E. coli attachment[71]. Furthermore, pre-administration of Lactobacillus rhamnosus GG in a mouse model of periodontitis provided protection against F. nucleatum and Porphyromonas gingivalis-induced cecal dysbiosis, significantly mitigating intestinal inflammation[72].
Probiotics are recognized for their immunomodulatory functions in the gut, where they play a crucial role in mitigating colonic inflammation. In an azoxymethane and dextran sulfate sodium-induced CRC murine model, Lactobacillus species were found to suppress Wnt/β-catenin signaling, leading to reduced inflammation and tumor growth[73]. Additionally, a specific strain of Lactobacillus rhamnosus demonstrated anti-tumor properties by reducing inflammatory responses and promoting apoptosis, ultimately decreasing tumor incidence[74]. In genetic models of CRC, probiotics have exhibited protective effects. In the APC Min/+ mouse model of colon cancer, Lactobacillus plantarum strain YYC-3 was shown to inhibit tumor formation, likely by downregulating inflammatory cytokine production and limiting inflammatory cell infiltration[75]. Furthermore, the administration of a probiotic mixture containing Lactobacillus acidophilus, L. rhamnosus, and Bifidobacterium bifidum significantly reduced colitis and resulted in a 40% decrease in tumor development compared to the control group[76].
Disruptions in tight junction proteins - such as occludin, claudins, junctional adhesion molecules, and zona occludens (ZO) - can lead to increased intestinal permeability, fostering chronic inflammation and heightening the risk of IBD-associated CRC[77]. Probiotics have been shown to enhance gut barrier integrity, providing a potential preventive mechanism. Notably, E. coli nissle 1917 improved intestinal barrier function by upregulating and redistributing tight junction proteins, including ZO-1, ZO-2, and claudin-14[78]. The beneficial effects of probiotics on gut barrier integrity have also been observed in colitis models. A probiotic formulation containing Bifidobacterium, L. acidophilus, and Enterococcus ameliorated colitis in mice by upregulating occludin and claudin-4 expression[79]. Additionally, in a 1,2-dimethylhydrazine dihydrochloride-induced CRC mouse model, supplementation with L. acidophilus, B. bifidum, and Bifidobacterium infantum strengthened gut mucosal barrier integrity through TLR2 signaling, which was associated with a lower incidence of tumor formation[80].
While some strains of LAB have been reported to generate ROS or contribute to nitrate metabolism under specific conditions, it is important to recognize that most probiotic formulations - particularly those used in clinical and nutritional contexts - employ LAB for their protective effects[81]. For instance, some studies[82-84], highlight the role of LAB in safeguarding the gastric mucosa, modulating inflammation, and attenuating the toxicity of chemotherapy agents. Thus, while emerging data suggest that certain LAB strains may possess pro-carcinogenic potential under select circumstances, the broader evidence base supports their beneficial role in GI health and cancer therapy support. A nuanced interpretation is warranted to avoid overgeneralizing LAB as solely pro-carcinogenic.
Prebiotics, such as inulin, fructo-oligosaccharides (FOS), and galactooligosaccharides are non-digestible carbohydrates that selectively stimulate the proliferation of beneficial anaerobic bacteria within the colon. These fibers bypass digestion in the small intestine and undergo fermentation by colonic microbiome, promoting the growth of beneficial bacterial genera, particularly Bifidobacterium spp.[85]. Elevated levels of Bifidobacterium spp. have been associated with a reduced incidence and progression of tumors[86]. Additionally, inulin and its oligofructose derivatives - characterized by a degree of polymerization ≤ 10 - were observed to decrease the formation of aberrant crypt foci in chemically induced colon carcinogenesis models in mice[87]. Research by Taper et al[88-90] explored the impact of dietary supplementation with 15% inulin or oligofructose on chemotherapy efficacy in experimental animals. Findings demonstrated that these prebiotics significantly enhanced the therapeutic efficacy of six commonly used chemotherapeutic agents, including 5-fluorouracil (5-FU) doxorubicin, vincristine, cyclophosphamide, methotrexate, and cytarabine[90].
Consistent with prior investigations from the same research group, inulin notably amplified the therapeutic response to cyclophosphamide by 47%[88]. Moreover, no detrimental interactions or adverse effects were observed when inulin or oligofructose was administered as an adjuvant therapy, highlighting their potential role in optimizing cancer treatment outcomes[90]. A randomized, double-blind, placebo-controlled trial evaluated the effects of a fiber blend (50% inulin and 50% FOS) on gut microbiome in gynecological cancer patients undergoing radiotherapy[91]. Patients receiving the prebiotic mixture exhibited a quicker restoration of Lactobacillus spp. and Bifidobacterium spp. populations within two weeks after radiotherapy, leading to improved stool consistency compared to the placebo group[91,92]. Diarrhea is a frequent side effect of enteral nutrition, often delaying recovery and prolonging hospital stays, particularly in postoperative GC patients. A study of 120 GC patients, divided into three groups (fiber-free nutrition, fiber-enriched nutrition, and fiber- and probiotic-enriched nutrition), found that hospital stays were shorter in the fiber-supplemented groups, with a significant reduction in diarrhea occurrence compared to the fiber-free group[93].
Recently, a phase II randomized controlled trial was initiated to explore the interactions between gut microbiome and prebiotics in patients with localized anal canal squamous cell carcinoma receiving radiotherapy, aiming to assess their impact on treatment outcomes[94]. Another randomized, double-blind clinical trial examined the effects of prebiotics (FOS, xylooligosaccharides, polydextrose, and resistant dextrin) on gut microbiome and immune function in 140 perioperative CRC patients[95]. In the preoperative phase, prebiotic consumption led to increased levels of Bifidobacterium and Enterococcus, along with a decrease in Bacteroides abundance compared to placebo[95]. Postoperatively, the control group exhibited a rise in Enterococcus, Bacillus, Lactococcus, and Streptococcus, whereas the prebiotic group experienced an increase in non-pathogenic Escherichia-Shigella. Additionally, microbial richness declined from pre- to post-surgery in patients who did not receive prebiotics[95].
Beyond microbiome modulation, prebiotics significantly influenced immune function. In the preoperative phase, serum levels of immunoglobulin (Ig) G, IgM, and transferrin were elevated, while in the postoperative phase, levels of IgG, IgA, suppressor/cytotoxic T cells (CD3+ CD8+), and total B lymphocytes increased compared to controls[95]. Rowland et al[87] examined changes in biological markers following three months of daily FOS supplementation (10 g) in 74 French patients, grouped into those with small colorectal adenomas, large adenomas, or no adenomas. Patients with adenomas exhibited significantly higher butyrate levels after FOS intake, while in patients without adenomas, FOS consumption led to increased levels of cholic acid, chenodeoxycholic acid, total primary bile acids, and ursodeoxycholic acid, as well as decreased fecal lithocholic acid, highlighting its potential role in modulating the colonic environment[96].
Inulin supplementation has been shown to enhance the growth of Bifidobacterium spp. and Akkermansia muciniphila[97,98]. An in vitro study demonstrated that culturing fecal samples with inulin and mucin increased the abundance of microbial species involved in suppressing tumor growth in Rnf5-/- mice[99]. Another study found that inulin consumption supports the enrichment of bacterial taxa associated with anti-tumor immune responses[89]. Remarkably, dietary supplementation with inulin or mucin in C57BL/6 mice triggered anti-tumor immunity, yet mucin alone failed to curb tumor growth in germ-free mice, emphasizing the essential role of gut microbiome in tumor-related immune activation[99]. Preclinical studies suggest that inulin may suppress tumor progression in colon cancer and NRAS-mutant melanoma models. Additionally, it has been found to enhance the effectiveness of mitogen-activated protein kinase inhibitors targeting the MAPK/extracellular signal-regulated kinase pathway, which is frequently dysregulated in melanoma[99]. These findings indicate that dietary interventions using inulin or oligofructose could enhance che
The ketogenic diet (KD) is a high-fat, isocaloric diet that significantly reduces carbohydrate intake, originally designed for the treatment of epilepsy and glucose transporter type 1 deficiency syndrome deficiency syndrome[101]. As a therapeutic approach, KD aims to trigger ketosis, and it has recently emerged as a potential strategy for cancer treatment[102]. Vander Heiden et al[103] found that tumor cells consume glucose at higher rates than surrounding tissue and rely on the aerobic glycolytic pathway to produce lactate. By limiting glucose availability, KD could deprive tumor cells of their primary energy source, potentially inhibiting tumor growth and proliferation. In vitro and in vivo animal models have shown that KD suppresses glycolysis and cancer cell proliferation, leading to promising anti-tumor effects[104,105]. However, the specific role of the gut microbiome in mediating these anti-tumor effects in cancer therapy remains unclear. The influence of the microbiome in such effects has been explored in other conditions like autism and epilepsy[106,107]. For example, in mice with autism spectrum disorder, KD was shown to normalize high levels of Akkermansia muciniphila and promote a higher Firmicutes/Bacteroidetes ratio[107]. In infants with refractory epilepsy, KD not only reduced seizure frequency but also increased the levels of Bacteroides and Prevotella, while decreasing Cronobacter abundance[106]. Although similar microbiome shifts could be beneficial for cancer patients undergoing treatment, this remains an area for further investigation.
Dietary strategies include: (1) Caloric restriction (CR), which entails a reduction in energy intake by 20%-50% without causing malnutrition or depleting essential nutrients; (2) Time-restricted feeding, where eating is confined to a specific 4- to 12-hour window; (3) Intermittent fasting, which alternates between 24-hour fasting periods and 24-hour ad libitum eating phases; and (4) The fasting-mimicking diet, a five-day period of CR with a low-calorie, vegetable-based diet, followed by a return to regular eating patterns once a month[108]. In recent years, there has been a growing body of research exploring the potential of these dietary approaches to delay the onset and progression of cancer, as well as other chronic diseases[109,110]. A key factor in how dietary restrictions may positively impact metabolism and immune function is the gut microbiome. As previously discussed, the microbiome plays an important role in cancer development and therapy response by influencing gut homeostasis, which in turn affects both the gut barrier and immune system. Therefore, dietary restrictions may help restore gut dysbiosis, improving metabolism and immune responses. Specifically, disruptions in gut permeability and the translocation of bacteria to the mucosal layer can trigger immune dysfunction. Immune homeostasis in the intestines is largely regulated by interactions between epithelial cells and dendritic cells[111]. Thus, by modulating the microbiome, these dietary changes could enhance cancer treatment outcomes. However, reliable data confirming these effects are still lacking. Moreover, it is important to note that interventions like fasting may worsen cachexia syndrome, which impacts nearly half of all cancer patients[112]. Future research should focus on determining the most effective timing and approach for these dietary interventions, considering factors such as gut microbiome composition, cancer type, and the individual characteristics of patients.
Dietary fiber serves as a key substrate for gut microbial fermentation, leading to the production of short-chain fatty acids, such as butyrate, which have been shown to exert anti-inflammatory and anticancer properties[113]. High-fiber diets have been associated with increased microbiota diversity and favorable shifts in microbial composition, which may enhance antitumor immunity and improve treatment responses[114].
Spencer et al[115] reported that melanoma patients consuming high-fiber diets had improved progression-free survival while on immune checkpoint inhibitor (ICI) therapy, potentially mediated by a more favorable gut microbiome profile[115]. Additionally, dietary fiber has been shown to increase microbial metabolites that promote mucosal immunity and inhibit tumor growth in preclinical models[116].
However, the interplay between diet, microbiota, and host immune responses is complex and influenced by multiple factors, including baseline microbiome composition and genetic background. Ongoing studies are investigating the optimal dietary patterns and fiber types for therapeutic benefit in different cancer types[117].
Fecal microbiota transplantation (FMT) involves the transfer of fecal material from a healthy donor to the GI tract of a recipient, aiming to restore microbial diversity and function[118]. Initially developed for treating recurrent Clostridioides difficile infection[119], FMT is now being explored as an adjunct in cancer therapy[120]. Preclinical studies and early-phase clinical trials suggest that FMT can modulate the gut microbiome in a way that enhances the efficacy of ICIs, particularly in melanoma and other solid tumors[121].
For instance, Baruch et al[122] demonstrated that FMT from ICI responders could induce clinical responses in melanoma patients previously refractory to anti- programmed death 1 (PD-1) therapy. Similarly, Davar et al[123] showed that microbial reprogramming through FMT could overcome primary resistance to immunotherapy in metastatic melanoma. While these findings are promising, further large-scale trials are needed to validate the safety, donor selection criteria, and long-term outcomes of FMT in oncology settings.
CRC is the third most prevalent cancer in men and the second in women and the predominant histological subtype is adenocarcinoma[2]. CRC is a multifactorial disease and the main etiology remains complex, environmental factors - including smoking, diet, and lifestyle - significantly influencing individual susceptibility and sporadic cases constitute approximately 80% of cases[124]. Some well-known risk factors are: Age, genetic predispositions, familial CRC history, IBD[125-127]. Gut microbiome alterations and microbial infections have shown an association with CRC pathogenesis and treatment response[128,129].
The human gut hosts over 500 bacterial species, with the densest microbial populations residing in the colon[130]. The microbiome primarily comprises obligate anaerobes such as Bacteroides, Eubacterium, Bifidobacterium, Fusobacterium, Peptostreptococcus, and Atopobium, while facultative anaerobes like Enterococci, Lactobacilli, Enterobacteriaceae, and Strep
The F. nucleatum protein Fap2 binds to Gal-GalNAc on CRC cells, facilitating bacterial colonization[135]. F. nucleatum also interacts with E-cadherin via FadA, activating the β-catenin and NF-κB pathways, increasing proinflammatory cytokines (TNF-α, IL-6, IL-8, IL-1β), and promoting immune evasion by binding to TIGIT receptors on natural killer cells and tumor-infiltrating lymphocytes[136,137]. It further enhances CRC progression through TLR4/MyD88 signaling[135,136,138,139].
Research has identified F. nucleatum in CRC primary tumors and liver metastases, while Bacteroides fragilis and E. coli disrupt the colonic mucus barrier. Their toxins cause DNA damage, inflammation, and mutations. E. coli also impairs DNA repair via EspF and increases ROS, contributing to genetic instability in CRC[140-144].
Other bacteria linked to CRC include Peptostreptococcus anaerobius, which increases ROS via TLR2/TLR4, and seven species (B. fragilis, F. nucleatum, Porphyromonas asaccharolytica, Parvimonas micra, Prevotella intermedia, Alistipes finegoldii, Thermanaerovibrio acidaminovorans) associated with lipopolysaccharide (LPS)-induced inflammation and energy biosynthesis pathways. Atopobium parvulum and Actinomyces odontolyticus are enriched in early CRC and may serve as biomarkers[145,146]. Enterotoxigenic B. fragilis (ETBF) drives CRC by inducing IL-17, activating NF-κB, and recruiting myeloid-derived suppressor cells, which suppress CD8+ T-cell activity. ETBF also upregulates matrix metalloproteinase 9 and vascular endothelial growth factor A, further promoting tumor progression via CXCL1 and CXCL2[147,148]. Dysbiosis is linked to colorectal diseases, including IBD, colitis, and CRC[149,150]. Several bacteria contribute to CRC development.
Streptococcus gallolyticus: Streptococcus gallolyticus (S. gallolyticus) is associated with enterococcal endocarditis and cecal carcinoma, with up to 40% of bacteremia cases linked to CRC[151-153]. It promotes tumorigenesis by increasing inflammatory markers and oxidative stress, adhering to colorectal tissues via collagen-binding proteins. Colonoscopic screening is recommended for infected patients[153]. While earlier reports suggested an association rate as high as 80%, more recent analyses indicate a lower prevalence. A retrospective cohort study conducted in the Skane Region of Sweden from 2003 to 2018 found that among patients with S. gallolyticus subsp. Gallolyticus bacteremia, 7% were diagnosed with CRC within 12 months. This study highlighted a strong association between this specific subspecies and CRC, with a standardized incidence ratio of 59.8 compared to the general population[154]. Additionally, a 2022 study from Iran reported that 60% of patients with CRC had S. gallolyticus detected in tumor samples, compared to 29.7% in control subjects without CRC[155].
Bacteroides fragilis: Bacteroides fragilis produces fragilysin, which disrupts E-cadherin, activating oncogenic pathways [β-catenin, cellular MYC (c-MYC), cyclin D1][156-159]. Its tumorigenic effects involve signal transducer and activator of transcription 3 activation and a TH17 immune response, which can be mitigated by IL-17/IL-23 blockade[160].
E. coli (pks-positive strains): E. coli produce colibactin, a genotoxin that induces DNA damage, mutations, and genomic instability, driving CRC development in mouse models[161,162].
F. nucleatum: F. nucleatum is enriched in CRC and correlates with poor survival and metastasis. Its virulence factors, Fap2 and FadA, inhibit immune responses and activate oncogenic signaling (β-catenin, TLR4/NF-κB, miR-21), promoting tumor growth and invasion[136,139,163].
Enterococcus faecalis: Enterococcus faecalis (E. faecalis) generates reactive oxygen/nitrogen species, leading to DNA damage and chromosomal instability. H. pylori may contribute to CRC via hypergastrinemia and cytotoxin-associated gene A (CagA)-mediated oncogenic effects, though its role remains unclear[164,165].
Microbial composition shifts in CRC patients include increased Bacteroides/Prevotella, Fusobacterium, Roseburia, and Faecalibacterium in tumors, with reduced Enterobacteriaceae levels[131,132]. Host-microbiome interactions via fecal microRNAs influence bacterial gene expression, potentially driving CRC progression[166]. Case-control studies show CRC-associated bacteria (F. nucleatum, E. faecalis, S. gallolyticus, ETBF) are enriched in adenomatous polyps, while beneficial bacteria (Lactobacillus, Roseburia, Bifidobacterium) are reduced[167,168]. Emerging research suggests gut microbiome profiling combined with machine learning could serve as a non-invasive CRC screening tool, offering accuracy comparable to colonoscopy[150]. A summarized table of the therapeutic approaches related to the gut microbiome in CRC can be found in Table 1[20,133,140,166,169-184].
| Therapeutic approach | Description | Key findings |
| Bacterial eradication | Targeting CRC-associated bacteria using antimicrobial agents | Colibactin-producing E. coli inhibited by ClbP inhibitors, reducing tumor growth in mice[169] |
| F. nucleatum targeted with metronidazole, lowering bacterial load and tumor proliferation[140] | ||
| Probiotics | Using beneficial bacteria to modulate gut microbiome for CRC prevention and chemotherapy support[20,133,170] | Lactobacillus and Bifidobacterium species reduced preneoplastic lesions in animal models[171,172] |
| VSL3 probiotic mix reduced adenoma formation and chemotherapy side effects[173] | ||
| Human studies show mixed results on probiotics’ protective effects against CRC[174-176] | ||
| FMT | Transplanting healthy microbiome to modify gut composition and potentially prevent CRC[177] | FMT from wild mice improved CRC resistance and inflammation markers in recipient mice[177] |
| Clinical evidence remains limited, requiring further studies[178] | ||
| Microbiome and chemotherapy resistance | Gut bacteria influence drug resistance and efficacy in CRC treatment[166] | F. nucleatum linked to resistance against 5-FU and oxaliplatin via immune signaling[166] |
| Mycoplasma hyorhinis and Gammaproteobacteria impair gemcitabine by enzymatic deactivation[179] | ||
| Ciprofloxacin restored gemcitabine sensitivity[179] | ||
| Antibiotic use may reduce chemotherapy efficacy in some cases[180] | ||
| Microbiome and immunotherapy | Gut bacteria influence immune responses to CRC therapies[27] | Microbiome impact responses to immune checkpoint inhibitors (e.g., Ipilimumab)[181] |
| Antibiotic-treated mice showed reduced immunotherapy effectiveness[181] | ||
| Microbiome and drug toxicity | Bacteria alter drug metabolism, affecting side effects and toxicity | E. coli β-glucuronidase reactivates irinotecan, increasing toxicity[182,183] |
| Irinotecan alters microbiome, increasing inflammatory bacteria (Fusobacteria, Proteobacteria)[182] | ||
| Antibiotic treatment reduced oxaliplatin-induced neuropathy in mice[184] |
Pancreatic cancer is a rapidly progressing and highly fatal disease, with only about 25% of patients surviving beyond one year after diagnosis[185]. Due to late-stage detection in most cases, treatment is often palliative, relying on chemotherapy or radiotherapy rather than surgical intervention. Among the different types, pancreatic ductal adenocarcinoma stands out as the most common and aggressive subtype[186]. Chronic pancreatitis is a well-established risk factor, with incidence rates significantly higher in affected individuals[187]. Studies suggest a strong association between bacterial infections and pancreatic cancer, not necessarily as a direct cause but as contributors to tumor progression, immune modulation, and inflammatory responses[188,189].
A growing body of research has associated H. pylori infection to pancreatic cancer development[190,191]. Initial case-control studies reported a twofold increased risk in infected individuals, with prospective cohort studies confirming a higher susceptibility in male smokers carrying H. pylori antibodies, particularly the CagA-positive strains[192]. Further meta-analyses reinforced these findings, with large cohort studies also establishing a correlation between gastric ulcers (often caused by H. pylori) and pancreatic cancer[193,194].
H. pylori is believed to promote pancreatic diseases through its production of ammonia, lipopolysaccharides, and inflammatory mediators. In vitro experiments demonstrated increased levels of IL-8 and vascular endothelial growth factor (VEGF) in pancreatic cancer cell lines co-cultured with H. pylori, contributing to inflammation and tumor growth[195]. Furthermore, H. pylori LPS has been shown to enhance KRAS mutations—present in over 90% of pancreatic adenocarcinomas - thereby initiating carcinogenesis[196,197]. Additionally, the infection was found to activate signal transducer and activator of transcription 3, a signaling pathway that promotes cancer progression by upregulating anti-apoptotic and pro-proliferative proteins such as B-cell lymphoma-extra large, myeloid cell leukemia 1, survivin, c-MYC, and cyclin D1[198,199]. While a clear causal relationship has yet to be established, further clinical intervention studies using H. pylori eradication therapy may provide insights into its role in pancreatic cancer initiation[200].
Periodontal pathogens have also been implicated in pancreatic cancer risk. Porphyromonas gingivalis (P. gingivalis), a major bacterium associated with periodontitis, has been extensively studied in this context[201,202]. The European Prospective Investigation into Cancer cohort reported that individuals with high P. gingivalis antibody levels had more than double the risk of developing pancreatic cancer[202]. Though the precise mechanisms remain unclear, P. gingivalis’ LPS is believed to stimulate the TLR4 signaling pathway, which plays a key role in pancreatic ductal adenocarcinoma (PDAC) by promoting tumor growth, angiogenesis, invasion, and apoptosis suppression.
Additionally, Fusobacterium species have been detected in 8.8% of pancreatic cancer tissues, with their presence linked to poor prognosis[203-205]. These findings suggest Fusobacterium as a potential negative prognostic biomarker for pancreatic cancer[206]. Recent studies have reported a higher prevalence of F. nucleatum in PDAC than previously estimated indicating a range of approximately 20% to 30%[207-209]. For instance, a 2024 review highlighted that F. nucleatum is present in a significant subset of PDAC tumors, with prevalence rates reaching up to 30% in certain cohorts[209]. This bacterium has been associated with tumor progression and poorer patient outcomes. F. nucleatum can promote PDAC cell proliferation and migration through the induction of cytokines such as granulocyte-macrophage colony-stimulating factor, CXCL1, and IL-8, which act via autocrine and paracrine signaling pathways[208]. Additionally, it can influence the tumor microenvironment by modulating immune responses, potentially contributing to an immunosuppressive milieu that favors tumor growth[208]. These findings underscore the importance of considering F. nucleatum in the context of PDAC pathogenesis and suggest potential avenues for therapeutic intervention targeting the tumor microbiome.
Another emerging bacterial player is Pseudomonas aeruginosa, which has been shown to enhance the expression of the ABCB1 gene, contributing to drug resistance, tumor invasion, and metastasis[210]. Table 2 summarizes recent findings on microbiome-targeted therapies for PDAC[179,189,211,212].
| Therapeutic approach | Description | Key findings |
| Antibiotic therapy | Targeting tumor-associated bacteria to reduce tumor burden and enhance immunotherapy | Pushalkar et al[211] found a 50% tumor burden reduction in PDAC mouse models after oral antibiotic treatment |
| Tumor microenvironment reprogrammed, reducing myeloid-derived suppressor cells and increasing M1 macrophages[211] | ||
| Antibiotics improved PD-1 checkpoint inhibitor response, enhancing T-cell activation and reducing tumor size[211] | ||
| A clinical trial is evaluating antibiotic-pembrolizumab combination in locally advanced PDAC[189] | ||
| Probiotics and PDAC prevention | Investigating probiotics’ role in mitigating PDAC risk factors (e.g., pancreatitis, obesity, diabetes) | Lactobacillus plantarum 299 reduced pancreatic sepsis and surgical need in acute pancreatitis patients[212] |
| Probiotics reduced fibrosis, inflammation, necrosis, ductal damage, and atypical cellular regeneration, potentially lowering PDAC risk | ||
| Microbiome and chemotherapy resistance | Examining bacterial interference with chemotherapy efficacy | Gammaproteobacteria enriched in 76% of PDAC samples, degrading gemcitabine into an inactive form[179] |
| Microbiome-targeted approaches could enhance chemotherapy effectiveness |
GC is a multifactorial disease influenced by environmental, dietary, host-related, genetic, and epigenetic factors[213,214]. H. pylori, designated as a group I carcinogen by the World Health Organization, is a major contributor to GC pa
Chronic H. pylori-induced inflammation, as described in the Correa model[217], promotes gastric epithelial turnover, increasing the probability of mitotic errors and malignant transformation[218,219]. This pathological cascade progresses from atrophic gastritis to intestinal metaplasia (IM), dysplasia, and ultimately gastric adenocarcinoma[218-220]. The inflammatory response involves complex interactions between the bacterial pathogen, gastric acid secretion, immune cells, and reactive oxygen and nitrogen species, leading to oxidative DNA damage and upregulation of pro-inflammatory mediators[221,222]. Elevated expression of cytokines (IL-1β, IL-6, IL-8, and TNF-α) and cyclooxygenase-2 has been implicated in promoting atrophic mucosal alterations and oncogenic intracellular signaling[223,224]. Furthermore, H. pylori infection is associated with epigenetic modifications, including aberrant DNA methylation and gene silencing, affecting key regulatory pathways involved in cell adhesion, proliferation, DNA repair, and immune modulation[223].
Recent high-quality evidence reinforces the protective effect of H. pylori eradication therapy in reducing the risk of GC, particularly when administered before the onset of precancerous changes such as IM or dysplasia. A recent meta-analysis published in Frontiers in Microbiology in 2025 evaluated the impact of H. pylori eradication on GC risk among patients with IM or dysplasia. The study included 16 randomized controlled trials involving 15027 participants. The findings indicated that H. pylori eradication was associated with a 45% reduction in the relative risk (RR) of developing GC compared to no treatment [RR = 0.55; 95% confidence interval (CI): 0.46-0.67; P < 0.001]. Subgroup analyses revealed that eradication therapy significantly lowered GC risk in patients with dysplasia (RR = 0.51; 95%CI: 0.32-0.82; P = 0.005) and IM (RR = 0.61; 95%CI: 0.40-0.93; P = 0.022)[225]. Another recent systematic review and meta-analysis published in Gastroenterology in January 2025 evaluated the impact of H. pylori eradication on GC incidence and mortality[226]. The study included 11 randomized controlled trials and 13 observational studies, encompassing a total of 58828 participants. The findings revealed that H. pylori eradication was associated with a 36% reduction in the RR of developing GC (RR = 0.64; 95%CI: 0.48-0.84) and a 22% reduction in GC mortality (RR = 0.78; 95%CI: 0.62-0.98). These results underscore the potential of H. pylori eradication as a preventive strategy against GC.
In addition to inflammation-driven carcinogenesis, H. pylori exerts direct tumorigenic effects through its virulence factors, particularly CagA and vacuolating cytotoxin A (VacA)[227,228]. CagA disrupts intracellular signaling networks such as the MAPK cascade, NF-κB, phosphoinositide 3-kinase/protein kinase B, and epithelial-mesenchymal transition pathways via oncogenic yes-associated protein activation[228,229]. The cag pathogenicity island has also been implicated in H. pylori-induced carcinogenesis through mutations in tumor suppressor genes such as TP53[230,231]. Meanwhile, VacA facilitates immune evasion by impairing T-cell responses and disrupting epithelial integrity, thereby creating a permissive environment for persistent infection and neoplastic progression[232,233]. Table 3 summarizes recent findings on microbiome-targeted treatment strategies in gastroesophageal cancer[234-244].
| Topic | Findings | Key insights |
| Microbiomes in esophageal cancer treatment | Disrupting microbiome with antibiotics decreases xenograft tumor response to CpG oligonucleotide immunotherapy and platinum-based chemotherapy (e.g., oxaliplatin)[180] | A stable commensal microbiome may enhance cancer therapy effectiveness[180] |
| Similar effects observed in germ-free mice[180,234] | Microbiome regulates myeloid-derived cell functions within the tumor microenvironment[234] | |
| Further clinical research is needed | ||
| H. pylori and GC risk | H. pylori eradication reduces GC risk but does not guarantee complete prevention[235,236] | Other factors contribute to GC beyond H. pylori infection[235] |
| A Colombian clinical trial showed no significant GC incidence difference between treated and untreated individuals over six years, but treated individuals had higher precancerous lesion regression rates[235] | Eradication reduces GC risk by about 44% in asymptomatic, infected individuals[236] | |
| Long-term impact of H. pylori eradication | A 15-year study showed a 39% reduction in precancerous lesions[237] | Protective effects are strongest in non-atrophic/atrophic gastritis patients[239,240] |
| An 8-year study found that atrophic body gastritis reversed in 50% of treated patients[238] | Intestinal metaplasia and dysplasia patients do not experience the same benefit | |
| Meta-analyses confirmed a significant reduction in GC risk post-eradication[239] | Early eradication is crucial as intestinal metaplasia is irreversible | |
| Epigenetic modifications in GC prevention | DNA demethylating agent (5-azadC) suppresses aberrant methylation and reduces GC incidence in animal models[241] | Developing safer DNA demethylating agents could benefit high-risk individuals[223] |
| Clinical use is limited due to high toxicity | ||
| Probiotics and H. pylori eradication | Probiotics inhibit H. pylori in animal studies[242] | Probiotics may play a role in GC prevention[243,244] |
| Clinical trials/meta-analyses show probiotics improve eradication rates, patient compliance, and reduce treatment side effects | Used alongside antibiotics, they enhance treatment efficacy |
Biliary tract cancers include cancers of the bile duct, gallbladder, and ampulla of Vater, with gallbladder cancer (GBC) being the most prevalent[245,246]. While rare in the Western world, GBC is more common in Chile, Central Europe, Thailand, Japan, and parts of India and Pakistan[247,248]. The main risk factors include gallstones, obesity, chronic gallbladder inflammation, genetic predisposition, and environmental factors. Additionally, bacterial infections have been linked to malignant transformation in GBC.
Salmonella enterica serovar Typhi (S. Typhi): Chronic typhoid carriers have a significantly higher risk of GBC, with some studies reporting up to a 167-fold increased risk. However, recent metagenomic studies in South America found other bacteria (e.g., E. coli, F. nucleatum, Enterobacter species) predominant in GBC patients instead of S. Typhi[249-251].
Helicobacter species (H. bilis, H. hepaticus, H. pylori)[11,252]: These bile-resistant bacteria can cause chronic inflammation and gallstone formation, contributing to carcinogenesis. Some studies report a 2-3-fold increased risk of GBC in infected individuals. However, more epidemiological studies are needed to confirm causality.
Mixed bacterial infections: E. coli, Enterobacter, Klebsiella, and E. faecalis have been found in high levels in GBC patients. Given their known role in colon cancer, it is hypothesized they might contribute to GBC through similar mechanisms[253,254].
Gallstones facilitate bacterial colonization, increasing the likelihood of persistent infection. S. Typhi infection triggers oncogenic pathways (protein kinase B/MAPK), leading to mutated p53 and amplified c-MYC - both commonly observed in GBC patients[255,256]. Bacterial glucuronidase enzymes produce carcinogenic metabolites from bile, potentially leading to DNA damage and tumor development[256]. Bacterial enzymes increase secondary bile acids, which are known tumor initiators and promoters in GBC[257].
Hepatocellular carcinoma (HCC), the most common primary liver cancer and a leading cause of cancer-related deaths, is often associated with underlying liver diseases such as cirrhosis, fibrosis, and chronic hepatitis B and C infections[258,259]. Other recognized risk factors include alcoholic liver disease, hemochromatosis, obesity, and non-alcoholic fatty liver disease (NAFLD)[260,261]. While bacterial infections are not traditionally considered a primary cause of HCC, growing evidence suggests that gut and hepatic microbiome may influence its development and progression[262,263]. The liver, though generally sterile, is directly connected to the gut through the hepatic portal system. Disruptions in the gut microbiome can lead to increased intestinal permeability, allowing bacteria and their metabolites - such as lipopolysaccharides - to translocate to the liver[264]. This process contributes to chronic inflammation, immune dysregulation, and liver disease progression[265,266].
Certain bacterial species have been implicated in HCC development. E. coli, Atopobium, Bacteroides, Clostridium, and Desulfovibrio were found enriched in the gut microbiome of HCC patients, correlating with increased LPS levels and disease severity[267]. Additionally, obesity-related alterations in gut microbiome contribute to hepatocarcinogenesis through increased production of secondary bile acids, such as deoxycholic acid, which can activate the mammalian target of rapamycin (mTOR) pathway and promote liver tumorigenesis[268]. In high-fat diet-fed mice, elevated deoxycholic acid levels were associated with a higher incidence of HCC, while antibiotic treatment reduced tumor formation. Helicobacter species, particularly Helicobacter hepaticus and H. pylori, have also been linked to HCC[269,270]. In vivo studies showed that H. hepaticus disrupts enterohepatic homeostasis and activates NF-κB and Wnt signaling pathways, leading to oxidative stress and hepatocyte turnover. Furthermore, H. pylori virulence factors such as VacA and CagA were detected in HCC tissues, and its LPS was found to enhance liver cancer cell growth and migration[271,272].
Beyond the gut-liver axis, alterations in oral microbiome have also been observed in HCC patients[273]. A me
| Topic | Findings | Key insights |
| Antibiotics, probiotics, and GBC | Limited experimental and clinical evidence on their use for GBC prevention or treatment[255] | Salmonella infection may trigger lasting oncogenic changes |
| A study by Scanu et al[255] found that Salmonella-infected mouse embryonic fibroblasts formed tumors even after bacterial eradication with ciprofloxacin | ||
| Salmonella infection activates Akt/MAPK signaling pathways, potentially sustaining carcinogenesis post-eradication[255] | Further research is needed to explore microbiome-targeted GBC therapies | |
| Cholecystectomy remains the most effective preventive measure for chronic typhoid carriers at risk of GBC[251] | ||
| Probiotics and HCC prevention | Probiotic-fermented milk and chlorophyllin reduced tumor incidence in AFB1-induced HCC in rats[276] | Probiotics show potential in reducing HCC risk and progression in animal models |
| VSL3 probiotic formula inhibited chemically induced HCC, reducing LPS levels, tumor size, and tumor count[267] | ||
| Prohep (novel probiotic mix) decreased tumor growth by 40%, enhanced anti-inflammatory responses, promoted T-cell activation, and reduced pro-angiogenic factors in mice[277] | Clinical evidence remains limited, requiring large-scale human trials | |
| Probiotics and aflatoxin exposure | Some studies suggest probiotics reduce aflatoxin exposure and toxic DNA adduct formation, potentially lowering HCC risk[278] | Needs further validation in human studies |
| FMT and liver disease | FMT has shown promise in treating[279]: (1) High-fat diet and alcohol-induced liver injury (animal models)[280]; (2) Severe alcoholic hepatitis (clinical studies)[281]; (3) Chronic hepatitis B[282]; and (4) Advanced liver cirrhosis and hepatic encephalopathy | FMT may help restore gut-liver axis balance[279] |
| Its role in HCC prevention and therapy remains unclear, requiring further clinical trials |
Understanding the distinct microbial signatures and dysbiosis patterns across GI cancers such as colorectal, gastric, and pancreatic cancer highlights the intricate role the gut microbiome plays in tumorigenesis and progression. However, the interplay between the microbiome and cancer extends beyond pathogenesis - it also critically influences how patients respond to treatment. The next section explores how common cancer therapies, including chemotherapy, radiotherapy, and immunotherapy, interact with and are modulated by the gut microbiome, shaping both efficacy and toxicity profiles.
One of the major side effects of irinotecan, a key anti-cancer drug used for metastatic CRC, is severe diarrhea[283]. A study demonstrated that irinotecan chemotherapy alters the intestinal microbiome in tumor-bearing rats, leading to an increased abundance of Clostridium clusters I, XI, and Enterobacteriaceae, particularly following dose-intensive therapy[284]. Similarly, 5-FU, an anti-metabolite used to treat several cancers, including colorectal, breast, and liver, also induces diarrhea as a frequent side effect[283]. An observational study on breast cancer patients receiving 5-FU, epirubicin, and cyclophosphamide found alterations in intestinal permeability and changes in gut peptides such as glucagon-like peptide-2, ghrelin, and epidermal growth factor. Notably, patients who experienced diarrhea had higher intestinal permeability than those who did not[285].
van Vliet et al[286] showed that in pediatric patients with acute myeloid leukemia, chemotherapy combined with anti-microbial prophylaxis caused a reduction in anaerobic bacteria and an increase in potentially pathogenic aerobic enterococci. These gut microbiome disturbances may increase the risk of gram-positive infections. Furthermore, a study on paclitaxel-induced neuropathic pain in C57BL/6 (B6) and 129SvEv (129) mice, which display greater neuropathic sensitivity, revealed significant changes in gut microbiome. In particular, paclitaxel chemotherapy led to a decrease in Akkermansia muciniphila abundance in the B6 microbiome. The reduction of this beneficial bacterium was linked to gut barrier dysfunction, which might increase systemic exposure to bacterial products and metabolites, contributing to systemic inflammation and pain sensitivity[165,287].
Given the close association between gut microbiome and the immune system, immunotherapeutic interventions also exert significant effects on microbial composition and function. In a cohort of HCC patients receiving anti-PD1 therapy, microbial shifts were observed by week 3, with a notable increase in E. coli abundance in non-responders' stool samples, while Lactobacillus, Ruminococcaceae, and Akkermansia muciniphila were enriched in responders’ microbiome[288]. Si
Recent advancements in cancer surgery have highlighted the significant impact of surgical interventions on gut microbiome, especially in patients with GI cancers. A study examining gastrectomy in GC patients assessed its effects on the gut microbiome and metabolome, exploring the association between microbial changes and post-gastrectomy outcomes. The analysis showed substantial shifts in microbial diversity and richness in patients who underwent gastrectomy, likely due to the reconstruction of the GI tract following surgery. These microbial alterations could influence microbial functions, including nutrient transport and organic compound biosynthesis, which may contribute to changes in post-surgical metabolism[294]. Similarly, a study conducted in China investigated the fecal microbiome of GC patients before and after radical distal gastrectomy[295]. Significant changes were observed in the microbial composition during the perioperative period, with variations in the abundances of Akkermansia muciniphila, Escherichia/Shigella, Lactobacillus, and Dialister. Post-surgery, there was an increase in Escherichia/Shigella, Veillonella, and Clostridium XVIII, as well as a decrease in Bacteroides, when compared to healthy controls. These shifts are likely driven by surgical stress and other perioperative factors[295]. Additionally, research has shown that a modified microbiota-accessible carbohydrate diet can positively influence the gut microbiome and clinical symptoms in CRC patients following surgical resection[296].
In CRC surgery, animal studies have shown significant alterations in the gut microbiome following colorectal resection, with a marked reduction in Bacteroidetes and Proteobacteria, such as Enterobacteriaceae and Rhodospirillaceae[297]. Moreover, these microbial changes were associated with altered gene expression patterns, including an increase in IL10 and IL12 and a decrease in TNF expression after surgery[297]. In CRC patients, numerous studies have reported post-surgical changes in the gut microbiome, comparing post-surgery samples to those from preoperative CRC patients and healthy controls. These changes include a decrease in Bacteroides, Bifidobacterium, Clostridium, Prevotella, Klebsiella, Faecalibacterium, and Parabacteroides, while Enterococcus, Pseudomonas, Staphylococcus, Escherichia-Shigella, Enterobacteriaceae, and Streptococcus increased post-surgery[298,299]. In contrast, some studies have shown a decrease in Escherichia-Shigella and an increase in Enterococcus and Parabacteroides[300]. At the phylum level, a reduction in Firmicutes and Bacteroidetes was observed, alongside a decrease in the Firmicutes/Bacteroides ratio and an increase in Proteobacteria[301,302]. These microbial changes are likely a consequence of perioperative antibiotic administration, which can alter the gut microbiome, impair immune function, and disrupt metabolic pathways[303]. The resulting disruption in gut barrier integrity, coupled with surgical stress, may contribute to the development of inflammation and other postoperative complications in cancer patients. Recent evidence underscores the benefits of early enteral nutrition and probiotic supplementation in enhancing postoperative recovery for CRC patients.
Early enteral nutrition: Initiating enteral nutrition within 48 hours post-CRC surgery has been associated with improved GI recovery and nutritional status. A retrospective comparative study involving 227 patients in each group demonstrated that early enteral nutrition led to a shorter time to first postoperative flatus, quicker achievement of nutritional goals, and reduced hospital stay, without increasing postoperative complications[304]. Similarly, a randomized controlled trial reported that patients receiving early oral feeding after colorectal surgery experienced faster return of intestinal movements, earlier defecation, and shorter hospital stays compared to those on a regular diet[305].
Probiotic supplementation: Probiotic use in the perioperative period has been shown to reduce postoperative infections and restore beneficial gut microbiota. A systematic review and meta-analysis of randomized controlled trials found that probiotic supplementation significantly decreased the incidence of diarrhea, surgical site infections, urinary infections, and pulmonary infections in CRC patients undergoing surgery[306].
Furthermore, probiotics have been associated with a reduction in postoperative complications such as ileus, abdominal collections, sepsis, and pneumonia. These benefits are attributed to the modulation of gut microbiota, enhancement of intestinal barrier function, and attenuation of inflammatory responses[307].
Radiotherapy stimulates anti-tumor responses through immune activation[308], and like other cancer treatments, it induces alterations in gut microbiome composition. In an animal study, irradiated mice exhibited luminal and mucosa-associated dysbiosis compared to controls at two post-radiation time points, which was linked to increased expression of TNF-α, IL-1β, and IL-6[71]. Another recent study on irradiated mice revealed an increase in the Alistipes genus and a decrease in Prevotella in the large intestine[309]. In human studies, Mitra et al[310] reported a reduction in gut microbiome diversity in cervical cancer patients following chemoradiation therapy, while a study of gynecological cancer patients receiving pelvic radiotherapy found a decrease in Firmicutes and an increase in Fusobacterium[311]. Though further validation is needed, these findings suggest that radiation-induced changes in gut microbiome could lead to alterations in intestinal mucosa, resulting in inflammation[312].
These observations open the door to novel strategies aimed at modulating the gut microbiome to improve cancer treatment outcomes. One such approach is dietary interventions, known to influence gut microbiome composition and functionality[313]. A 2024 study demonstrated that dietary fiber supplementation not only improved tumor control during radiotherapy[314] but also alleviated intestinal radiation toxicity, highlighting the dual benefits of such interventions in cancer therapy[315]. As a result, research into the effects of nutritional interventions during cancer therapy on gut microbiome and clinical outcomes is on the rise. In the following section, we will examine various nutritional strategies that may help optimize gut microbiome and enhance the efficacy of cancer therapies.
The dynamic interplay between cancer therapies and the gut microbiome underscores the bidirectional nature of this relationship - therapies can disrupt microbial homeostasis, while the microbiota, in turn, modulates treatment efficacy and toxicity. These insights not only deepen our mechanistic understanding but also prompt critical questions about how best to leverage the microbiome to optimize clinical outcomes. The following discussion synthesizes these findings, evaluates emerging therapeutic interventions such as dietary modulation and FMT, and highlights future directions for integrating microbiome science into oncology practice.
In addition to standard chemotherapy, several targeted therapies have become integral to the treatment of GI malignancies, including monoclonal antibodies and antibody-drug conjugates. Emerging evidence suggests that the gut microbiota and local inflammatory milieu may significantly influence the efficacy, resistance, and toxicity of these therapies.
Bevacizumab, an anti-VEGF antibody, inhibits tumor angiogenesis and is widely used in combination regimens for metastatic CRC[316,317] and HCC[318]. A recent study investigated the role of the gut microbiota in modulating treatment response by analyzing pretreatment fecal samples from 37 patients[318]. Participants were classified as responders (n = 28) or non-responders (n = 9). Although overall microbial diversity did not differ significantly between the groups, the relative abundance of Bacteroides stercoris and Parabacteroides merdae was higher in responders. Notably, patients lacking both bacterial species had significantly worse prognoses. These findings suggest that specific gut microbiome may influence the therapeutic efficacy of atezolizumab and bevacizumab in advanced HCC.
While the gut microbiota’s impact on chemotherapy in CRC has been studied, its role in targeted therapy remains less understood. Chen et al[318] evaluated the association between gut microbiota composition and treatment outcomes in 110 patients with metastatic CRC undergoing combined chemotherapy and targeted therapy. Patients were stratified into anti- epidermal growth factor receptor (EGFR) (cetuximab) and anti-VEGF (bevacizumab) subgroups. Higher gut microbial α-diversity was observed in the partial response group and specifically in the bevacizumab partial response subgroup. β-diversity significantly differed between the bevacizumab partial response and progressive disease groups (P = 0.029)[319]. Notably, Klebsiella quasipneumoniae showed the highest fold increase in the progressive disease group, while F. nucleatum was about 32 times more abundant in progressive disease vs partial response patients. Despite higher diversity being associated with better outcomes, the bacterial profiles were heterogeneous, underscoring the need for further microbiome-based investigations, particularly in East Asian populations.
EGFR inhibitors are effective in RAS wild-type metastatic CRC. However, the gut microbiota may modulate EGFR pathway activity through inflammation-related mechanisms[319]. As of now, there is limited research directly linking panitumumab or cetuximab - monoclonal antibodies targeting the EGFR - to specific alterations in the gut microbiota in CRC patients[320]. For instance, dysbiosis-induced IL-17 and TNF-α production can interfere with EGFR inhibition and contribute to resistance. Moreover, cetuximab-induced skin toxicity has been linked to microbial alterations, raising the potential for microbiota-based strategies to mitigate adverse effects. A study by Liu et al[321] investigated the characteristics of the skin microbiome in patients experiencing severe skin toxicities induced by EGFR inhibitors, such as afatinib. The research revealed that patients with systemic drug-related intertriginous and flexural exanthema-like lesions exhibited significant skin microbial dysbiosis, characterized by reduced microbial diversity and an increased abundance of pathogenic bacteria. These findings suggest that alterations in the skin microbiota are associated with EGFR inhibitor-induced skin toxicities, highlighting the potential for microbiota-based strategies to mitigate such adverse effects. A review article by Ghidini et al[322] discusses the impact of tumor sidedness on the efficacy of anti-EGFR agents like panitumumab in metastatic CRC. While the article does not delve deeply into microbiota interactions, it acknowledges that the prognostic role of microbiota in CRC requires further clarification before being integrated into clinical practice.
These findings highlight the bidirectional relationship between the gut microbiota and targeted therapies. Microbial composition may affect drug pharmacokinetics, immune activation, and inflammation, thereby altering clinical outcomes. Integrating microbiome profiling into treatment planning and exploring microbiota-modulating strategies (e.g., pro
Emerging evidence supports the idea that an altered gut microbiome contributes to the initiation and progression of GI cancers[29]. Inflammation, a hallmark of cancer, plays a significant role in promoting tumorigenesis, and the gut microbiome is crucial in modulating the inflammatory processes within the GI tract[29]. The relationship between gut microbiome and chronic inflammation has been well-documented, with studies showing that dysbiosis can enhance inflammation by disrupting gut barrier integrity, leading to the translocation of bacteria and their metabolites to the systemic circulation[29,206]. This can activate immune cells and trigger inflammatory cascades that ultimately contribute to the development of cancer.
For instance, certain microbial species such as F. nucleatum in CRC[163] or H. pylori in GC[216] have been associated with chronic inflammation that drives carcinogenesis. These microorganisms can either directly or indirectly manipulate immune responses in the gut, promoting an environment conducive to tumor growth. In CRC, F. nucleatum has been shown to induce an inflammatory response that favors the progression of tumors through mechanisms like the activation of NF-κB[163] and the production of pro-inflammatory cytokines. While inflammation is a key driver of cancer progression, it is also an essential component of the immune response to tumors[288]. The immune system relies on a controlled inflammatory response to detect and destroy cancer cells. However, chronic inflammation, induced by dysbiosis or other factors, can impair immune surveillance and promote tumorigenesis[291]. Thus, the goal of nutritional interventions is to strike a balance - modulating inflammation to prevent cancer progression while ensuring that the immune system remains active and capable of combating tumor cells[108].
One of the major challenges in leveraging dietary interventions is understanding how they can modulate the gut microbiome to produce beneficial effects without compromising immune function. For example, while probiotics can be effective in reducing inflammation, overuse of certain strains or imbalanced supplementation could potentially exacerbate dysbiosis and impair immune responses[24]. Similarly, while fasting can induce beneficial changes in the gut microbiome, excessive or prolonged fasting could worsen conditions like cachexia or malnutrition, which are common in cancer patients[104].
This article has explored the multifaceted role of the gut microbiome in cancer initiation, progression, and response to therapy, emphasizing not only well-established mechanisms but also emerging therapeutic strategies[123,323,324]. A novel contribution of this review is the integration of cutting-edge evidence on interventions such as FMT[120,123] and high-fiber dietary modulation, which have shown promising potential to reshape oncologic outcomes via microbiome-targeted approaches. Specifically, the discussion of FMT in immune checkpoint blockade-refractory melanoma offers a timely update on how microbial reprogramming may overcome therapeutic resistance[122,123]. Recent phase I clinical trials have demonstrated that FMT from ICI responders can induce partial and complete responses in previously nonresponsive melanoma patients, marking a significant shift toward microbiome-informed personalized medicine[122,123]. These trials also provide preliminary data on safety and feasibility, although larger, controlled studies are needed to establish standardized protocols, including donor selection and long-term follow-up.
Additionally, this article highlights the underappreciated role of dietary interventions in modulating gut microbial communities to promote favorable cancer immunosurveillance[28,104,115]. Evidence from recent studies suggests that increased fiber intake correlates with enhanced progression-free survival in melanoma patients treated with immunotherapy[115]. These findings are supported by preclinical work indicating that microbial-derived short-chain fatty acids, such as butyrate, can enhance antitumor immunity[116]. However, interindividual variability in microbiota composition underscores the need for personalized dietary recommendations and further interventional trials.
Although the existing evidence on the role of the gut microbiome in cancer and the potential of nutritional interventions to modulate inflammation is promising, there are several challenges that need to be addressed. First, the precise mechanisms by which the gut microbiome influences cancer progression and response to treatment remain unclear. Further research is needed to identify the specific microbial species and metabolic pathways involved in cancer-related inflammation and how dietary interventions can target these pathways. Second, the heterogeneity of cancer types and patient populations makes it difficult to generalize findings across different cancers. The effects of dietary strategies may vary depending on the cancer type, stage, and the individual’s microbiome composition. Personalized nutrition, which takes into account the unique gut microbiome and metabolic profile of each patient, could offer a more tailored approach to cancer treatment. Finally, larger clinical trials are needed to validate the findings from preclinical studies and assess the long-term effects of dietary interventions on cancer outcomes. Only through well-designed, rigorous studies can we determine the most effective strategies for using the gut microbiome as a therapeutic target in cancer care.
While there is substantial interest in leveraging the gut microbiome for the prevention and treatment of GI cancers, current findings across studies are not always consistent, highlighting the complexity and nascent stage of this field[236,239,324]. One notable example is the ongoing controversy surrounding H. pylori eradication in GC prevention[239,240]. The role of H. pylori eradication in GC prevention remains an area of ongoing debate, particularly in relation to the histological stage of gastric mucosal damage. A meta-analysis of randomized controlled trials involving 7955 participants[240] demonstrated that H. pylori treatment significantly reduced GC risk overall (RR = 0.64), but the protective effect was notably confined to individuals without advanced mucosal changes - specifically those with non-atrophic or atrophic gastritis. In contrast, patients with IM or dysplasia did not experience a statistically significant reduction in GC risk following eradication, suggesting a “point of no return” beyond which eradication offers limited benefit. These findings were corroborated by a larger systematic review and meta-analysis of 26 studies (52363 subjects), which confirmed a significant reduction in GC risk with H. pylori eradication (pooled RR = 0.56)[239]. Subgroup analysis again emphasized that the benefit was primarily observed in patients without IM or dysplasia. Collectively, these studies highlight the critical importance of early H. pylori detection and treatment, as the chemopreventive benefit appears to diminish once precancerous lesions are established.
Similarly, the clinical applicability of microbiome-targeted strategies, including FMT, remains hampered by unresolved concerns. Although FMT has shown promise in restoring microbial diversity and enhancing immunotherapy response[122], its long-term safety - particularly in cancer patients with immunocompromised states - remains inadequately studied. Additionally, the efficacy and safety profiles of individual probiotic strains differ markedly[325].
While nutritional interventions hold promise in modulating the gut microbiome to improve cancer outcomes, their practical application in clinical settings faces significant challenges. The American Society of Clinical Oncology highlights that the heterogeneity of interventions, patient populations, and cancer stages complicates the translation of evidence into practice, and the benefits of nutritional interventions in advanced cancer and cachexia remain inconsistent and modest at best[326].
Cancer patients, particularly those experiencing cachexia or treatment-induced GI complications, often struggle with dietary compliance due to symptoms like anorexia, nausea, diarrhea, and mucositis[327-329]. These symptoms can limit the tolerance for high-fiber or microbiota-targeted diets, narrowing the therapeutic window for nutritional modulation.
Recent studies highlight these challenges. For instance, a systematic review analyzing appetite and dietary intake endpoints in cancer cachexia clinical trials found considerable variability in assessment methods, underscoring the need for standardized and validated tools to effectively monitor nutritional interventions in this population[330]. Additionally, a randomized controlled trial investigating preoperative immunonutrition in GC patients with cachexia demonstrated that while immunonutrition reduced postoperative infectious complications and improved inflammatory and immune parameters, its implementation requires careful consideration of patient-specific factors and potential GI intolerance[331].
Practical barriers include limited access to registered dietitians, lack of standardized protocols, and patient reluctance or inability to adhere to dietary recommendations due to symptom burden or treatment side effects. Additionally, up to half of patients may pursue unproven or restrictive diets and supplements, increasing the risk of adverse interactions and further nutritional compromise. The American Society of Clinical Oncology recommends referral to a registered dietitian to mitigate these risks and provide individualized support, but acknowledges that such resources are not universally available[326].
For patients with severe GI dysfunction, such as those with nonfunctioning alimentary tracts, parenteral nutrition may be considered, but this approach requires careful patient selection, clear goal-setting, and ongoing evaluation due to its risks, costs, and limited indications1. Multidisciplinary collaboration, early identification of at-risk patients, and patient/caregiver education are essential to optimize feasibility and outcomes in real-world settings[326,332,333].
These findings emphasize the necessity for personalized nutrition approaches that account for individual patient conditions and the integration of dietary planning with oncologic care. Moreover, the development of evidence-based guidelines and the inclusion of dietetic support within multidisciplinary teams are crucial to address the heterogeneity of patient needs and the complexities of clinical practice.
Despite growing interest in the gut microbiome and nutritional strategies for GI tumors, several limitations constrain current research. A major challenge lies in the significant individual variability of the gut microbiome, which is shaped by host genetics, diet, environment, medications, and lifestyle factors[324]. This heterogeneity complicates efforts to identify universal microbial biomarkers or design broadly applicable nutritional interventions. Furthermore, while numerous studies have established associations between specific microbial taxa or dietary components and cancer outcomes[12,293,294,334,335], the underlying molecular mechanisms remain largely unresolved. The complexity of the gut ecosystem, involving dynamic interactions between microbes, host immunity, and metabolic pathways, makes it difficult to establish clear causal relationships.
Methodological inconsistencies also limit the comparability of findings across studies[323,324,335]. Variations in sample collection methods, sequencing technologies, bioinformatics pipelines, and data analysis approaches introduce biases and reduce reproducibility. Moreover, much of the current evidence derives from observational studies or preclinical models[150,288,294], with a scarcity of large-scale, well-controlled clinical trials evaluating microbiome-modulating nutritional interventions in GI cancers. These factors highlight the need for more rigorous and standardized research designs to validate initial findings and support clinical translation.
Despite significant advances in understanding the gut microbiome’s role in GI cancers, critical gaps remain that limit translational progress. First, mechanistic insights into microbiota-nutrition-immunity interactions are uneven across GI cancer types. While CRC has been extensively studied, with clear links between diet-modulated microbial metabolites (e.g., short-chain fatty acids) and immune modulation, other GI cancers such as pancreatic[189] and esophageal cancers[334] remain poorly characterized in this context. Detailed mechanistic studies are needed to elucidate how microbiota-derived signals influence tumor immunity and metabolism in these less-studied cancers.
Second, promising microbiota targets for therapeutic intervention - such as Akkermansia muciniphila and Faecalibacterium prausnitzii - have shown potential in preclinical models for enhancing immunotherapy response and reducing inflammation, yet they lack robust clinical validation[239]. Moreover, FMT trials in GI cancer patients are still limited in scale and often lack standardized protocols, impeding definitive conclusions on efficacy and safety[118,119]. Third, although dietary interventions such as high-fiber diets and microbiota-accessible carbohydrates show promise in modulating the gut ecosystem, the heterogeneity in patient responses and challenges in adherence highlight the need for personalized nutritional strategies guided by microbiome profiling[113].
Addressing these gaps will require multidisciplinary efforts integrating microbiology, immunology, nutrition science, and clinical oncology, alongside longitudinal and interventional clinical trials to translate mechanistic insights into effective therapies. Finally, fostering interdisciplinary collaboration among microbiologists, oncologists, nutritionists, and computational biologists will be essential to translate laboratory discoveries into clinical practice. Collectively, these efforts will pave the way for integrating microbiome-informed nutritional strategies into precision oncology for GI tumors.
In conclusion, the interplay between the gut microbiome, inflammation, and cancer development is a complex and evolving area of research. Nutritional strategies that modulate the gut microbiome hold significant potential in enhancing cancer therapy and improving patient outcomes. Probiotics, prebiotics, CR, and fasting-mimicking diets are all promising approaches to managing gut inflammation and promoting a healthy microbiome. However, further research is required to fully understand the mechanisms underlying these effects and to identify the most effective and personalized approaches for cancer patients. By leveraging the gut microbiome’s role in inflammation, we may uncover new avenues for improving cancer treatment efficacy and overall patient quality of life.
In summary, the gut microbiome represents a highly dynamic and therapeutically targetable axis in cancer care. By synthesizing recent advances in FMT and diet-based modulation, this review offers a translational perspective that bridges mechanistic microbiome science with actionable clinical strategies. Future directions should prioritize randomized controlled trials, multi-omics integration, and microbiome-informed precision nutrition to fully realize the promise of microbiome-based interventions in oncology.
| 1. | Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev. 2016;25:16-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2004] [Cited by in RCA: 2569] [Article Influence: 233.5] [Reference Citation Analysis (0)] |
| 2. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 12653] [Article Influence: 6326.5] [Reference Citation Analysis (6)] |
| 3. | Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 664] [Cited by in RCA: 6016] [Article Influence: 1504.0] [Reference Citation Analysis (2)] |
| 4. | Debora C, Gerardo Antonio Pio N. The bacteria-hypothesis of colorectal cancer: pathogenetic and therapeutic implications. Transl Gastrointest Cancer. 2014;3:44-53. |
| 5. | Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804-810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4573] [Cited by in RCA: 3855] [Article Influence: 202.9] [Reference Citation Analysis (0)] |
| 6. | Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 865] [Cited by in RCA: 1171] [Article Influence: 117.1] [Reference Citation Analysis (0)] |
| 7. | Thomas S, Izard J, Walsh E, Batich K, Chongsathidkiet P, Clarke G, Sela DA, Muller AJ, Mullin JM, Albert K, Gilligan JP, DiGuilio K, Dilbarova R, Alexander W, Prendergast GC. The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists. Cancer Res. 2017;77:1783-1812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 264] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 8. | Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, Trinchieri G. Microbes and Cancer. Annu Rev Immunol. 2017;35:199-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 215] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 9. | Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25:377-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 444] [Cited by in RCA: 848] [Article Influence: 121.1] [Reference Citation Analysis (0)] |
| 10. | World Health Organization. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Biological agents. Volume 100 B. Lyon: IARC Publications, 2012. |
| 11. | de Martel C, Franceschi S. Infections and cancer: established associations and new hypotheses. Crit Rev Oncol Hematol. 2009;70:183-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 175] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 12. | Baba Y, Iwatsuki M, Yoshida N, Watanabe M, Baba H. Review of the gut microbiome and esophageal cancer: Pathogenesis and potential clinical implications. Ann Gastroenterol Surg. 2017;1:99-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 93] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 13. | Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin. 2017;67:326-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 456] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 14. | Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1054] [Cited by in RCA: 1283] [Article Influence: 98.7] [Reference Citation Analysis (2)] |
| 15. | Fulbright LE, Ellermann M, Arthur JC. The microbiome and the hallmarks of cancer. PLoS Pathog. 2017;13:e1006480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 108] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 16. | Johnson CH, Spilker ME, Goetz L, Peterson SN, Siuzdak G. Metabolite and Microbiome Interplay in Cancer Immunotherapy. Cancer Res. 2016;76:6146-6152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 17. | Dzutsev A, Goldszmid RS, Viaud S, Zitvogel L, Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur J Immunol. 2015;45:17-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 217] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 18. | Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1198] [Cited by in RCA: 1566] [Article Influence: 120.5] [Reference Citation Analysis (0)] |
| 19. | Poutahidis T, Erdman SE. Commensal bacteria modulate the tumor microenvironment. Cancer Lett. 2016;380:356-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 20. | Panebianco C, Andriulli A, Pazienza V. Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome. 2018;6:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 229] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 21. | Li W, Deng Y, Chu Q, Zhang P. Gut microbiome and cancer immunotherapy. Cancer Lett. 2019;447:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 190] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 22. | Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33:570-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 677] [Cited by in RCA: 1017] [Article Influence: 127.1] [Reference Citation Analysis (0)] |
| 23. | Fessler J, Matson V, Gajewski TF. Exploring the emerging role of the microbiome in cancer immunotherapy. J Immunother Cancer. 2019;7:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 228] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 24. | Javanmard A, Ashtari S, Sabet B, Davoodi SH, Rostami-Nejad M, Esmaeil Akbari M, Niaz A, Mortazavian AM. Probiotics and their role in gastrointestinal cancers prevention and treatment; an overview. Gastroenterol Hepatol Bed Bench. 2018;11:284-295. [PubMed] |
| 25. | Meng C, Bai C, Brown TD, Hood LE, Tian Q. Human Gut Microbiota and Gastrointestinal Cancer. Genomics Proteomics Bioinformatics. 2018;16:33-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 285] [Cited by in RCA: 303] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 26. | Wong SH, Kwong TNY, Wu CY, Yu J. Clinical applications of gut microbiota in cancer biology. Semin Cancer Biol. 2019;55:28-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 27. | Ma W, Mao Q, Xia W, Dong G, Yu C, Jiang F. Gut Microbiota Shapes the Efficiency of Cancer Therapy. Front Microbiol. 2019;10:1050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 143] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 28. | David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8117] [Cited by in RCA: 7170] [Article Influence: 597.5] [Reference Citation Analysis (1)] |
| 29. | Chattopadhyay I, Gundamaraju R, Jha NK, Gupta PK, Dey A, Mandal CC, Ford BM. Interplay between Dysbiosis of Gut Microbiome, Lipid Metabolism, and Tumorigenesis: Can Gut Dysbiosis Stand as a Prognostic Marker in Cancer? Dis Markers. 2022;2022:2941248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 30. | Cianci R, Franza L, Schinzari G, Rossi E, Ianiro G, Tortora G, Gasbarrini A, Gambassi G, Cammarota G. The Interplay between Immunity and Microbiota at Intestinal Immunological Niche: The Case of Cancer. Int J Mol Sci. 2019;20:501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 31. | Pagliari D, Gambassi G, Piccirillo CA, Cianci R. The Intricate Link among Gut "Immunological Niche," Microbiota, and Xenobiotics in Intestinal Pathology. Mediators Inflamm. 2017;2017:8390595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 32. | Al-Asmakh M, Zadjali F. Use of Germ-Free Animal Models in Microbiota-Related Research. J Microbiol Biotechnol. 2015;25:1583-1588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 177] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 33. | Geem D, Medina-Contreras O, McBride M, Newberry RD, Koni PA, Denning TL. Specific microbiota-induced intestinal Th17 differentiation requires MHC class II but not GALT and mesenteric lymph nodes. J Immunol. 2014;193:431-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | Yoon K, Kim N. The Effect of Microbiota on Colon Carcinogenesis. J Cancer Prev. 2018;23:117-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 35. | Cianci R, Pagliari D, Pietroni V, Landolfi R, Pandolfi F. Tissue infiltrating lymphocytes: the role of cytokines in their growth and differentiation. J Biol Regul Homeost Agents. 2010;24:239-249. [PubMed] |
| 36. | Backert S, Neddermann M, Maubach G, Naumann M. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2016;21 Suppl 1:19-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 89] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 37. | Vinasco K, Mitchell HM, Kaakoush NO, Castaño-Rodríguez N. Microbial carcinogenesis: Lactic acid bacteria in gastric cancer. Biochim Biophys Acta Rev Cancer. 2019;1872:188309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 38. | Ohta K, Kawano R, Ito N. Lactic acid bacteria convert human fibroblasts to multipotent cells. PLoS One. 2012;7:e51866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 39. | Dalmasso G, Cougnoux A, Delmas J, Darfeuille-Michaud A, Bonnet R. The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes. 2014;5:675-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 223] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 40. | Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1355] [Cited by in RCA: 1396] [Article Influence: 174.5] [Reference Citation Analysis (0)] |
| 41. | Chattopadhyay I, Nandi D, Nag A. The pint- sized powerhouse: Illuminating the mighty role of the gut microbiome in improving the outcome of anti- cancer therapy. Semin Cancer Biol. 2021;70:98-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 42. | Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 501] [Cited by in RCA: 480] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 43. | Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858-20863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 829] [Cited by in RCA: 782] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 44. | Menendez A, Willing BP, Montero M, Wlodarska M, So CC, Bhinder G, Vallance BA, Finlay BB. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell α-defensins. J Innate Immun. 2013;5:39-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 45. | Schnare M, Barton GM, Holt AC, Takeda K, Akira S, Medzhitov R. Toll-like receptors control activation of adaptive immune responses. Nat Immunol. 2001;2:947-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1047] [Cited by in RCA: 1027] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 46. | Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA Jr. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998;2:253-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1188] [Cited by in RCA: 1215] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 47. | Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 460] [Article Influence: 24.2] [Reference Citation Analysis (1)] |
| 48. | Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3061] [Cited by in RCA: 3213] [Article Influence: 146.0] [Reference Citation Analysis (0)] |
| 49. | Apidianakis Y, Pitsouli C, Perrimon N, Rahme L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc Natl Acad Sci U S A. 2009;106:20883-20888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 179] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 50. | von Frieling J, Fink C, Hamm J, Klischies K, Forster M, Bosch TCG, Roeder T, Rosenstiel P, Sommer F. Grow With the Challenge - Microbial Effects on Epithelial Proliferation, Carcinogenesis, and Cancer Therapy. Front Microbiol. 2018;9:2020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 51. | Fukata M, Chen A, Klepper A, Krishnareddy S, Vamadevan AS, Thomas LS, Xu R, Inoue H, Arditi M, Dannenberg AJ, Abreu MT. Cox-2 is regulated by Toll-like receptor-4 (TLR4) signaling: Role in proliferation and apoptosis in the intestine. Gastroenterology. 2006;131:862-877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 350] [Cited by in RCA: 354] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 52. | Kopp E, Medzhitov R. Recognition of microbial infection by Toll-like receptors. Curr Opin Immunol. 2003;15:396-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 406] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 53. | Das M, Garlick DS, Greiner DL, Davis RJ. The role of JNK in the development of hepatocellular carcinoma. Genes Dev. 2011;25:634-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 162] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 54. | Kennedy NJ, Davis RJ. Role of JNK in tumor development. Cell Cycle. 2003;2:199-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 55. | Niccolai E, Ricci F, Russo E, Nannini G, Emmi G, Taddei A, Ringressi MN, Melli F, Miloeva M, Cianchi F, Bechi P, Prisco D, Amedei A. The Different Functional Distribution of "Not Effector" T Cells (Treg/Tnull) in Colorectal Cancer. Front Immunol. 2017;8:1900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 56. | Zhang M, Wen B, Anton OM, Yao Z, Dubois S, Ju W, Sato N, DiLillo DJ, Bamford RN, Ravetch JV, Waldmann TA. IL-15 enhanced antibody-dependent cellular cytotoxicity mediated by NK cells and macrophages. Proc Natl Acad Sci U S A. 2018;115:E10915-E10924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 57. | Pandolfi F, Cianci R, Pagliari D, Landolfi R, Cammarota G. Cellular mediators of inflammation: tregs and TH17 cells in gastrointestinal diseases. Mediators Inflamm. 2009;2009:132028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 58. | Shahmarvand N, Nagy A, Shahryari J, Ohgami RS. Mutations in the signal transducer and activator of transcription family of genes in cancer. Cancer Sci. 2018;109:926-933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 59. | Hurtado CG, Wan F, Housseau F, Sears CL. Roles for Interleukin 17 and Adaptive Immunity in Pathogenesis of Colorectal Cancer. Gastroenterology. 2018;155:1706-1715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 60. | Francescone R, Hou V, Grivennikov SI. Microbiome, inflammation, and cancer. Cancer J. 2014;20:181-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 196] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 61. | Pandolfi F, Cianci R, Pagliari D, Casciano F, Bagalà C, Astone A, Landolfi R, Barone C. The immune response to tumors as a tool toward immunotherapy. Clin Dev Immunol. 2011;2011:894704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 62. | Kryczek I, Wei S, Szeliga W, Vatan L, Zou W. Endogenous IL-17 contributes to reduced tumor growth and metastasis. Blood. 2009;114:357-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 326] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 63. | McCaw TR, Li M, Starenki D, Cooper SJ, Liu M, Meza-Perez S, Arend RC, Buchsbaum DJ, Forero A, Randall TD. The expression of MHC class II molecules on murine breast tumors delays T-cell exhaustion, expands the T-cell repertoire, and slows tumor growth. Cancer Immunol Immunother. 2019;68:175-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 64. | Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2481] [Cited by in RCA: 3193] [Article Influence: 212.9] [Reference Citation Analysis (1)] |
| 65. | Gupta PK, Godec J, Wolski D, Adland E, Yates K, Pauken KE, Cosgrove C, Ledderose C, Junger WG, Robson SC, Wherry EJ, Alter G, Goulder PJ, Klenerman P, Sharpe AH, Lauer GM, Haining WN. CD39 Expression Identifies Terminally Exhausted CD8+ T Cells. PLoS Pathog. 2015;11:e1005177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 211] [Cited by in RCA: 331] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 66. | Pagliari D, Piccirillo CA, Larbi A, Cianci R. The Interactions between Innate Immunity and Microbiota in Gastrointestinal Diseases. J Immunol Res. 2015;2015:898297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 67. | Reddavide R, Rotolo O, Caruso MG, Stasi E, Notarnicola M, Miraglia C, Nouvenne A, Meschi T, De' Angelis GL, Di Mario F, Leandro G. The role of diet in the prevention and treatment of Inflammatory Bowel Diseases. Acta Biomed. 2018;89:60-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 49] [Reference Citation Analysis (0)] |
| 68. | Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. 2020;39:4925-4943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 424] [Article Influence: 70.7] [Reference Citation Analysis (0)] |
| 69. | Piewngam P, Zheng Y, Nguyen TH, Dickey SW, Joo HS, Villaruz AE, Glose KA, Fisher EL, Hunt RL, Li B, Chiou J, Pharkjaksu S, Khongthong S, Cheung GYC, Kiratisin P, Otto M. Pathogen elimination by probiotic Bacillus via signalling interference. Nature. 2018;562:532-537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 450] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 70. | Shin DS, Rhee KJ, Eom YB. Effect of Probiotic Clostridium butyricum NCTC 7423 Supernatant on Biofilm Formation and Gene Expression of Bacteroides fragilis. J Microbiol Biotechnol. 2020;30:368-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 71. | Gharbi Y, Fhoula I, Ruas-madiedo P, Afef N, Boudabous A, Gueimonde M, Ouzari H. In-vitro characterization of potentially probiotic Lactobacillus strains isolated from human microbiota: interaction with pathogenic bacteria and the enteric cell line HT29. Ann Microbiol. 2019;69:61-72. [RCA] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 72. | Gatej SM, Bright R, Weyrich LS, Marino V, Christophersen CT, Gibson RJ, Gully N, Zilm P, Bartold PM. Probiotic Lactobacillus Rhamnosus GG Protects Against P. Gingivalis And F. Nucleatum Gut Dysbiosis. J Int Acad Periodontol. 2020;22:18-27. [PubMed] |
| 73. | Ghanavati R, Akbari A, Mohammadi F, Asadollahi P, Javadi A, Talebi M, Rohani M. Lactobacillus species inhibitory effect on colorectal cancer progression through modulating the Wnt/β-catenin signaling pathway. Mol Cell Biochem. 2020;470:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 74. | Gamallat Y, Meyiah A, Kuugbee ED, Hago AM, Chiwala G, Awadasseid A, Bamba D, Zhang X, Shang X, Luo F, Xin Y. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomed Pharmacother. 2016;83:536-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 162] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 75. | Yue Y, Ye K, Lu J, Wang X, Zhang S, Liu L, Yang B, Nassar K, Xu X, Pang X, Lv J. Probiotic strain Lactobacillus plantarum YYC-3 prevents colon cancer in mice by regulating the tumour microenvironment. Biomed Pharmacother. 2020;127:110159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 76. | Mendes MCS, Paulino DS, Brambilla SR, Camargo JA, Persinoti GF, Carvalheira JBC. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World J Gastroenterol. 2018;24:1995-2008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 101] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (1)] |
| 77. | Landy J, Ronde E, English N, Clark SK, Hart AL, Knight SC, Ciclitira PJ, Al-Hassi HO. Tight junctions in inflammatory bowel diseases and inflammatory bowel disease associated colorectal cancer. World J Gastroenterol. 2016;22:3117-3126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 294] [Cited by in RCA: 390] [Article Influence: 39.0] [Reference Citation Analysis (12)] |
| 78. | Alvarez CS, Badia J, Bosch M, Giménez R, Baldomà L. Outer Membrane Vesicles and Soluble Factors Released by Probiotic Escherichia coli Nissle 1917 and Commensal ECOR63 Enhance Barrier Function by Regulating Expression of Tight Junction Proteins in Intestinal Epithelial Cells. Front Microbiol. 2016;7:1981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 160] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 79. | Zhang Y, Zhao X, Zhu Y, Ma J, Ma H, Zhang H. Probiotic Mixture Protects Dextran Sulfate Sodium-Induced Colitis by Altering Tight Junction Protein Expressions and Increasing Tregs. Mediators Inflamm. 2018;2018:9416391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 80. | Kuugbee ED, Shang X, Gamallat Y, Bamba D, Awadasseid A, Suliman MA, Zang S, Ma Y, Chiwala G, Xin Y, Shang D. Structural Change in Microbiota by a Probiotic Cocktail Enhances the Gut Barrier and Reduces Cancer via TLR2 Signaling in a Rat Model of Colon Cancer. Dig Dis Sci. 2016;61:2908-2920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 113] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 81. | Feng T, Wang J. Oxidative stress tolerance and antioxidant capacity of lactic acid bacteria as probiotic: a systematic review. Gut Microbes. 2020;12:1801944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 327] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 82. | Feng J, Gao M, Zhao C, Yang J, Gao H, Lu X, Ju R, Zhang X, Zhang Y. Oral Administration of Probiotics Reduces Chemotherapy-Induced Diarrhea and Oral Mucositis: A Systematic Review and Meta-Analysis. Front Nutr. 2022;9:823288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 83. | Shen SR, Chen WJ, Chu HF, Wu SH, Wang YR, Shen TL. Amelioration of 5-fluorouracil-induced intestinal mucositis by Streptococcus thermophilus ST4 in a mouse model. PLoS One. 2021;16:e0253540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 84. | López-Gómez L, Alcorta A, Abalo R. Probiotics and Probiotic-like Agents against Chemotherapy-Induced Intestinal Mucositis: A Narrative Review. J Pers Med. 2023;13:1487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 85. | Gibson GR, Beatty ER, Wang X, Cummings JH. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology. 1995;108:975-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1062] [Cited by in RCA: 898] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 86. | Reddy BS, Rivenson A. Inhibitory effect of Bifidobacterium longum on colon, mammary, and liver carcinogenesis induced by 2-amino-3-methylimidazo[4,5-f]quinoline, a food mutagen. Cancer Res. 1993;53:3914-3918. [PubMed] |
| 87. | Rowland IR, Rumney CJ, Coutts JT, Lievense LC. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis. 1998;19:281-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 259] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 88. | Taper HS, Roberfroid MB. Possible adjuvant cancer therapy by two prebiotics--inulin or oligofructose. In Vivo. 2005;19:201-204. [PubMed] |
| 89. | Taper HS, Roberfroid MB. Inulin/oligofructose and anticancer therapy. Br J Nutr. 2002;87 Suppl 2:S283-S286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 90. | Taper HS, Roberfroid MB. Nontoxic potentiation of cancer chemotherapy by dietary oligofructose or inulin. Nutr Cancer. 2000;38:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 91. | García-Peris P, Velasco C, Lozano MA, Moreno Y, Paron L, de la Cuerda C, Bretón I, Camblor M, García-Hernández J, Guarner F, Hernández M. Effect of a mixture of inulin and fructo-oligosaccharide on Lactobacillus and Bifidobacterium intestinal microbiota of patients receiving radiotherapy: a randomised, double-blind, placebo-controlled trial. Nutr Hosp. 2012;27:1908-1915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 92. | Garcia-Peris P, Velasco C, Hernandez M, Lozano MA, Paron L, de la Cuerda C, Breton I, Camblor M, Guarner F. Effect of inulin and fructo-oligosaccharide on the prevention of acute radiation enteritis in patients with gynecological cancer and impact on quality-of-life: a randomized, double-blind, placebo-controlled trial. Eur J Clin Nutr. 2016;70:170-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 93. | Zhao R, Wang Y, Huang Y, Cui Y, Xia L, Rao Z, Zhou Y, Wu X. Effects of fiber and probiotics on diarrhea associated with enteral nutrition in gastric cancer patients: A prospective randomized and controlled trial. Medicine (Baltimore). 2017;96:e8418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 94. | Straus Farber R, Walker EL, Diallo F, Onomichi K, Riley C, Zhang L, Zhu W, De Jager PL, Xia Z. A randomized cross-over trial of prebiotics and probiotics in multiple sclerosis: Trial feasibility, supplement tolerability and symptom abatement. Mult Scler Relat Disord. 2024;89:105762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 95. | Xie X, He Y, Li H, Yu D, Na L, Sun T, Zhang D, Shi X, Xia Y, Jiang T, Rong S, Yang S, Ma X, Xu G. Effects of prebiotics on immunologic indicators and intestinal microbiota structure in perioperative colorectal cancer patients. Nutrition. 2019;61:132-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 96. | Boutron-Ruault MC, Marteau P, Lavergne-Slove A, Myara A, Gerhardt MF, Franchisseur C, Bornet F; Eripolyp Study Group. Effects of a 3-mo consumption of short-chain fructo-oligosaccharides on parameters of colorectal carcinogenesis in patients with or without small or large colorectal adenomas. Nutr Cancer. 2005;53:160-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 41] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 97. | Fehlbaum S, Prudence K, Kieboom J, Heerikhuisen M, van den Broek T, Schuren FHJ, Steinert RE, Raederstorff D. In Vitro Fermentation of Selected Prebiotics and Their Effects on the Composition and Activity of the Adult Gut Microbiota. Int J Mol Sci. 2018;19:3097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 98. | Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110:9066-9071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2639] [Cited by in RCA: 3482] [Article Influence: 267.8] [Reference Citation Analysis (1)] |
| 99. | Li Y, Tinoco R, Elmén L, Segota I, Xian Y, Fujita Y, Sahu A, Zarecki R, Marie K, Feng Y, Khateb A, Frederick DT, Ashkenazi SK, Kim H, Perez EG, Day CP, Segura Muñoz RS, Schmaltz R, Yooseph S, Tam MA, Zhang T, Avitan-Hersh E, Tzur L, Roizman S, Boyango I, Bar-Sela G, Orian A, Kaufman RJ, Bosenberg M, Goding CR, Baaten B, Levesque MP, Dummer R, Brown K, Merlino G, Ruppin E, Flaherty K, Ramer-Tait A, Long T, Peterson SN, Bradley LM, Ronai ZA. Gut microbiota dependent anti-tumor immunity restricts melanoma growth in Rnf5(-/-) mice. Nat Commun. 2019;10:1492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 100. | Mazraeh R, Azizi-Soleiman F, Jazayeri SMHM, Noori SMA. Effect of inulin-type fructans in patients undergoing cancer treatments: A systematic review. Pak J Med Sci. 2019;35:575-580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 101. | Tagliabue A, Ferraris C, Uggeri F, Trentani C, Bertoli S, de Giorgis V, Veggiotti P, Elli M. Short-term impact of a classical ketogenic diet on gut microbiota in GLUT1 Deficiency Syndrome: A 3-month prospective observational study. Clin Nutr ESPEN. 2017;17:33-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 104] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 102. | Chung HY, Park YK. Rationale, Feasibility and Acceptability of Ketogenic Diet for Cancer Treatment. J Cancer Prev. 2017;22:127-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 103. | Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029-1033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12491] [Cited by in RCA: 12314] [Article Influence: 724.4] [Reference Citation Analysis (0)] |
| 104. | Weber DD, Aminazdeh-Gohari S, Kofler B. Ketogenic diet in cancer therapy. Aging (Albany NY). 2018;10:164-165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 113] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 105. | Klement RJ. Beneficial effects of ketogenic diets for cancer patients: a realist review with focus on evidence and confirmation. Med Oncol. 2017;34:132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 107] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 106. | Xie G, Zhou Q, Qiu CZ, Dai WK, Wang HP, Li YH, Liao JX, Lu XG, Lin SF, Ye JH, Ma ZY, Wang WJ. Ketogenic diet poses a significant effect on imbalanced gut microbiota in infants with refractory epilepsy. World J Gastroenterol. 2017;23:6164-6171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 228] [Cited by in RCA: 223] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 107. | Newell C, Bomhof MR, Reimer RA, Hittel DS, Rho JM, Shearer J. Ketogenic diet modifies the gut microbiota in a murine model of autism spectrum disorder. Mol Autism. 2016;7:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 214] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 108. | Rinninella E, Raoul P, Cintoni M, Palombaro M, Pulcini G, Gasbarrini A, Mele MC. Nutritional Interventions Targeting Gut Microbiota during Cancer Therapies. Microorganisms. 2021;9:1469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 109. | Cox LM, Schafer MJ, Sohn J, Vincentini J, Weiner HL, Ginsberg SD, Blaser MJ. Calorie restriction slows age-related microbiota changes in an Alzheimer's disease model in female mice. Sci Rep. 2019;9:17904. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 108] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 110. | Wei S, Han R, Zhao J, Wang S, Huang M, Wang Y, Chen Y. Intermittent administration of a fasting-mimicking diet intervenes in diabetes progression, restores β cells and reconstructs gut microbiota in mice. Nutr Metab (Lond). 2018;15:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 81] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 111. | Rimoldi M, Chieppa M, Salucci V, Avogadri F, Sonzogni A, Sampietro GM, Nespoli A, Viale G, Allavena P, Rescigno M. Intestinal immune homeostasis is regulated by the crosstalk between epithelial cells and dendritic cells. Nat Immunol. 2005;6:507-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 601] [Cited by in RCA: 608] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 112. | Tisdale MJ. Mechanisms of cancer cachexia. Physiol Rev. 2009;89:381-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 789] [Cited by in RCA: 852] [Article Influence: 50.1] [Reference Citation Analysis (1)] |
| 113. | Barber TM, Kabisch S, Pfeiffer AFH, Weickert MO. The Health Benefits of Dietary Fibre. Nutrients. 2020;12:3209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 434] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 114. | Sun L, Zhang Z, Xu J, Xu G, Liu X. Dietary fiber intake reduces risk for Barrett's esophagus and esophageal cancer. Crit Rev Food Sci Nutr. 2017;57:2749-2757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 115. | Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, Khan MAW, Zhang X, White MG, Peterson CB, Wong MC, Morad G, Rodgers T, Badger JH, Helmink BA, Andrews MC, Rodrigues RR, Morgun A, Kim YS, Roszik J, Hoffman KL, Zheng J, Zhou Y, Medik YB, Kahn LM, Johnson S, Hudgens CW, Wani K, Gaudreau PO, Harris AL, Jamal MA, Baruch EN, Perez-Guijarro E, Day CP, Merlino G, Pazdrak B, Lochmann BS, Szczepaniak-Sloane RA, Arora R, Anderson J, Zobniw CM, Posada E, Sirmans E, Simon J, Haydu LE, Burton EM, Wang L, Dang M, Clise-Dwyer K, Schneider S, Chapman T, Anang NAS, Duncan S, Toker J, Malke JC, Glitza IC, Amaria RN, Tawbi HA, Diab A, Wong MK, Patel SP, Woodman SE, Davies MA, Ross MI, Gershenwald JE, Lee JE, Hwu P, Jensen V, Samuels Y, Straussman R, Ajami NJ, Nelson KC, Nezi L, Petrosino JF, Futreal PA, Lazar AJ, Hu J, Jenq RR, Tetzlaff MT, Yan Y, Garrett WS, Huttenhower C, Sharma P, Watowich SS, Allison JP, Cohen L, Trinchieri G, Daniel CR, Wargo JA. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. 2021;374:1632-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 661] [Article Influence: 132.2] [Reference Citation Analysis (0)] |
| 116. | Zitvogel L, Daillère R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15:465-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 384] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 117. | Munteanu C, Schwartz B. Interactions between Dietary Antioxidants, Dietary Fiber and the Gut Microbiome: Their Putative Role in Inflammation and Cancer. Int J Mol Sci. 2024;25:8250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 118. | Porcari S, Benech N, Valles-Colomer M, Segata N, Gasbarrini A, Cammarota G, Sokol H, Ianiro G. Key determinants of success in fecal microbiota transplantation: From microbiome to clinic. Cell Host Microbe. 2023;31:712-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 180] [Reference Citation Analysis (0)] |
| 119. | Minkoff NZ, Aslam S, Medina M, Tanner-Smith EE, Zackular JP, Acra S, Nicholson MR, Imdad A. Fecal microbiota transplantation for the treatment of recurrent Clostridioides difficile (Clostridium difficile). Cochrane Database Syst Rev. 2023;4:CD013871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 58] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 120. | Yadegar A, Bar-Yoseph H, Monaghan TM, Pakpour S, Severino A, Kuijper EJ, Smits WK, Terveer EM, Neupane S, Nabavi-Rad A, Sadeghi J, Cammarota G, Ianiro G, Nap-Hill E, Leung D, Wong K, Kao D. Fecal microbiota transplantation: current challenges and future landscapes. Clin Microbiol Rev. 2024;37:e0006022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 181] [Article Influence: 90.5] [Reference Citation Analysis (0)] |
| 121. | Routy B, Lenehan JG, Miller WH Jr, Jamal R, Messaoudene M, Daisley BA, Hes C, Al KF, Martinez-Gili L, Punčochář M, Ernst S, Logan D, Belanger K, Esfahani K, Richard C, Ninkov M, Piccinno G, Armanini F, Pinto F, Krishnamoorthy M, Figueredo R, Thebault P, Takis P, Magrill J, Ramsay L, Derosa L, Marchesi JR, Parvathy SN, Elkrief A, Watson IR, Lapointe R, Segata N, Haeryfar SMM, Mullish BH, Silverman MS, Burton JP, Maleki Vareki S. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat Med. 2023;29:2121-2132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 320] [Article Influence: 106.7] [Reference Citation Analysis (0)] |
| 122. | Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S, Bloch N, Rotin D, Anafi L, Avivi C, Melnichenko J, Steinberg-Silman Y, Mamtani R, Harati H, Asher N, Shapira-Frommer R, Brosh-Nissimov T, Eshet Y, Ben-Simon S, Ziv O, Khan MAW, Amit M, Ajami NJ, Barshack I, Schachter J, Wargo JA, Koren O, Markel G, Boursi B. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 1150] [Article Influence: 191.7] [Reference Citation Analysis (0)] |
| 123. | Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, Deblasio RN, Menna C, Ding Q, Pagliano O, Zidi B, Zhang S, Badger JH, Vetizou M, Cole AM, Fernandes MR, Prescott S, Costa RGF, Balaji AK, Morgun A, Vujkovic-Cvijin I, Wang H, Borhani AA, Schwartz MB, Dubner HM, Ernst SJ, Rose A, Najjar YG, Belkaid Y, Kirkwood JM, Trinchieri G, Zarour HM. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 843] [Cited by in RCA: 1200] [Article Influence: 240.0] [Reference Citation Analysis (0)] |
| 124. | Tan J, Chen YX. Dietary and Lifestyle Factors Associated with Colorectal Cancer Risk and Interactions with Microbiota: Fiber, Red or Processed Meat and Alcoholic Drinks. Gastrointest Tumors. 2016;3:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 125. | Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1014] [Cited by in RCA: 1187] [Article Influence: 107.9] [Reference Citation Analysis (0)] |
| 126. | Axelrad JE, Lichtiger S, Yajnik V. Inflammatory bowel disease and cancer: The role of inflammation, immunosuppression, and cancer treatment. World J Gastroenterol. 2016;22:4794-4801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 285] [Cited by in RCA: 354] [Article Influence: 35.4] [Reference Citation Analysis (4)] |
| 127. | Vacante M, Borzì AM, Basile F, Biondi A. Biomarkers in colorectal cancer: Current clinical utility and future perspectives. World J Clin Cases. 2018;6:869-881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 81] [Cited by in RCA: 114] [Article Influence: 14.3] [Reference Citation Analysis (6)] |
| 128. | Montalban-Arques A, Scharl M. Intestinal microbiota and colorectal carcinoma: Implications for pathogenesis, diagnosis, and therapy. EBioMedicine. 2019;48:648-655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 129. | Garrett WS. The gut microbiota and colon cancer. Science. 2019;364:1133-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 242] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 130. | Canny GO, McCormick BA. Bacteria in the intestine, helpful residents or enemies from within? Infect Immun. 2008;76:3360-3373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 122] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 131. | Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996-1047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 765] [Cited by in RCA: 814] [Article Influence: 67.8] [Reference Citation Analysis (0)] |
| 132. | Baliou S, Adamaki M, Spandidos D, Kyriakopoulos A, Christodoulou I, Zoumpourlis V. The microbiome, its molecular mechanisms and its potential as a therapeutic strategy against colorectal carcinogenesis (Review). World Acad Sci J. 2018;1:3-19. [DOI] [Full Text] |
| 133. | Jahani-Sherafat S, Alebouyeh M, Moghim S, Ahmadi Amoli H, Ghasemian-Safaei H. Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol Hepatol Bed Bench. 2018;11:101-109. [PubMed] |
| 134. | Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell. 2018;33:954-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 582] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 135. | Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1085] [Article Influence: 98.6] [Reference Citation Analysis (1)] |
| 136. | Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1511] [Cited by in RCA: 1818] [Article Influence: 139.8] [Reference Citation Analysis (1)] |
| 137. | Kashani N, Bezmin Abadi AT, Rahimi F, Forootan M. FadA-positive Fusobacterium nucleatum is prevalent in biopsy specimens of Iranian patients with colorectal cancer. New Microbes New Infect. 2020;34:100651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 138. | Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 645] [Article Influence: 71.7] [Reference Citation Analysis (0)] |
| 139. | Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, Li H, Guo B, Zhu Q, Wei Q, Moyer MP, Wang P, Cai S, Goel A, Qin H, Ma Y. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851-866.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 786] [Article Influence: 87.3] [Reference Citation Analysis (1)] |
| 140. | Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 1169] [Article Influence: 129.9] [Reference Citation Analysis (1)] |
| 141. | Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, Paganelli FL, Geurts MH, Beumer J, Mizutani T, Miao Y, van der Linden R, van der Elst S; Genomics England Research Consortium, Garcia KC, Top J, Willems RJL, Giannakis M, Bonnet R, Quirke P, Meyerson M, Cuppen E, van Boxtel R, Clevers H. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. 2020;580:269-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 831] [Article Influence: 138.5] [Reference Citation Analysis (0)] |
| 142. | Allen-Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett. 2014;162:54-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 143. | Maddocks OD, Scanlon KM, Donnenberg MS. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio. 2013;4:e00152-e00113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 144. | Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, Anders RA, Giardiello FM, Wick EC, Wang H, Wu S, Pardoll DM, Housseau F, Sears CL. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 761] [Cited by in RCA: 849] [Article Influence: 106.1] [Reference Citation Analysis (17)] |
| 145. | Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, Chan FKL, Kristiansen K, Sung JJY, Wong SH, Yu J. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 250] [Cited by in RCA: 377] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 146. | Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 971] [Article Influence: 138.7] [Reference Citation Analysis (0)] |
| 147. | Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S, Chung L, Finard BB, Wu X, Fathi P, Ganguly S, Fu J, Pardoll DM, Sears CL, Housseau F. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017;10:421-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 169] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 148. | Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, Tam AJ, McAllister F, Fan H, Wu X, Ganguly S, Lebid A, Metz P, Van Meerbeke SW, Huso DL, Wick EC, Pardoll DM, Wan F, Wu S, Sears CL, Housseau F. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. 2018;23:203-214.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 418] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 149. | Rezasoltani S, Asadzadeh Aghdaei H, Dabiri H, Akhavan Sepahi A, Modarressi MH, Nazemalhosseini Mojarad E. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb Pathog. 2018;124:244-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 150. | Dadkhah E, Sikaroodi M, Korman L, Hardi R, Baybick J, Hanzel D, Kuehn G, Kuehn T, Gillevet PM. Gut microbiome identifies risk for colorectal polyps. BMJ Open Gastroenterol. 2019;6:e000297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 151. | Pasquereau-Kotula E, Martins M, Aymeric L, Dramsi S. Significance of Streptococcus gallolyticus subsp. gallolyticus Association With Colorectal Cancer. Front Microbiol. 2018;9:614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 152. | Abdulamir AS, Hafidh RR, Abu Bakar F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its etiological role. J Exp Clin Cancer Res. 2011;30:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 232] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 153. | Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis. 2011;53:870-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 273] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 154. | Öberg J, Rasmussen M, Buchwald P, Nilson B, Inghammar M. Streptococcus bovis-bacteremia: subspecies distribution and association with colorectal cancer: a retrospective cohort study. Epidemiol Infect. 2021;150:e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 155. | Kamali N, Talebi Bezmin Abadi A, Abadi B, Rahimi F, Forootan M. Identification of Streptococcus gallolyticus in tumor samples of Iranian patients diagnosed with colorectal cancer. BMC Res Notes. 2022;15:316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 156. | Purcell RV, Pearson J, Aitchison A, Dixon L, Frizelle FA, Keenan JI. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017;12:e0171602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 174] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 157. | Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124:392-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 307] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 158. | Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, Soyletir G. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 341] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 159. | Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, Platz EA, Pardoll DM, Sears CL. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 479] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 160. | Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1113] [Cited by in RCA: 1362] [Article Influence: 80.1] [Reference Citation Analysis (0)] |
| 161. | Maddocks OD, Short AJ, Donnenberg MS, Bader S, Harrison DJ. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One. 2009;4:e5517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 109] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 162. | Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010;107:11537-11542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 651] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 163. | Wu J, Li Q, Fu X. Fusobacterium nucleatum Contributes to the Carcinogenesis of Colorectal Cancer by Inducing Inflammation and Suppressing Host Immunity. Transl Oncol. 2019;12:846-851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 182] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 164. | Pillar CM, Gilmore MS. Enterococcal virulence--pathogenicity island of E. Faecalis. Front Biosci. 2004;9:2335-2346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 165. | Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23:1298-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 230] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 166. | Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548-563.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 814] [Cited by in RCA: 1615] [Article Influence: 179.4] [Reference Citation Analysis (0)] |
| 167. | Shah MS, DeSantis T, Yamal JM, Weir T, Ryan EP, Cope JL, Hollister EB. Re-purposing 16S rRNA gene sequence data from within case paired tumor biopsy and tumor-adjacent biopsy or fecal samples to identify microbial markers for colorectal cancer. PLoS One. 2018;13:e0207002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 168. | Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, Comstock LE, Gandhi R, Weiner HL. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe. 2016;19:32-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 609] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 169. | Faïs T, Delmas J, Cougnoux A, Dalmasso G, Bonnet R. Targeting colorectal cancer-associated bacteria: A new area of research for personalized treatments. Gut Microbes. 2016;7:329-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 170. | Drago L. Probiotics and Colon Cancer. Microorganisms. 2019;7:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (1)] |
| 171. | Bassaganya-Riera J, Viladomiu M, Pedragosa M, De Simone C, Hontecillas R. Immunoregulatory mechanisms underlying prevention of colitis-associated colorectal cancer by probiotic bacteria. PLoS One. 2012;7:e34676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 172. | Molska M, Reguła J. Potential Mechanisms of Probiotics Action in the Prevention and Treatment of Colorectal Cancer. Nutrients. 2019;11:2453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 108] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 173. | Bowen JM, Stringer AM, Gibson RJ, Yeoh AS, Hannam S, Keefe DM. VSL#3 probiotic treatment reduces chemotherapy-induced diarrhea and weight loss. Cancer Biol Ther. 2007;6:1449-1454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 155] [Article Influence: 8.2] [Reference Citation Analysis (1)] |
| 174. | Malhotra SL. Dietary factors in a study of cancer colon from Cancer Registry, with special reference to the role of saliva, milk and fermented milk products and vegetable fibre. Med Hypotheses. 1977;3:122-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 45] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 175. | Peters RK, Pike MC, Garabrant D, Mack TM. Diet and colon cancer in Los Angeles County, California. Cancer Causes Control. 1992;3:457-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 195] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 176. | Young TB, Wolf DA. Case-control study of proximal and distal colon cancer and diet in Wisconsin. Int J Cancer. 1988;42:167-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 150] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 177. | Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, Trinchieri G, Pardo-Manuel de Villena F, Yewdell JW, Rehermann B. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171:1015-1028.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 618] [Article Influence: 68.7] [Reference Citation Analysis (10)] |
| 178. | Filip M, Tzaneva V, Dumitrascu DL. Fecal transplantation: digestive and extradigestive clinical applications. Clujul Med. 2018;91:259-265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 179. | Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, Thaiss CA, Reuben A, Livny J, Avraham R, Frederick DT, Ligorio M, Chatman K, Johnston SE, Mosher CM, Brandis A, Fuks G, Gurbatri C, Gopalakrishnan V, Kim M, Hurd MW, Katz M, Fleming J, Maitra A, Smith DA, Skalak M, Bu J, Michaud M, Trauger SA, Barshack I, Golan T, Sandbank J, Flaherty KT, Mandinova A, Garrett WS, Thayer SP, Ferrone CR, Huttenhower C, Bhatia SN, Gevers D, Wargo JA, Golub TR, Straussman R. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156-1160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1467] [Cited by in RCA: 1322] [Article Influence: 146.9] [Reference Citation Analysis (1)] |
| 180. | Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, Dai RM, Kiu H, Cardone M, Naik S, Patri AK, Wang E, Marincola FM, Frank KM, Belkaid Y, Trinchieri G, Goldszmid RS. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1253] [Cited by in RCA: 1758] [Article Influence: 135.2] [Reference Citation Analysis (0)] |
| 181. | Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, Poirier-Colame V, Roux A, Becharef S, Formenti S, Golden E, Cording S, Eberl G, Schlitzer A, Ginhoux F, Mani S, Yamazaki T, Jacquelot N, Enot DP, Bérard M, Nigou J, Opolon P, Eggermont A, Woerther PL, Chachaty E, Chaput N, Robert C, Mateus C, Kroemer G, Raoult D, Boneca IG, Carbonnel F, Chamaillard M, Zitvogel L. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1834] [Cited by in RCA: 2695] [Article Influence: 245.0] [Reference Citation Analysis (0)] |
| 182. | Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, Redinbo MR. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831-835. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 863] [Cited by in RCA: 839] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 183. | Xu Y, Villalona-Calero MA. Irinotecan: mechanisms of tumor resistance and novel strategies for modulating its activity. Ann Oncol. 2002;13:1841-1851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 293] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 184. | Shen S, Lim G, You Z, Ding W, Huang P, Ran C, Doheny J, Caravan P, Tate S, Hu K, Kim H, McCabe M, Huang B, Xie Z, Kwon D, Chen L, Mao J. Gut microbiota is critical for the induction of chemotherapy-induced pain. Nat Neurosci. 2017;20:1213-1216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 227] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 185. | Weledji EP, Enoworock G, Mokake M, Sinju M. How Grim is Pancreatic Cancer? Oncol Rev. 2016;10:294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 186. | Adamska A, Domenichini A, Falasca M. Pancreatic Ductal Adenocarcinoma: Current and Evolving Therapies. Int J Mol Sci. 2017;18:1338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 382] [Cited by in RCA: 454] [Article Influence: 50.4] [Reference Citation Analysis (1)] |
| 187. | Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 1394] [Article Influence: 107.2] [Reference Citation Analysis (1)] |
| 188. | Wang C, Li J. Pathogenic Microorganisms and Pancreatic Cancer. Gastrointest Tumors. 2015;2:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 189. | Wei MY, Shi S, Liang C, Meng QC, Hua J, Zhang YY, Liu J, Zhang B, Xu J, Yu XJ. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer. 2019;18:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 210] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 190. | Raderer M, Wrba F, Kornek G, Maca T, Koller DY, Weinlaender G, Hejna M, Scheithauer W. Association between Helicobacter pylori infection and pancreatic cancer. Oncology. 1998;55:16-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 101] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 191. | Gravina AG, Zagari RM, De Musis C, Romano L, Loguercio C, Romano M. Helicobacter pylori and extragastric diseases: A review. World J Gastroenterol. 2018;24:3204-3221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 244] [Cited by in RCA: 215] [Article Influence: 26.9] [Reference Citation Analysis (7)] |
| 192. | Stolzenberg-Solomon RZ, Blaser MJ, Limburg PJ, Perez-Perez G, Taylor PR, Virtamo J, Albanes D; ATBC Study. Helicobacter pylori seropositivity as a risk factor for pancreatic cancer. J Natl Cancer Inst. 2001;93:937-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 130] [Article Influence: 5.2] [Reference Citation Analysis (1)] |
| 193. | Bao Y, Spiegelman D, Li R, Giovannucci E, Fuchs CS, Michaud DS. History of peptic ulcer disease and pancreatic cancer risk in men. Gastroenterology. 2010;138:541-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 194. | Luo J, Nordenvall C, Nyrén O, Adami HO, Permert J, Ye W. The risk of pancreatic cancer in patients with gastric or duodenal ulcer disease. Int J Cancer. 2007;120:368-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 195. | Manes G, Balzano A, Vaira D. Helicobacter pylori and pancreatic disease. JOP. 2003;4:111-116. [PubMed] |
| 196. | Huang H, Daniluk J, Liu Y, Chu J, Li Z, Ji B, Logsdon CD. Oncogenic K-Ras requires activation for enhanced activity. Oncogene. 2014;33:532-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 197. | Daniluk J, Liu Y, Deng D, Chu J, Huang H, Gaiser S, Cruz-Monserrate Z, Wang H, Ji B, Logsdon CD. An NF-κB pathway-mediated positive feedback loop amplifies Ras activity to pathological levels in mice. J Clin Invest. 2012;122:1519-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 239] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 198. | Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2935] [Cited by in RCA: 3502] [Article Influence: 206.0] [Reference Citation Analysis (21)] |
| 199. | Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Klöppel G, Yoshimura A, Reindl W, Sipos B, Akira S, Schmid RM, Algül H. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 618] [Cited by in RCA: 709] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 200. | Michaud DS. Role of bacterial infections in pancreatic cancer. Carcinogenesis. 2013;34:2193-2197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 201. | Tribble GD, Kerr JE, Wang BY. Genetic diversity in the oral pathogen Porphyromonas gingivalis: molecular mechanisms and biological consequences. Future Microbiol. 2013;8:607-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 202. | Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjønneland A, Dahm CC, Overvad K, Jenab M, Fedirko V, Boutron-Ruault MC, Clavel-Chapelon F, Racine A, Kaaks R, Boeing H, Foerster J, Trichopoulou A, Lagiou P, Trichopoulos D, Sacerdote C, Sieri S, Palli D, Tumino R, Panico S, Siersema PD, Peeters PH, Lund E, Barricarte A, Huerta JM, Molina-Montes E, Dorronsoro M, Quirós JR, Duell EJ, Ye W, Sund M, Lindkvist B, Johansen D, Khaw KT, Wareham N, Travis RC, Vineis P, Bueno-de-Mesquita HB, Riboli E. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62:1764-1770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 323] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 203. | Zhang JJ, Wu HS, Wang L, Tian Y, Zhang JH, Wu HL. Expression and significance of TLR4 and HIF-1alpha in pancreatic ductal adenocarcinoma. World J Gastroenterol. 2010;16:2881-2888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 64] [Cited by in RCA: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 204. | Ikebe M, Kitaura Y, Nakamura M, Tanaka H, Yamasaki A, Nagai S, Wada J, Yanai K, Koga K, Sato N, Kubo M, Tanaka M, Onishi H, Katano M. Lipopolysaccharide (LPS) increases the invasive ability of pancreatic cancer cells through the TLR4/MyD88 signaling pathway. J Surg Oncol. 2009;100:725-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 140] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 205. | Ochi A, Nguyen AH, Bedrosian AS, Mushlin HM, Zarbakhsh S, Barilla R, Zambirinis CP, Fallon NC, Rehman A, Pylayeva-Gupta Y, Badar S, Hajdu CH, Frey AB, Bar-Sagi D, Miller G. MyD88 inhibition amplifies dendritic cell capacity to promote pancreatic carcinogenesis via Th2 cells. J Exp Med. 2012;209:1671-1687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 246] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 206. | Zhang H, Sun L. When human cells meet bacteria: precision medicine for cancers using the microbiota. Am J Cancer Res. 2018;8:1157-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 207. | Udayasuryan B, Ahmad RN, Nguyen TTD, Umaña A, Monét Roberts L, Sobol P, Jones SD, Munson JM, Slade DJ, Verbridge SS. Fusobacterium nucleatum induces proliferation and migration in pancreatic cancer cells through host autocrine and paracrine signaling. Sci Signal. 2022;15:eabn4948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 104] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 208. | Hayashi M, Ikenaga N, Nakata K, Luo H, Zhong P, Date S, Oyama K, Higashijima N, Kubo A, Iwamoto C, Torata N, Abe T, Yamada Y, Ohuchida K, Oda Y, Nakamura M. Intratumor Fusobacterium nucleatum promotes the progression of pancreatic cancer via the CXCL1-CXCR2 axis. Cancer Sci. 2023;114:3666-3678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 54] [Reference Citation Analysis (0)] |
| 209. | D'Antonio DL, Zenoniani A, Umme S, Piattelli A, Curia MC. Intratumoral Fusobacterium nucleatum in Pancreatic Cancer: Current and Future Perspectives. Pathogens. 2024;14:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 210. | Gaida MM, Mayer C, Dapunt U, Stegmaier S, Schirmacher P, Wabnitz GH, Hänsch GM. Expression of the bitter receptor T2R38 in pancreatic cancer: localization in lipid droplets and activation by a bacteria-derived quorum-sensing molecule. Oncotarget. 2016;7:12623-12632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 211. | Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, Werba G, Zhang K, Guo Y, Li Q, Akkad N, Lall S, Wadowski B, Gutierrez J, Kochen Rossi JA, Herzog JW, Diskin B, Torres-Hernandez A, Leinwand J, Wang W, Taunk PS, Savadkar S, Janal M, Saxena A, Li X, Cohen D, Sartor RB, Saxena D, Miller G. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018;8:403-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1016] [Cited by in RCA: 1073] [Article Influence: 134.1] [Reference Citation Analysis (0)] |
| 212. | Oláh A, Romics L Jr. Early enteral nutrition in acute pancreatitis--benefits and limitations. Langenbecks Arch Surg. 2008;393:261-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 213. | Zabaleta J. Multifactorial etiology of gastric cancer. Methods Mol Biol. 2012;863:411-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 214. | Ishaq S, Nunn L. Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol Hepatol Bed Bench. 2015;8:S6-S14. [PubMed] |
| 215. | Pucułek M, Machlowska J, Wierzbicki R, Baj J, Maciejewski R, Sitarz R. Helicobacter pylori associated factors in the development of gastric cancer with special reference to the early-onset subtype. Oncotarget. 2018;9:31146-31162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 216. | Zhang RG, Duan GC, Fan QT, Chen SY. Role of Helicobacter pylori infection in pathogenesis of gastric carcinoma. World J Gastrointest Pathophysiol. 2016;7:97-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 44] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (2)] |
| 217. | Correa P, Haenszel W, Cuello C, Tannenbaum S, Archer M. A model for gastric cancer epidemiology. Lancet. 1975;2:58-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 725] [Cited by in RCA: 746] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 218. | Watari J, Chen N, Amenta PS, Fukui H, Oshima T, Tomita T, Miwa H, Lim KJ, Das KM. Helicobacter pylori associated chronic gastritis, clinical syndromes, precancerous lesions, and pathogenesis of gastric cancer development. World J Gastroenterol. 2014;20:5461-5473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 202] [Cited by in RCA: 211] [Article Influence: 17.6] [Reference Citation Analysis (6)] |
| 219. | Moss SF. The Clinical Evidence Linking Helicobacter pylori to Gastric Cancer. Cell Mol Gastroenterol Hepatol. 2017;3:183-191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 223] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 220. | Song H, Ekheden IG, Zheng Z, Ericsson J, Nyrén O, Ye W. Incidence of gastric cancer among patients with gastric precancerous lesions: observational cohort study in a low risk Western population. BMJ. 2015;351:h3867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 219] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 221. | Bhattacharyya A, Chattopadhyay R, Mitra S, Crowe SE. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol Rev. 2014;94:329-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1089] [Cited by in RCA: 1667] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 222. | Tsai HF, Hsu PN. Interplay between Helicobacter pylori and immune cells in immune pathogenesis of gastric inflammation and mucosal pathology. Cell Mol Immunol. 2010;7:255-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 223. | Maeda M, Moro H, Ushijima T. Mechanisms for the induction of gastric cancer by Helicobacter pylori infection: aberrant DNA methylation pathway. Gastric Cancer. 2017;20:8-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 224. | Muhammad JS, Eladl MA, Khoder G. Helicobacter pylori-induced DNA Methylation as an Epigenetic Modulator of Gastric Cancer: Recent Outcomes and Future Direction. Pathogens. 2019;8:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 225. | Fu Q, Yu H, Liu M, Chen L, Chen W, Wang Z, Li W. Effect of Helicobacter pylori eradication on gastric cancer risk in patients with intestinal metaplasia or dysplasia: a meta-analysis of randomized controlled trials. Front Microbiol. 2025;16:1530549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 226. | Ford AC, Yuan Y, Park JY, Forman D, Moayyedi P. Eradication Therapy to Prevent Gastric Cancer in Helicobacterpylori-Positive Individuals: Systematic Review and Meta-Analysis of Randomized Controlled Trials and Observational Studies. Gastroenterology. 2025;169:261-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 33] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 227. | Kikuchi S, Crabtree JE, Forman D, Kurosawa M. Association between infections with CagA-positive or -negative strains of Helicobacter pylori and risk for gastric cancer in young adults. Research Group on Prevention of Gastric Carcinoma Among Young Adults. Am J Gastroenterol. 1999;94:3455-3459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 228. | Yong X, Tang B, Li BS, Xie R, Hu CJ, Luo G, Qin Y, Dong H, Yang SM. Helicobacter pylori virulence factor CagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun Signal. 2015;13:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 169] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 229. | Li N, Feng Y, Hu Y, He C, Xie C, Ouyang Y, Artim SC, Huang D, Zhu Y, Luo Z, Ge Z, Lu N. Helicobacter pylori CagA promotes epithelial mesenchymal transition in gastric carcinogenesis via triggering oncogenic YAP pathway. J Exp Clin Cancer Res. 2018;37:280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 230. | Wei J, Noto J, Zaika E, Romero-Gallo J, Correa P, El-Rifai W, Peek RM, Zaika A. Pathogenic bacterium Helicobacter pylori alters the expression profile of p53 protein isoforms and p53 response to cellular stresses. Proc Natl Acad Sci U S A. 2012;109:E2543-E2550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 231. | Shibata A, Parsonnet J, Longacre TA, Garcia MI, Puligandla B, Davis RE, Vogelman JH, Orentreich N, Habel LA. CagA status of Helicobacter pylori infection and p53 gene mutations in gastric adenocarcinoma. Carcinogenesis. 2002;23:419-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 232. | Torres VJ, VanCompernolle SE, Sundrud MS, Unutmaz D, Cover TL. Helicobacter pylori vacuolating cytotoxin inhibits activation-induced proliferation of human T and B lymphocyte subsets. J Immunol. 2007;179:5433-5440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 82] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 233. | Palframan SL, Kwok T, Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front Cell Infect Microbiol. 2012;2:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (2)] |
| 234. | Goldszmid RS, Dzutsev A, Viaud S, Zitvogel L, Restifo NP, Trinchieri G. Microbiota modulation of myeloid cells in cancer therapy. Cancer Immunol Res. 2015;3:103-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 235. | Correa P, Fontham ET, Bravo JC, Bravo LE, Ruiz B, Zarama G, Realpe JL, Malcom GT, Li D, Johnson WD, Mera R. Chemoprevention of gastric dysplasia: randomized trial of antioxidant supplements and anti-helicobacter pylori therapy. J Natl Cancer Inst. 2000;92:1881-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 490] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 236. | Ford AC, Forman D, Hunt RH, Yuan Y, Moayyedi P. Helicobacter pylori eradication therapy to prevent gastric cancer in healthy asymptomatic infected individuals: systematic review and meta-analysis of randomised controlled trials. BMJ. 2014;348:g3174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 456] [Cited by in RCA: 450] [Article Influence: 37.5] [Reference Citation Analysis (1)] |
| 237. | Ma JL, Zhang L, Brown LM, Li JY, Shen L, Pan KF, Liu WD, Hu Y, Han ZX, Crystal-Mansour S, Pee D, Blot WJ, Fraumeni JF Jr, You WC, Gail MH. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality. J Natl Cancer Inst. 2012;104:488-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 360] [Article Influence: 25.7] [Reference Citation Analysis (1)] |
| 238. | Vannella L, Lahner E, Bordi C, Pilozzi E, Di Giulio E, Corleto VD, Osborn J, Delle Fave G, Annibale B. Reversal of atrophic body gastritis after H. pylori eradication at long-term follow-up. Dig Liver Dis. 2011;43:295-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 239. | Chen HN, Wang Z, Li X, Zhou ZG. Helicobacter pylori eradication cannot reduce the risk of gastric cancer in patients with intestinal metaplasia and dysplasia: evidence from a meta-analysis. Gastric Cancer. 2016;19:166-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 172] [Article Influence: 17.2] [Reference Citation Analysis (1)] |
| 240. | Rokkas T, Rokka A, Portincasa P. A systematic review and meta-analysis of the role of Helicobacter pylori eradication in preventing gastric cancer. Ann Gastroenterol. 2017;30:414-423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 241. | Niwa T, Toyoda T, Tsukamoto T, Mori A, Tatematsu M, Ushijima T. Prevention of Helicobacter pylori-induced gastric cancers in gerbils by a DNA demethylating agent. Cancer Prev Res (Phila). 2013;6:263-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 242. | Araya M, Morelli L, Reid G, Sanders ME, Stanton C, Pineiro M, Ben Embarek P. Guidelines for the evaluation of probiotics in food. 2002. [cited 3 August 2021]. Available from: https://www.researchgate.net/publication/284392003_Guidelines_for_the_evaluation_of_probiotics_in_food. |
| 243. | Losurdo G, Cubisino R, Barone M, Principi M, Leandro G, Ierardi E, Di Leo A. Probiotic monotherapy and Helicobacter pylori eradication: A systematic review with pooled-data analysis. World J Gastroenterol. 2018;24:139-149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 82] [Cited by in RCA: 89] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 244. | Zhu R, Chen K, Zheng YY, Zhang HW, Wang JS, Xia YJ, Dai WQ, Wang F, Shen M, Cheng P, Zhang Y, Wang CF, Yang J, Li JJ, Lu J, Zhou YQ, Guo CY. Meta-analysis of the efficacy of probiotics in Helicobacter pylori eradication therapy. World J Gastroenterol. 2014;20:18013-18021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 75] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 245. | Marsh Rde W, Alonzo M, Bajaj S, Baker M, Elton E, Farrell TA, Gore RM, Hall C, Nowak J, Roy H, Shaikh A, Talamonti MS. Comprehensive review of the diagnosis and treatment of biliary tract cancer 2012. Part I: diagnosis-clinical staging and pathology. J Surg Oncol. 2012;106:332-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 246. | Rakić M, Patrlj L, Kopljar M, Kliček R, Kolovrat M, Loncar B, Busic Z. Gallbladder cancer. Hepatobiliary Surg Nutr. 2014;3:221-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 105] [Reference Citation Analysis (0)] |
| 247. | Andia ME, Hsing AW, Andreotti G, Ferreccio C. Geographic variation of gallbladder cancer mortality and risk factors in Chile: a population-based ecologic study. Int J Cancer. 2008;123:1411-1416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 74] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 248. | Hundal R, Shaffer EA. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol. 2014;6:99-109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 517] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 249. | Ruby T, McLaughlin L, Gopinath S, Monack D. Salmonella's long-term relationship with its host. FEMS Microbiol Rev. 2012;36:600-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 250. | Koshiol J, Wozniak A, Cook P, Adaniel C, Acevedo J, Azócar L, Hsing AW, Roa JC, Pasetti MF, Miquel JF, Levine MM, Ferreccio C; Gallbladder Cancer Chile Working Group. Salmonella enterica serovar Typhi and gallbladder cancer: a case-control study and meta-analysis. Cancer Med. 2016;5:3310-3235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 251. | Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014;22:648-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 218] [Article Influence: 18.2] [Reference Citation Analysis (1)] |
| 252. | Tsuchiya Y, Loza E, Villa-Gomez G, Trujillo CC, Baez S, Asai T, Ikoma T, Endoh K, Nakamura K. Metagenomics of Microbial Communities in Gallbladder Bile from Patients with Gallbladder Cancer or Cholelithiasis. Asian Pac J Cancer Prev. 2018;19:961-967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 253. | Kumar S, Kumar S, Kumar S. Infection as a risk factor for gallbladder cancer. J Surg Oncol. 2006;93:633-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 75] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 254. | Csendes A, Becerra M, Burdiles P, Demian I, Bancalari K, Csendes P. Bacteriological studies of bile from the gallbladder in patients with carcinoma of the gallbladder, cholelithiasis, common bile duct stones and no gallstones disease. Eur J Surg. 1994;160:363-367. [PubMed] |
| 255. | Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, Broeks A, Shukla VK, Kumar M, Janssen H, Song JY, Neefjes-Borst EA, te Riele H, Holden DW, Nath G, Neefjes J. Salmonella Manipulation of Host Signaling Pathways Provokes Cellular Transformation Associated with Gallbladder Carcinoma. Cell Host Microbe. 2015;17:763-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 192] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 256. | Kinoshita H, Nagata E, Hirohashi K, Sakai K, Kobayashi Y. Carcinoma of the gallbladder with an anomalous connection between the choledochus and the pancreatic duct. Report of 10 cases and review of the literature in Japan. Cancer. 1984;54:762-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 257. | Sharma V, Chauhan VS, Nath G, Kumar A, Shukla VK. Role of bile bacteria in gallbladder carcinoma. Hepatogastroenterology. 2007;54:1622-1625. [PubMed] |
| 258. | White DL, Kanwal F, Jiao L, El-serag HB. Epidemiology of Hepatocellular Carcinoma. In: Carr B, editor. Hepatocellular Carcinoma. Current Clinical Oncology. New York: Springer, 2016. |
| 259. | Wu CH, Chiu NC, Yeh YC, Kuo Y, Yu SS, Weng CY, Liu CA, Chou YH, Chiou YY. Uncommon liver tumors: Case report and literature review. Medicine (Baltimore). 2016;95:e4952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 260. | Blonski W, Kotlyar DS, Forde KA. Non-viral causes of hepatocellular carcinoma. World J Gastroenterol. 2010;16:3603-3615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 115] [Cited by in RCA: 127] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 261. | Balogh J, Victor D 3rd, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour HP Jr. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 755] [Cited by in RCA: 835] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 262. | Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl:S2-S6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 894] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 263. | Sanduzzi Zamparelli M, Rocco A, Compare D, Nardone G. The gut microbiota: A new potential driving force in liver cirrhosis and hepatocellular carcinoma. United European Gastroenterol J. 2017;5:944-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 264. | Gupta H, Youn GS, Shin MJ, Suk KT. Role of Gut Microbiota in Hepatocarcinogenesis. Microorganisms. 2019;7:121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 265. | Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513-1524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 615] [Cited by in RCA: 777] [Article Influence: 64.8] [Reference Citation Analysis (2)] |
| 266. | Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14:527-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 444] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 267. | Zhang HL, Yu LX, Yang W, Tang L, Lin Y, Wu H, Zhai B, Tan YX, Shan L, Liu Q, Chen HY, Dai RY, Qiu BJ, He YQ, Wang C, Zheng LY, Li YQ, Wu FQ, Li Z, Yan HX, Wang HY. Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarcinogenesis in rats. J Hepatol. 2012;57:803-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 223] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 268. | Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1318] [Cited by in RCA: 1722] [Article Influence: 132.5] [Reference Citation Analysis (1)] |
| 269. | Huang Y, Fan XG, Wang ZM, Zhou JH, Tian XF, Li N. Identification of helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J Clin Pathol. 2004;57:1273-1277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 122] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 270. | Dore MP, Realdi G, Mura D, Graham DY, Sepulveda AR. Helicobacter infection in patients with HCV-related chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Dig Dis Sci. 2002;47:1638-1643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 56] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 271. | Queiroz DM, Rocha AM, Rocha GA, Cinque SM, Oliveira AG, Godoy A, Tanno H. Association between Helicobacter pylori infection and cirrhosis in patients with chronic hepatitis C virus. Dig Dis Sci. 2006;51:370-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 272. | Liu X, Liang J, Li G. Lipopolysaccharide promotes adhesion and invasion of hepatoma cell lines HepG2 and HepG2.2.15. Mol Biol Rep. 2010;37:2235-2239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 273. | Lu H, Ren Z, Li A, Zhang H, Jiang J, Xu S, Luo Q, Zhou K, Sun X, Zheng S, Li L. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci Rep. 2016;6:33142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 274. | Grąt M, Wronka KM, Krasnodębski M, Masior Ł, Lewandowski Z, Kosińska I, Grąt K, Stypułkowski J, Rejowski S, Wasilewicz M, Gałęcka M, Szachta P, Krawczyk M. Profile of Gut Microbiota Associated With the Presence of Hepatocellular Cancer in Patients With Liver Cirrhosis. Transplant Proc. 2016;48:1687-1691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 152] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 275. | Ponziani FR, Bhoori S, Castelli C, Putignani L, Rivoltini L, Del Chierico F, Sanguinetti M, Morelli D, Paroni Sterbini F, Petito V, Reddel S, Calvani R, Camisaschi C, Picca A, Tuccitto A, Gasbarrini A, Pompili M, Mazzaferro V. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology. 2019;69:107-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 524] [Article Influence: 74.9] [Reference Citation Analysis (1)] |
| 276. | Wan MLY, El-Nezami H. Targeting gut microbiota in hepatocellular carcinoma: probiotics as a novel therapy. Hepatobiliary Surg Nutr. 2018;7:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 277. | Li J, Sung CY, Lee N, Ni Y, Pihlajamäki J, Panagiotou G, El-Nezami H. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Natl Acad Sci U S A. 2016;113:E1306-E1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 445] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 278. | El-Nezami HS, Polychronaki NN, Ma J, Zhu H, Ling W, Salminen EK, Juvonen RO, Salminen SJ, Poussa T, Mykkänen HM. Probiotic supplementation reduces a biomarker for increased risk of liver cancer in young men from Southern China. Am J Clin Nutr. 2006;83:1199-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 279. | Bajaj JS, Kassam Z, Fagan A, Gavis EA, Liu E, Cox IJ, Kheradman R, Heuman D, Wang J, Gurry T, Williams R, Sikaroodi M, Fuchs M, Alm E, John B, Thacker LR, Riva A, Smith M, Taylor-Robinson SD, Gillevet PM. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology. 2017;66:1727-1738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 481] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 280. | Ferrere G, Wrzosek L, Cailleux F, Turpin W, Puchois V, Spatz M, Ciocan D, Rainteau D, Humbert L, Hugot C, Gaudin F, Noordine ML, Robert V, Berrebi D, Thomas M, Naveau S, Perlemuter G, Cassard AM. Fecal microbiota manipulation prevents dysbiosis and alcohol-induced liver injury in mice. J Hepatol. 2017;66:806-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 259] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 281. | Philips CA, Pande A, Shasthry SM, Jamwal KD, Khillan V, Chandel SS, Kumar G, Sharma MK, Maiwall R, Jindal A, Choudhary A, Hussain MS, Sharma S, Sarin SK. Healthy Donor Fecal Microbiota Transplantation in Steroid-Ineligible Severe Alcoholic Hepatitis: A Pilot Study. Clin Gastroenterol Hepatol. 2017;15:600-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 280] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 282. | Ren YD, Ye ZS, Yang LZ, Jin LX, Wei WJ, Deng YY, Chen XX, Xiao CX, Yu XF, Xu HZ, Xu LZ, Tang YN, Zhou F, Wang XL, Chen MY, Chen LG, Hong MZ, Ren JL, Pan JS. Fecal microbiota transplantation induces hepatitis B virus e-antigen (HBeAg) clearance in patients with positive HBeAg after long-term antiviral therapy. Hepatology. 2017;65:1765-1768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 283. | Stein A, Voigt W, Jordan K. Chemotherapy-induced diarrhea: pathophysiology, frequency and guideline-based management. Ther Adv Med Oncol. 2010;2:51-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 317] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 284. | Lin XB, Dieleman LA, Ketabi A, Bibova I, Sawyer MB, Xue H, Field CJ, Baracos VE, Gänzle MG. Irinotecan (CPT-11) chemotherapy alters intestinal microbiota in tumour bearing rats. PLoS One. 2012;7:e39764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 285. | Russo F, Linsalata M, Clemente C, D'Attoma B, Orlando A, Campanella G, Giotta F, Riezzo G. The effects of fluorouracil, epirubicin, and cyclophosphamide (FEC60) on the intestinal barrier function and gut peptides in breast cancer patients: an observational study. BMC Cancer. 2013;13:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 286. | van Vliet MJ, Tissing WJ, Dun CA, Meessen NE, Kamps WA, de Bont ES, Harmsen HJ. Chemotherapy treatment in pediatric patients with acute myeloid leukemia receiving antimicrobial prophylaxis leads to a relative increase of colonization with potentially pathogenic bacteria in the gut. Clin Infect Dis. 2009;49:262-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 206] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 287. | Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, de Vos WM, Satokari R. Akkermansia muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl Environ Microbiol. 2015;81:3655-3662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 431] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 288. | Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, Jiang W, Cai S, Zhao P, Song R, Li P, Qin N, Fang W. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer. 2019;7:193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 366] [Cited by in RCA: 422] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 289. | Wortelboer K, Nieuwdorp M, Herrema H. Fecal microbiota transplantation beyond Clostridioides difficile infections. EBioMedicine. 2019;44:716-729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 118] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 290. | Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, Shiota A, Takeshita K, Yasuma-Mitobe K, Riethmacher D, Kaisho T, Norman JM, Mucida D, Suematsu M, Yaguchi T, Bucci V, Inoue T, Kawakami Y, Olle B, Roberts B, Hattori M, Xavier RJ, Atarashi K, Honda K. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 864] [Article Influence: 123.4] [Reference Citation Analysis (1)] |
| 291. | Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, Koh AY. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia. 2017;19:848-855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 307] [Cited by in RCA: 530] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 292. | Liu L, Shah K. The Potential of the Gut Microbiome to Reshape the Cancer Therapy Paradigm: A Review. JAMA Oncol. 2022;8:1059-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 293. | Gamrath L, Pedersen TB, Møller MV, Volmer LM, Holst-Christensen L, Vestermark LW, Donskov F. Role of the Microbiome and Diet for Response to Cancer Checkpoint Immunotherapy: A Narrative Review of Clinical Trials. Curr Oncol Rep. 2025;27:45-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 294. | Erawijantari PP, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Saito Y, Fukuda S, Yachida S, Yamada T. Influence of gastrectomy for gastric cancer treatment on faecal microbiome and metabolome profiles. Gut. 2020;69:1404-1415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 127] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 295. | Liang W, Yang Y, Wang H, Wang H, Yu X, Lu Y, Shen S, Teng L. Gut microbiota shifts in patients with gastric cancer in perioperative period. Medicine (Baltimore). 2019;98:e16626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 296. | Hu Y, Zhou P, Deng K, Zhou Y, Hu K. Targeting the gut microbiota: a new strategy for colorectal cancer treatment. J Transl Med. 2024;22:915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 297. | Lapthorne S, Bines JE, Fouhy F, Dellios NL, Wilson G, Thomas SL, Scurr M, Stanton C, Cotter PD, Pereira-Fantini PM. Changes in the colon microbiota and intestinal cytokine gene expression following minimal intestinal surgery. World J Gastroenterol. 2015;21:4150-4158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 298. | Kong C, Gao R, Yan X, Huang L, He J, Li H, You J, Qin H. Alterations in intestinal microbiota of colorectal cancer patients receiving radical surgery combined with adjuvant CapeOx therapy. Sci China Life Sci. 2019;62:1178-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 299. | Ohigashi S, Sudo K, Kobayashi D, Takahashi T, Nomoto K, Onodera H. Significant changes in the intestinal environment after surgery in patients with colorectal cancer. J Gastrointest Surg. 2013;17:1657-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 99] [Article Influence: 7.6] [Reference Citation Analysis (3)] |
| 300. | Liu CJ, Zhang YL, Shang Y, Wu B, Yang E, Luo YY, Li XR. Intestinal bacteria detected in cancer and adjacent tissue from patients with colorectal cancer. Oncol Lett. 2019;17:1115-1127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 301. | Cong J, Zhu H, Liu D, Li T, Zhang C, Zhu J, Lv H, Liu K, Hao C, Tian Z, Zhang J, Zhang X. A Pilot Study: Changes of Gut Microbiota in Post-surgery Colorectal Cancer Patients. Front Microbiol. 2018;9:2777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 302. | Deng Y, Ding X, Song Q, Zhao G, Han L, Ding B, Wang X, Hao X, Li H. Alterations in mucosa-associated microbiota in the stomach of patients with gastric cancer. Cell Oncol (Dordr). 2021;44:701-714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 303. | Francino MP. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front Microbiol. 2015;6:1543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 340] [Cited by in RCA: 544] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 304. | Wang WY, Chen CW, Wang TJ, Lin KL, Liu CY. Outcomes of early enteral feeding in patients after curative colorectal cancer surgery: A retrospective comparative study. Eur J Oncol Nurs. 2021;54:101970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 305. | Dag A, Colak T, Turkmenoglu O, Gundogdu R, Aydin S. A randomized controlled trial evaluating early versus traditional oral feeding after colorectal surgery. Clinics (Sao Paulo). 2011;66:2001-2005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 306. | Persson JE, Viana P, Persson M, Relvas JH, Danielski LG. Perioperative or Postoperative Probiotics Reduce Treatment-Related Complications in Adult Colorectal Cancer Patients Undergoing Surgery: A Systematic Review and Meta-analysis. J Gastrointest Cancer. 2024;55:740-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 307. | Araújo MM, Montalvão-Sousa TM, Teixeira PDC, Figueiredo ACMG, Botelho PB. The effect of probiotics on postsurgical complications in patients with colorectal cancer: a systematic review and meta-analysis. Nutr Rev. 2023;81:493-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 308. | Zhang S, Wang Q, Zhou C, Chen K, Chang H, Xiao W, Gao Y. Colorectal cancer, radiotherapy and gut microbiota. Chin J Cancer Res. 2019;31:212-222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 309. | Kim YS, Kim J, Park SJ. High-throughput 16S rRNA gene sequencing reveals alterations of mouse intestinal microbiota after radiotherapy. Anaerobe. 2015;33:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 310. | Mitra A, Grossman Biegert GW, Delgado AY, Karpinets TV, Solley TN, Mezzari MP, Yoshida-Court K, Petrosino JF, Mikkelson MD, Lin L, Eifel P, Zhang J, Ramondetta LM, Jhingran A, Sims TT, Schmeler K, Okhuysen P, Colbert LE, Klopp AH. Microbial Diversity and Composition Is Associated with Patient-Reported Toxicity during Chemoradiation Therapy for Cervical Cancer. Int J Radiat Oncol Biol Phys. 2020;107:163-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 311. | Nam YD, Kim HJ, Seo JG, Kang SW, Bae JW. Impact of pelvic radiotherapy on gut microbiota of gynecological cancer patients revealed by massive pyrosequencing. PLoS One. 2013;8:e82659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 183] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 312. | Gerassy-Vainberg S, Blatt A, Danin-Poleg Y, Gershovich K, Sabo E, Nevelsky A, Daniel S, Dahan A, Ziv O, Dheer R, Abreu MT, Koren O, Kashi Y, Chowers Y. Radiation induces proinflammatory dysbiosis: transmission of inflammatory susceptibility by host cytokine induction. Gut. 2018;67:97-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 229] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 313. | Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1643] [Cited by in RCA: 1731] [Article Influence: 192.3] [Reference Citation Analysis (0)] |
| 314. | Then CK, Paillas S, Moomin A, Misheva MD, Moir RA, Hay SM, Bremner D, Roberts Nee Nellany KS, Smith EE, Heidari Z, Sescu D, Wang X, Suárez-Bonnet A, Hay N, Murdoch SL, Saito R, Collie-Duguid ESR, Richardson S, Priestnall SL, Wilson JM, Gurumurthy M, Royle JS, Samuel LM, Ramsay G, Vallis KA, Foster KR, McCullagh JSO, Kiltie AE. Dietary fibre supplementation enhances radiotherapy tumour control and alleviates intestinal radiation toxicity. Microbiome. 2024;12:89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 315. | Qu CY, Zheng Y, Zhou M, Zhang Y, Shen F, Cao J, Xu LM. Value of bevacizumab in treatment of colorectal cancer: A meta-analysis. World J Gastroenterol. 2015;21:5072-5080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 51] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 316. | Prager GW, Taieb J, Fakih M, Ciardiello F, Van Cutsem E, Elez E, Cruz FM, Wyrwicz L, Stroyakovskiy D, Pápai Z, Poureau PG, Liposits G, Cremolini C, Bondarenko I, Modest DP, Benhadji KA, Amellal N, Leger C, Vidot L, Tabernero J; SUNLIGHT Investigators. Trifluridine-Tipiracil and Bevacizumab in Refractory Metastatic Colorectal Cancer. N Engl J Med. 2023;388:1657-1667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 263] [Article Influence: 87.7] [Reference Citation Analysis (0)] |
| 317. | Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, Kudo M, Breder V, Merle P, Kaseb AO, Li D, Verret W, Xu DZ, Hernandez S, Liu J, Huang C, Mulla S, Wang Y, Lim HY, Zhu AX, Cheng AL; IMbrave150 Investigators. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med. 2020;382:1894-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2542] [Cited by in RCA: 5306] [Article Influence: 884.3] [Reference Citation Analysis (29)] |
| 318. | Chen YC, Chuang CH, Miao ZF, Yip KL, Liu CJ, Li LH, Wu DC, Cheng TL, Lin CY, Wang JY. Gut microbiota composition in chemotherapy and targeted therapy of patients with metastatic colorectal cancer. Front Oncol. 2022;12:955313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 319. | Martini G, Ciardiello D, Dallio M, Famiglietti V, Esposito L, Corte CMD, Napolitano S, Fasano M, Gravina AG, Romano M, Loguercio C, Federico A, Maiello E, Tuccillo C, Morgillo F, Troiani T, Di Maio M, Martinelli E, Ciardiello F. Gut microbiota correlates with antitumor activity in patients with mCRC and NSCLC treated with cetuximab plus avelumab. Int J Cancer. 2022;151:473-480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 320. | Liu W, Peng L, Chen L, Wan J, Lou S, Yang T, Shen Z. Skin microbial dysbiosis is a characteristic of systemic drug-related intertriginous and flexural exanthema-like lesions induced by EGFR inhibitor. Heliyon. 2023;9:e21690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 321. | Liu Y, Baba Y, Ishimoto T, Gu X, Zhang J, Nomoto D, Okadome K, Baba H, Qiu P. Gut microbiome in gastrointestinal cancer: a friend or foe? Int J Biol Sci. 2022;18:4101-4117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 322. | Ghidini M, Petrelli F, Tomasello G. Right Versus Left Colon Cancer: Resectable and Metastatic Disease. Curr Treat Options Oncol. 2018;19:31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 323. | Wieërs G, Belkhir L, Enaud R, Leclercq S, Philippart de Foy JM, Dequenne I, de Timary P, Cani PD. How Probiotics Affect the Microbiota. Front Cell Infect Microbiol. 2019;9:454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 343] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 324. | Roeland EJ, Bohlke K, Baracos VE, Bruera E, Del Fabbro E, Dixon S, Fallon M, Herrstedt J, Lau H, Platek M, Rugo HS, Schnipper HH, Smith TJ, Tan W, Loprinzi CL. Management of Cancer Cachexia: ASCO Guideline. J Clin Oncol. 2020;38:2438-2453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 154] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 325. | Sánchez-Sánchez E, Orúe I, Guerra JA, Estornell MA, Barragán B, Blanco M, Comellas M, Cancer E. Nutritional management of cancer patients in clinical practice in Spain: patients' and multidisciplinary health care professionals' perceptions. Eur J Clin Nutr. 2023;77:454-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 326. | Lidoriki I, Frountzas M, Karanikki E, Katsarlinou E, Tsikrikou I, Toutouzas KG, Schizas D. Adherence to Oral Nutrition Supplementation in Gastrointestinal Cancer Patients: A Systematic Review of the Literature. Nutr Cancer. 2024;76:31-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 327. | Bozzetti F, Cozzaglio L, Gavazzi C, Bidoli P, Bonfanti G, Montalto F, Soto Parra H, Valente M, Zucali R. Nutritional support in patients with cancer of the esophagus: impact on nutritional status, patient compliance to therapy, and survival. Tumori. 1998;84:681-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 328. | Vagnildhaug OM, Balstad TR, Ottestad I, Bye A, Greil C, Arends J, Baracos V, Brown LR, Dajani OF, Dolan RD, Fallon M, Fraser E, Grzyb A, Hjermstad MJ, Jakobsen G, Kaasa S, McDonald J, Philips I, Sayers J, Simpson MR, Sousa MS, Skipworth RJE, Laird BJA, Solheim TS; Cancer Cachexia Endpoints Working Group. Appetite and dietary intake endpoints in cancer cachexia clinical trials: Systematic Review 2 of the cachexia endpoints series. J Cachexia Sarcopenia Muscle. 2024;15:513-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 329. | Yu J, Yuan A, Liu Q, Wang W, Sun Y, Li Z, Meng C, Zhou Y, Cao S. Effect of preoperative immunonutrition on postoperative short-term clinical outcomes in patients with gastric cancer cachexia: a prospective randomized controlled trial. World J Surg Oncol. 2024;22:101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 330. | Bianchini C, Bonomo P, Bossi P, Caccialanza R, Fabi A. Bridging gaps in cancer cachexia Care: Current insights and future perspectives. Cancer Treat Rev. 2024;125:102717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 331. | Garutti M, Noto C, Pastò B, Cucciniello L, Alajmo M, Casirati A, Pedrazzoli P, Caccialanza R, Puglisi F. Nutritional Management of Oncological Symptoms: A Comprehensive Review. Nutrients. 2023;15:5068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 332. | Zhou J, Sun S, Luan S, Xiao X, Yang Y, Mao C, Chen L, Zeng X, Zhang Y, Yuan Y. Gut Microbiota for Esophageal Cancer: Role in Carcinogenesis and Clinical Implications. Front Oncol. 2021;11:717242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 333. | Elsalem L, Jum'ah AA, Alfaqih MA, Aloudat O. The Bacterial Microbiota of Gastrointestinal Cancers: Role in Cancer Pathogenesis and Therapeutic Perspectives. Clin Exp Gastroenterol. 2020;13:151-185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (2)] |
| 334. | Loke YL, Chew MT, Ngeow YF, Lim WWD, Peh SC. Colon Carcinogenesis: The Interplay Between Diet and Gut Microbiota. Front Cell Infect Microbiol. 2020;10:603086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 335. | Kok CR, Rose D, Hutkins R. Predicting Personalized Responses to Dietary Fiber Interventions: Opportunities for Modulation of the Gut Microbiome to Improve Health. Annu Rev Food Sci Technol. 2023;14:157-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/