SIX HOMEOBOX FAMILY MEMBERS
The first member of the SIX family, sine oculis (SO), was initially discovered in fruit flies, with its name originating from Latin, meaning “without eyes” as mutations in the SO gene lead to eye development defects in fruit flies[6]. Subsequent studies in fruit flies identified the presence of two more SIX genes, namely OPTIX and DSIX4. Similar to SO, Optix is also expressed in developing eyes, but its role in the retina differs from that of SO[7]. Dsix4, on the other hand, is not involved in eye development but plays a role in muscle and gonad development[8]. Gene duplications during evolution have expanded the SIX family, which in humans comprises six members organized into three subfamilies, that is SIX1/SIX2 (SO), SIX3/SIX6 (Optix), and SIX4/SIX5 (Dsix4)[2].
Structurally, all of these proteins possess two functional domains, the SIX domain (SD) and the homeobox nucleic acid recognition domain (HD)[2]. The SD is an evolutionarily conserved domain located immediately after the 5’ end and adjacent to the HD, playing a role in mediating protein-protein interactions. For instance, the eyes absent (Eya) protein, a coactivator for the SIX family, predominantly interacts with SD, facilitating gene activation[9]. In HD, the substitution of conserved amino acids leads to structural variations that influence DNA binding. In addition to the conserved SD and HD domains, SIX family proteins also feature variable-length regions on both sides, defined as the C-terminal and N-terminal domains[10].
SIX1
The SIX1 gene is located on human chromosome 14q23.1, spanning a length of 6057 bp and comprising five transcripts that encode two protein isoforms. The SIX1 protein consists of 284 amino acids, with serine or threonine residues replacing amino acids at positions 5 and 12 within the HD[11]. Despite lacking an activation domain, SIX1 collaborates with auxiliary factors to regulate gene expression. During transcriptional activation, SIX1 interacts with Eya, facilitating their nuclear translocation and participation in regulation [12].
SIX1, the earliest identified member of the SIX family, has been extensively studied[6]. During early development, SIX1 is closely associated with progenitor cell proliferation and intercellular communication, contributing significantly to tissue and organ formation. For instance, during the development of skeletal muscle, an analysis of gene expression in early mouse embryonic stages revealed that SIX1 is expressed in mesoderm-derived tissues, including skeletal muscles and dorsal root ganglia[13]. It was subsequently discovered that the expression of SIX1 and PAX3 precedes that of myogenic regulatory factors, which are critical determinants of the myogenic fate in muscle progenitor cells[14].
Additionally, studies have shown that SIX1 enhances and sustains MyoD expression in adult muscle satellite cells, promoting the regeneration of muscle[15]. Furthermore, SIX1 collaborates with EYA3 to bind to the MEF3 promoter site, thereby facilitating the transcription of myogenin[16]. In the development of sensory organs, SIX1 orchestrates the balance among the neural crest, epidermis, and pre-placodal ectoderm through transcriptional activation and repression mechanisms, playing a critical role in regulating cranial placode development[17]. During the early kidney development process, overlapping expression of PAX2, EYA1, SIX1, and SIX2 in the renal metanephric mesenchyme have been noted. The absence of SIX1 leads to a failure of ureteric bud invasion into the mesenchyme, followed by mesenchymal apoptosis. Additionally, research has found that PAX2 expression depends on EYA1 and SIX1, while SIX2 expression in the renal metanephric mesenchyme is also dependent on SIX1[18]. During embryonic gonadal development, the collaborative function of SIX1 and SIX4 is essential for male gonadal differentiation, affecting SRY activation and downstream targets, such as FOG2 and NR5A1, which are critical for sex determination and gonadal size regulation[19].
Research has shown that defects in the SIX1 gene are implicated in the etiology of branchiootic syndrome 3 and autosomal dominant deafness 23[20]. Branchiootic syndrome 3 is a genetic disorder characterized by developmental defects in the auditory system. SIX1 plays a pivotal role in the regulation of organogenesis by interacting with the EYA and PAX gene families[21]. On the other hand, dominant deafness 23 is an autosomal dominant genetic condition characterized by symmetric hearing loss. Mutations in SIX1 impair its normal function in the development of the auditory system, particularly in the structures of the inner ear[20].
SIX2
The SIX2 gene is located on human chromosome 2p21, spanning a total length of 4271 bp. Initially identified by Boucher et al[22], the gene was discovered through screening of the human genome library using human SIX1 cDNA. The predicted SIX2 protein comprises 291 amino acids and consists of two exons.
In craniofacial development, SIX2 is robustly expressed in the neural crest-derived frontonasal mesenchyme, and its absence is associated with frontonasal developmental defects[23]. In gastric development, the expression of SIX2 is linked to the formation of the pyloric sphincter[24]. In the maturation process of human pancreatic β cells, SIX2 is essential for the expression of multiple hallmark genes, and its absence significantly impairs the control of insulin processing and secretion in β cells[25].
During kidney development, SIX2 is expressed in the early stages within the uninduced metanephric mesenchyme of the nephrogenic cortex, where it has the potential to regulate ureteric bud outgrowth and kidney differentiation[26]. Furthermore, SIX2 is crucial for maintaining the undifferentiated state of renal pelvic mesenchymal progenitor cells, opposing the inductive signals from the ureteric bud to prevent premature and ectopic epithelial differentiation and preserve the progenitor pool[27]. It also inhibits nephron formation by directly or indirectly suppressing the expression of WNT4 and SFRP2 within the renal mesenchyme[27].
SIX3
The SIX3 gene is located on human chromosome 2p21, spanning a length of 4370 bp and comprising 2 exons. A single transcript encodes a protein of 332 amino acids in length[28]. Research on SIX3 binding sites has identified the DNA sequence ATTA as the motif that binds to SIX3[29].
During the process of eye development, SIX3 is primarily expressed in the lens and neuroretina, where it facilitates the formation of ectopic vesicle-like structures in mouse embryos[30]. It is indicated that conditional loss of SIX3 does not affect the initial development of the optic vesicle but impedes subsequent neuroretinal specification, as SIX3 inhibits the expression of WNT8B to promote neuroretinal formation[31]. Recent research highlighted the collaborative role of SIX3 and SIX6 in maintaining multipotent neuroretinal progenitors in the retina, regulating the expression of crucial markers and suppressing aberrant activation of Wnt/β-catenin signaling, thereby ensuring normal retinal development and differentiation[32].
During the maturation process of human pancreatic β cells, SIX3 prevents the inappropriate expression of genes typically active in fetal β cells, adult α cells, and other non-β cell types[25]. Additionally, SIX3 suppresses fetal gene expression programs and alternative islet cell fates, thereby enhancing insulin secretion and the proper regulation of β cell gene expression, ultimately strengthening the function of mature β cells[25].
Haploinsufficiency of SIX3 can lead to holoprosencephaly 2, a congenital brain malformation characterized by the failure of the forebrain to properly divide into the right and left hemispheres during early embryonic development[33]. Research indicated that SIX3 influences the expression of key developmental genes such as SHH and FOXG1, with variations in SIX3 dosage potentially resulting in different forms of holoprosencephaly[34].
SIX4
The SIX4 gene is located on human chromosome 14q23.1 and spans a total length of 14813 base pairs. It comprises five exons and three transcripts, encoding two protein variants. The SIX4 protein consists of 781 amino acids[35].
It was found that SIX4 mainly collaborates with other members of the SIX family during development. For instance, SIX4 and SIX5 have been identified to act together during mice abdominal wall development, crucial for abdominal wall growth and morphological changes, with their deficiency resulting in umbilical herniation[36]. SIX4, along with SIX1, has been implicated in mice myogenesis[37], kidney development[38], and gonadal development[19]. During embryonic development, SIX4 functionally serves as an auxiliary factor, collaborating with other family members, and playing a crucial supportive role.
SIX5
Originally known as myotonic dystrophy-associated homeodomain protein, SIX5 is located downstream of the dystrophia myotonica protein kinase gene on human chromosome 19q13.32, spanning three exons that encode three protein isoforms, with a total length of 4468 bp. The SIX5 protein consists of 739 amino acids and contains an intrinsic C-terminal activation domain[39].
The SIX5 gene is associated with myotonic dystrophy 1 (DM1) and branchio-oto-renal (BOR) syndrome. DM1 is a multisystem disorder characterized by myotonia, muscle wasting, testicular atrophy, and cataracts. The underlying genetic cause involves the repeat expansion downstream of the dystrophia myotonica protein kinase gene on chromosome 19, which impedes the expression of neighboring genes, including SIX5, leading to reduced expression in DM1[40]. For instance, knockout mice lacking SIX5, both heterozygous and homozygous, develop cataracts, implicating the involvement of SIX5 deficiency in the cataract phenotype of DM1[41]. Furthermore, loss of SIX5 leads to reproductive defects in mice, including testicular atrophy, infertility, and hormonal alterations, suggesting the significance of SIX5 in spermatogenesis and interstitial cell regulation[42]. Moreover, the muscle contractility, electromyographic insertional activity, and histology of SIX5-deficient mice are normal, contrasting with the muscle stiffness and wasting observed in human DM1[43].
Another disorder associated with SIX5 mutations is BOR. Similar to SIX1, mutations in SIX5 hinder the interaction between EYA1 and SIX5, leading to reduced transcriptional activity of the EYA1-SIX5 complex[44]. However, some studies have questioned the pathogenic impact of SIX5 mutations in BOR syndrome, as SIX5 mutations were not detected in their samples[45,46]. Further research is needed to explore the relationship between SIX5 mutations and BOR syndrome.
SIX6
The SIX6 gene is located on human chromosome 14q23.1, spanning a length of 3705 base pairs, and consists of two exons and one transcript encoding a protein of 246 amino acids[47].
During eye development, SIX6, similar to its subfamily member SIX3, is expressed in the optic vesicles and optic nerve[48]. Subsequent research has found that both SIX6 and SIX3 promote retinogenic factors to maintain the progenitor population of the neuroretina[49]. Furthermore, point mutations and allelic deletions in SIX6 are associated with various ocular malformations, including anophthalmia[47], microphthalmia[50], primary open-angle glaucoma[51,52], optic disc anomalies, and macular atrophy[53].
THE ROLE OF SIX FAMILY GENES IN GICS
Gastric cancer
Gastric cancer (GC) ranks as the fifth most prevalent cancer globally and stands as the third-leading cause of cancer-related mortality[4]. Its metastatic pathways, encompassing direct infiltration, hematogenous spread, transluminal dissemination, and lymphatic dissemination, significantly contribute to the unfavorable prognosis seen in advanced cases. Despite notable advancements in GC diagnosis and treatment, its incidence and mortality rates persist without effective control, with metastasis and chemotherapeutic resistance posing substantial challenges[54].
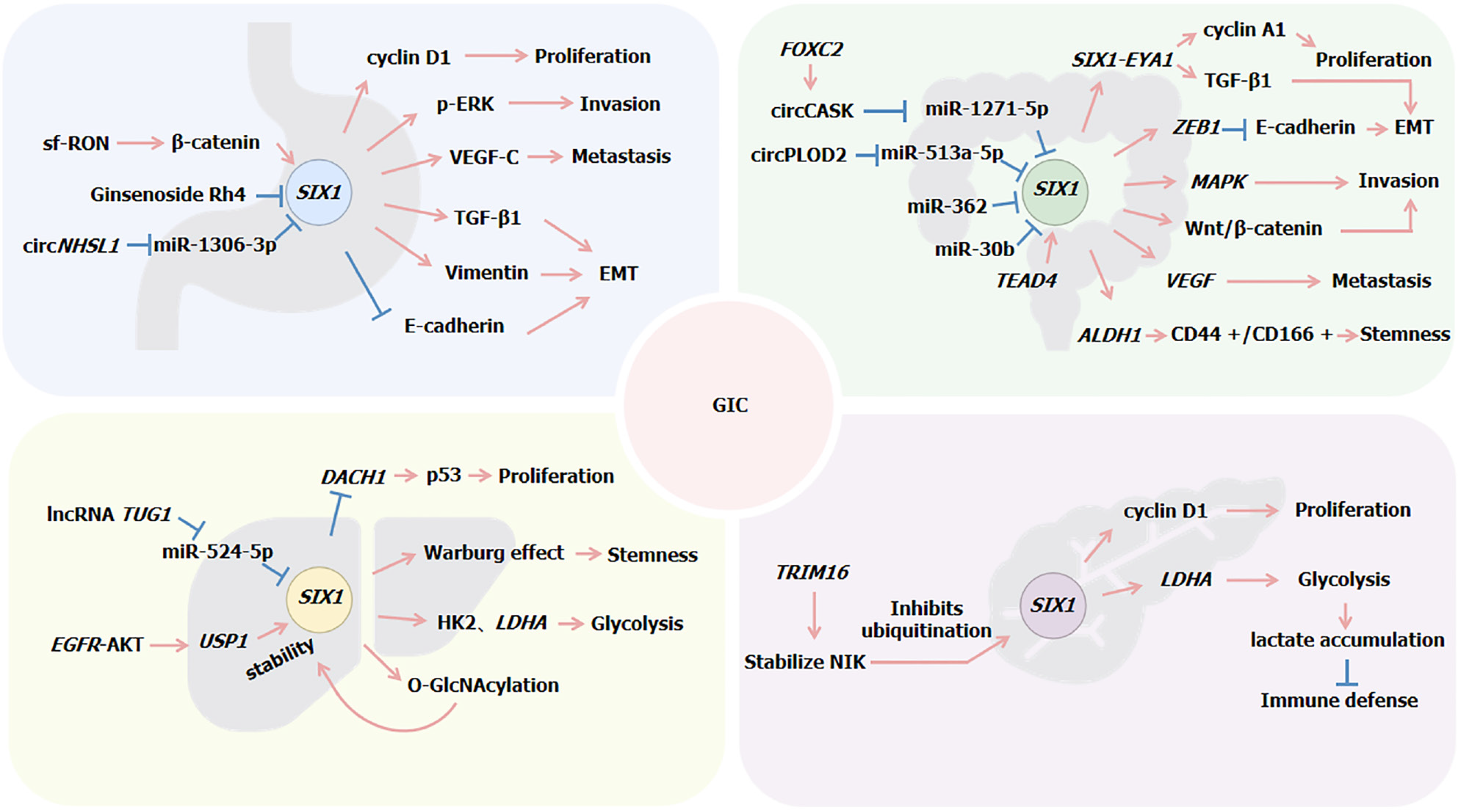
Lv et al[55] analyzed the correlation between SIX1 expression and clinicopathological parameters in GC, finding that overexpression of SIX1 is significantly associated with larger tumor size, serosal invasion, and lymph node metastasis as well as closely related to local recurrence and distant metastasis. High levels of SIX1 expression were found to decrease both overall and disease-free survival rates in patients with GC[55]. Furthermore, the induction of vascular endothelial growth factor-C (VEGF-C) by SIX1 is crucial for its ability to promote lymphatic and distant metastases. A positive correlation between SIX1 and VEGF-C has been established, with SIX1 acting as an upstream regulator that activates the expression of VEGF-C[56]. Du et al[57] observed elevated levels of SIX1 in GC tissues, where silencing of SIX1 expression could promote mitochondrial apoptosis by inhibiting the anti-apoptotic protein B-cell lymphoma-2 and activating caspase-7.
Further investigations revealed that SIX1 overexpression increased the expression of cyclin D1, MMP2, extracellular regulated protein kinases, and proteins associated with epithelial-mesenchymal transition (EMT). It promoted GC cell proliferation by targeting cyclin D1 and facilitated the EMT process as well as cell invasion by modulating MMP2 and E-cadherin[58]. Additionally, circNHSL1 acted as a “sponge” for miR-1306-3p to alleviate its inhibitory effect on the expression of its target SIX1, thereby promoting GC progression. Among them, SIX1 enhanced vimentin expression by directly binding to its promoter region, thus influencing the malignant progression of GC[59].
Wang et al[60] confirmed that short-form RON (sf-RON) activation of the β-catenin/SIX1 signaling pathway enhanced glucose metabolism in GC cells, leading to cell proliferation. Upregulation of SIX1 was observed upon overexpression of short-form RON or RON. Silencing expression of β-catenin decreased SIX1 levels as well as the expression levels of glycolytic proteins (glucose transporter type 1 and LDHA), while introduction of SIX1 cDNA rescued this process. Furthermore, SIX1 can induce the expression of transforming growth factor (TGF)-β1, which leads to the phosphorylation and activation of the receptors for TGF-β1. This activation initiates SMAD2 and SMAD3, which then bind with SMAD4 to form a Smad complex. This complex subsequently translocates to the nucleus to regulate the expression of specific genes. Within the nucleus, the Smad complex functions as a transcription factor, regulating the expression of various genes associated with EMT, such as E-cadherin, N-cadherin, vimentin, and snail. SIX1 promotes the EMT process by upregulating these genes through the TGF-β/Smad2/3 signaling pathway[61]. These findings underscore the critical role of SIX1 in GC cell migration and invasion (Figure 1).
Figure 1 Regulation mechanism of SIX1 expression in gastrointestinal cancers.
EMT: Epithelial-mesenchymal transition; GIC: Gastrointestinal cancer; TGF: Transforming growth factor; VEGF: Vascular endothelial growth factor.
Rajkumar et al[62] discovered that SIX3 is differentially expressed between patients with GC and matched normal controls. Recent research has revealed that SIX3 is involved in GC progression. The long noncoding RNA DLGAP1-AS2 interacts directly with SIX3, influencing the inhibitory effect of SIX3 on the expression of the WNT1 gene. Specifically, upon binding with SIX3, DLGAP1-AS2 impedes the binding of SIX3 to the promoter region of the WNT1 gene, thereby relieving the inhibition of WNT1 transcription and activating WNT1 transcription. Furthermore, the study found that Wnt/β-catenin signaling mediated by DLGAP1-AS2 depends on WNT1. This process leads to the malignant transformation of GC cells, including enhanced proliferation, migration, and invasion capabilities[63].
Elevated expression levels of SIX4 in GC tissues are positively correlated with the malignant behavior of GC cells and associated with poor prognosis in patients[64]. Additionally, miR-384 has been found to directly target the 3’ untranslated regions (3’UTR) of the SIX4 gene and inhibit its expression. Circ-0000670 acts as a “sponge” by binding to miR-384, releasing the inhibition on SIX4, which leads to increased expression of SIX4. This higher expression of SIX4 may promote the malignant behavior of GC cells[64]. These findings suggest that the SIX family and its related signaling pathways could be potential therapeutic targets for GC.
Colorectal cancer
Colorectal cancer (CRC) ranks as the third most prevalent cancer globally, constituting about 10% of all cancer cases and standing as the second-leading cause of cancer-related deaths worldwide[4]. CRC exhibits a diverse array of genetic and epigenetic characteristics, with its onset influenced by various internal and external factors, including mutation accumulation, susceptibility loci linked to family history, abnormal expression of non-coding RNA, and chronic or persistent inflammation[65]. Despite advancements in medical technology that have introduced new treatment methods, such as endoscopic and surgical local resection, preoperative radiotherapy and systemic therapy, extensive surgery for localized and metastatic disease, metastatic local ablation therapy, as well as palliative chemotherapy, targeted therapy, and immunotherapy, the cure rate is still limited by recurrence and chemotherapy resistance. Therefore, the treatment of CRC still faces several challenges, including a high recurrence rate, poor prognosis, impact on patients’ postoperative quality of life, and low 5-year survival rates for patients with metastasis patients[66].
Ono et al[67] discovered through gene expression analysis that SIX1 is expressed in CRC cells and is associated with EMT. SIX1 enhances the expression of ZEB1, a known inhibitor of E-cadherin, by activating it at the post-transcriptional level. ZEB1 can bind directly to the promoter region of the E-cadherin gene, inhibiting its transcriptional activity and leading to reduced E-cadherin expression. SIX1 also suppresses the transcriptional activity of the miR-200 family members, which negatively regulates the expression of ZEB1 and/or other EMT-related transcription factors by targeting their 3’UTR regions. The downregulation of the miR-200 family diminishes inhibition of ZEB1, allowing increased levels of ZEB1 that in turn inhibit E-cadherin expression[67]. Inhibition of E-cadherin is a key hallmark of EMT, marking the loss of epithelial characteristics and acquisition of mesenchymal traits.
Additionally, studies have confirmed that miR-30b can directly target the 3’UTR region of the SIX1 gene, causing degradation of SIX1 mRNA or preventing its translation into protein, thereby reducing SIX1 expression levels, indirectly increasing E-cadherin expression, and inhibiting EMT. Thus, miR-30b controls CRC cell migration and invasion by negatively regulating SIX1 expression[68]. Similarly, SIX1 is also a direct target of miR-362, with the expression of miR-362 being inversely correlated with that of SIX1 in CRC[69].
Kahlert et al[70] analyzed the expression pattern of SIX1 across normal mucosa, adenomas, and primary adenocarcinomas, establishing a correlation between progressive epithelial dedifferentiation and increasing SIX1 expression. Overexpression of SIX1 in HCT116 cells induced a mesenchymal-like phenotype and enhanced cell migration capabilities. Furthermore, both univariate and multivariate analyses confirmed that high SIX1 expression is associated with reduced overall survival.
SIX1 has been confirmed to promote the development and metastasis of CRC through multiple mechanisms. SIX1 is a direct target of TEAD4 in CRC, with TEAD4 binding to two MCAT motifs within the SIX1 promoter to enhance its transcription. TEAD4 facilitates EMT and CRC cell migration. The absence of SIX1 diminishes this effect, whereas its overexpression amplifies it, indicating that SIX1 mediates the role of TEAD4 in promoting CRC cell migration[71]. Moreover, in MC38 cells, overexpression of SIX1 increases the levels of the ALDH1 protein and expands the cluster of differentiation (CD) 44 +/CD166 + cell population, characteristics indicative of an increase in cancer stem cell traits. SIX1 stimulates angiogenesis by upregulating the expression of VEGF. Tumor cells with SIX1 overexpression recruit tumor-associated macrophages by increasing the expression of macrophage-specific colony-stimulating factor, chemokines C-C motif chemokine ligand 2/5, and VEGF, further promoting tumor growth and metastasis[72].
Additionally, research has found that SIX1 activates the mitogen-activated protein kinase signaling pathway in CRC cells[72]. Song et al[73] discovered that SIX1 promotes the malignant progression of CRC cells by activating the Wnt/β-catenin signaling pathway, a process confirmed by the localization of β-catenin in the cell nucleus. Similarly, FOXC2 enhances the expression of circCASK, which increases the sponging effect of circCASK on miR-1271-5p, reducing the inhibition of miR-1271-5p on SIX1. This, in turn, promotes SIX1 expression and further activates the Wnt/β-catenin signaling pathway[74].
Wu et al[75] discovered that the expression of SIX1 and EYA1 is upregulated in CRC tissues. The SIX1/EYA1 complex can regulate the expression of key genes, including cyclin A1 and TGF-β1, by binding to specific DNA sequences. Furthermore, the dual knockdown of SIX1 and EYA1 reduces cell proliferation, invasion, tumor growth, and in vivo tumor development. Li and Ma[76] discovered that circPLOD2 acts as a “sponge” for miR-513a-5p, enhancing the activity of its target gene SIX1, which in turn increases the transcriptional expression of the glycolytic enzyme LDHA, associated with the Warburg effect, thereby regulating the glycolysis process in CRC cells[76] (Figure 1).
SIX2 has been recognized as an important EMT-related gene associated with CRC prognosis. Its methylation status and mutation state may influence its regulatory role in EMT, and changes in its expression levels are closely linked to tumor invasiveness, immune evasion, and drug treatment responsiveness[77]. Wu et al[78] discovered that the RNA helicase DDX3 upregulates the transcription of SIX2 by enhancing the binding of the c-fos protein to the SIX2 promoter. This process involves the activation of YAP1 through the phosphatidylinositol 3 kinase (PI3K)/AKT signaling pathway, which increases the expression and phosphorylation of c-fos, thereby facilitating its binding to the SIX2 promoter[78]. Further studies indicated that DDX3-mediated invasiveness and cetuximab (CTX) resistance are regulated through the YAP1/SIX2 axis in KRAS-WT cells and were further validated in animal models[78]. Additionally, cells overexpressing DDX3 exhibited reduced sensitivity to CTX, but knockdown of YAP1 or SIX2 or the exogenous expression of E-cadherin significantly increased the sensitivity of DDX3-overexpressing cells to CTX, suggesting that the YAP1/SIX2 axis plays a role in DDX3-mediated CTX resistance. Then, in patients with KRAS-WT CRC, SIX2 expression levels were associated with overall survival and recurrence-free survival[78].
Jin et al[79] found that SIX3 can inhibit cell growth, suggesting that it may act as a tumor suppressor in CRC. This differs from its role in GC. Additionally, studies have shown that SIX3 is coregulated by DNMT3B and the PRC1/PRC2 complexes, indicating that its expression may be controlled by these protein complexes. SIX3 frequently undergoes DNA hypermethylation in CRC, leading to gene silencing[79]. Furthermore, the knockout of DNMT1 and DNMT3B can affect the methylation status and expression of the SIX3 gene. The absence of DNMT3B results in the loss of H2AK119 ubiquitination marks at specific gene promoter regions, potentially affecting the expression and function of the SIX3 gene[79].
The expression levels of SIX4 in CRC tissues are significantly higher than those in normal tissues. Furthermore, elevated SIX4 expression is associated with lymph node metastasis, advanced tumor-lymph node-metastasis (TNM) stage, and poor prognosis in patients with CRC, including overall survival and recurrence-free survival[80]. Further investigations have revealed that SIX4 promotes the metastasis of CRC cells by activating the PI3K-AKT signaling pathway. Rescue experiments further confirmed the regulatory role of SIX4 in the PI3K-AKT signaling pathway[80].
Additionally, SIX4 increases the expression of VEGF-A by interacting with hypoxia-inducible factor 1α (HIF-1α). VEGF-A is a key regulatory factor in tumor angiogenesis, and its expression is regulated by low oxygen conditions and HIF-1α. HIF-1α, a critical transcription factor under hypoxic conditions, can bind to the hypoxia response elements on the VEGF-A promoter, activating VEGF-A transcription[81]. Subsequently, SIX4 upregulates HIF-1α expression through the PI3K/Akt signaling pathway dependency. Akt, a protein kinase, enhances the stability and activity of HIF-1α through phosphorylation, thereby increasing VEGF-A expression to promote tumor growth and angiogenesis[81].
SIX5, together with EYA3 and p300, forms the EYA3-SIX5-p300 complex. This complex is formed after the induction of EYA3 expression under hypoxic conditions, with the involvement of hypoxia-inducible factors HIF-1α and HIF-2α. Additionally, SIX5 plays a role in recognizing and binding to the promoter regions of target genes within the complex, recruiting p300 to acetylate chromatin, leading to chromatin relaxation and further enhancement of gene expression. SIX5 also synergistically interacts with EYA3 to enhance transcriptional activation. SIX5 participates in the regulation of genes associated with tumor cell proliferation, invasion, and tumor growth through the aforementioned mechanisms, thereby playing a role in the development of CRC[82].
Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) constitutes the predominant form of primary liver cancer, ranks as the sixth most prevalent malignant tumor globally, and is the fourth-leading cause of cancer-related mortality worldwide, with chronic viral hepatitis, alcohol consumption, diabetes mellitus, and nonalcoholic steatohepatitis representing the principal risk factors for its onset[4]. While surgical resection and liver transplantation serve as primary treatments, long-term disease-free survival remains suboptimal. Tumor recurrence and metastasis stand as the primary causes of mortality among patients with HCC[83].
Overexpression of SIX1 is associated with tumor progression in HCC. Studies have found that approximately 85% of HCC tumor tissues exhibit increased SIX1 mRNA expression compared to non-tumor liver tissues, with about 60% of tumor tissues showing elevated SIX1 protein expression. High levels of SIX1 protein expression are observed in HCC cell lines with high metastatic potential. Furthermore, the overexpression of the SIX1 protein is significantly associated with the pathological TNM stage and vascular invasion in HCC. It is also correlated with poorer overall survival in patients with HCC after liver resection[84]. Further investigation has revealed that inhibition of SIX1 expression significantly reduces the growth and proliferation capabilities of HCC cells as well as diminishes their migration and invasion abilities. It also leads to a delay in the G2/M transition of cells, affecting the abnormal progression of the cell cycle and chemoresistance. Through cDNA microarray analysis, it has been found that SIX1 can regulate the expression of multiple genes associated with tumor development[85]. Similarly, Cheng et al[86] also confirmed the role of SIX1 in cell proliferation, apoptosis, and cell cycle regulation.
SIX1 influences the metabolic activities and stem cell characteristics of HCC cells by modulating metabolic processes linked to the Warburg effect, including sugar uptake, lactate production, adenosine triphosphate generation, and oxygen consumption rate[87]. Lu et al[88] found that SIX1 plays a role in glycolysis by directly regulating the expression of glycolytic genes such as hexokinase 2 and LDHA, which are involved in regulating cell proliferation and/or apoptosis. Additionally, miR-524-5p can inhibit the expression of SIX1 by targeting its 3’UTR. This regulatory action of miR-524-5p can suppress the positive regulation of glycolytic genes by SIX1, thereby reducing glycolysis levels. Conversely, the lncRNA TUG1 acts as a “sponge” for miR-524-5p, inhibiting its suppressive effect on SIX1 and thereby indirectly increasing SIX1 activity, which in turn promotes glycolysis[88].
Furthermore, Chu et al[89] discovered that SIX1 enhances the level of O-GlcNAcylation, a post-translational protein modification involving the addition of O-GlcNAc to serine/threonine residues of proteins. SIX1 undergoes O-GlcNAcylation modification at serine 276, inhibiting its ubiquitination degradation and thus enhancing the stability of the SIX1 protein. This enhancement promotes tumor growth and further exacerbates the malignant phenotype of HCC cells by increasing glucose uptake and O-GlcNAcylation levels. Subsequently, Liu et al[90] also identified a modification that enhances the stability of SIX1. They found that activation of the EGFR-AKT signaling pathway promotes the expression of ubiquitin-specific peptidase 1 (USP1). Ubiquitin-specific peptidase 1, a deubiquitinating enzyme, reduces the ubiquitination levels of SIX1 by removing ubiquitin chains from the protein, thereby stabilizing SIX1 expression and enhancing its stability.
Moreover, significant colocalization of SIX1, DACH1, and p53 was reported. Overexpression of SIX1 inhibited the expression of both DACH1 and p53, while overexpression of DACH1 enhanced the expression of p53, suggesting that SIX1 may indirectly decrease the expression of p53 by suppressing DACH1 and thereby promote tumor progression[86]. Li et al[91] found that SIX1 played a crucial role in drug resistance in HCC cells by modulating reactive oxygen species and autophagy. Treatment with SIX1 siRNA, paclitaxel alone, or their combination significantly increased reactive oxygen species levels in HCC cells, and it resulted in decreased expression of the autophagy-related protein light chain 3-I and increased expression of light chain 3-II, along with a significant reduction in p62 levels. Then, treatment with the reactive oxygen scavenger N-acetylcysteine and the autophagy inhibitor reversed these effects.
Similarly, research has also confirmed the relationship between SIX1 and cellular stemness and chemotherapy drug sensitivity. Knockdown of SIX1 enhances the sensitivity of HCC cells to 5-fluorouracil (5-FU). Additionally, SIX1 can directly bind to the promoter region of the SOX2 gene, enhancing its transcriptional activities and expression accordingly. SOX2 is a master regulator of stemness. Therefore, SIX1 affects the stemness of HCC cells by regulating SOX2. Overexpression of SIX1 can induce normal liver cells to acquire stem cell characteristics, including increased ability for sphere formation and expression of stemness markers. Furthermore, the decreased stemness and the increased sensitivity to 5-FU caused by SIX1 knockdown can be partially reversed by overexpression of SOX2[92] (Figure 1).
It was revealed that the expression of SIX2 was significantly elevated in HCC tumor tissues compared to adjacent non-tumorous liver tissue and normal liver tissue. High levels of SIX2 are associated with shorter overall survival and disease-free survival in patients with HCC. Additionally, the expression of SIX2 correlates with factors such as sex, tumor size, alpha-fetoprotein levels, and portal vein invasion, suggesting that SIX2 may enhance tumor growth and invasion clinically. In cases of HCC with portal vein tumor thrombosis, the expression levels of SIX2 are even higher, indicating that SIX2 may facilitate tumor metastasis[93]. Li et al[94] also found that elevated expression levels of SIX2 are associated with shorter overall survival in patients with HCC. Furthermore, SIX2 suppresses the expression of E-cadherin by stimulating methylation in the promoter region of the E-cadherin gene. This suppression reduces the sensitivity of HCC cells to 5-FU and enhances the stem cell-like properties of the HCC cells[94].
Moreover, recent research has revealed that NIK, an NF-κB-inducing kinase, enhances the stability of SIX2 protein by inhibiting its ubiquitination. Knockdown of NIK promotes the ubiquitination of SIX2, and this reduction in SIX2 protein stability can be rescued by treating with the proteasome inhibitor. The overexpression of SIX2 partially reverses the inhibitory effect of NIK knockdown on the stem cell-like properties of HCC cells. These findings suggest that NIK promotes the stem cell-like properties of HCC cells through SIX2, including self-renewal capacity and tumorigenicity[95].
It is noteworthy that the expression of SIX3 mRNA in HCC tissues is significantly lower compared to adjacent non-tumorous tissues, a contrast to the expression patterns of other members of the SIX family in HCC[96]. Studies indicated that SIX3 can bind to the promoter region of WDR26 gene, and the overexpression of lncWDR26 enhances the binding of SIX3 to this promoter region. This binding inhibits the transcription of the WDR26 gene, thereby inhibiting the growth and metastasis of HCC cells[97].
SIX4 is also found to be significantly upregulated in HCC tissues compared to adjacent non-tumor tissues, which is positively correlated with the absence of a tumor capsule, microvascular invasion, higher TNM staging, and poor prognosis[96]. Further studies revealed that SIX4 can directly bind to the promoter regions of the YAP1 and c-MET genes, activating their expression, which promotes the invasion and metastasis of HCC. The expression of SIX4 is positively regulated by hepatocyte growth factor, which activates the ERK/NF-κB signaling pathway through its receptor, c-MET, thus upregulating SIX4 expression. This created a novel positive feedback loop of hepatocyte growth factor-SIX4-c-MET that may enhance the malignant progression of HCC[96].
Pancreatic cancer
The incidence of pancreatic cancer (PC) is increasing and continues to be one of the most lethal cancers[4]. It is characterized by its highly aggressive tumor growth rate, high rate of metastasis, and notable resistance to chemotherapy. Various treatment modalities, such as surgical resection, chemotherapy, and radiation therapy, have been progressively advancing over time. Nevertheless, due to the advanced or metastatic stage of diagnosis in most patients, treatment options are constrained, leading to an unfavorable prognosis[98].
Research has found that SIX1 is expressed at significantly higher levels in PC tissues compared to normal pancreatic tissues. Overexpression is positively correlated with tumor size, TNM staging, lymph node metastasis, and tumor grading in PC, and it is closely associated with patient prognosis[99]. It is confirmed that SIX1 can bind to specific regions of the cyclin D1 promoter, enhancing its activity. Additionally, the expression of SIX1 is significantly correlated with cyclin D1 in human PC tissues. SIX1 promotes the proliferation and colony formation of PC cells, partly by directly activating the transcription of cyclin D1, facilitating cell cycle progression and enhancing the proliferative capacity[100]. Conversely, inhibiting SIX1 reduces the migratory capacity of PC cells in vitro, slows tumor growth in vivo, and diminishes the CD24-/ CD44 + tumor stem cell phenotype[101].
Additionally, SIX1 enhances glycolysis in PC cells by directly targeting and activating the expression of LDHA, leading to lactate accumulation. Natural killer cells co-cultured with PC cells overexpressing SIX1 exhibit a reduced proportion of activated surface receptors and diminished expression of cytotoxic mediators. This indicates that lactate accumulation caused by SIX1 overexpression inhibits natural killer cell function, weakening the immune defense against malignant tumor cells[102]. Zhou et al[103] also discovered that the expression of SIX1 in glycolysis is regulated by the E3 ubiquitin ligase TRIM16. TRIM16 promotes glycolysis and metastasis in PC cells by stabilizing NIK, which inhibits the ubiquitination and degradation of SIX1 (Figure 1).
The role of the SIX3 gene in various cancers has been extensively studied, particularly as a tumor suppressor gene[79,97]. Research has revealed that the methylation status of SIX3 is associated with early diagnosis of PC, showing significant differences in methylation levels between patients with PC and healthy controls. In PCs, the high methylation level of the SIX3 gene may lead to gene silencing, thereby impairing its function as a tumor suppressor[104].
Database analysis and quantitative real-time PCR techniques have confirmed the overexpression of SIX4 in PC tissues. Knockdown of SIX4 via siRNA technology significantly reduces the survival rate, colony formation ability, and mitochondrial membrane potential of PC cells. Silencing SIX4 alters the expression of apoptosis-related genes, thereby increasing apoptosis and autophagy in cancer cells[105]. Additionally, the knockdown of SIX4 leads to cell cycle arrest at the G1 and sub-G1 phases, which may be associated with the inhibition of cell proliferation and the initiation of apoptosis[105].
Furthermore, SIX1 and SIX4 act as downstream effectors of hepatocyte nuclear factor 4α (HNF4α). HNF4α inhibits tumor growth and steers tumor cells towards epithelial properties, while SIX1 and SIX4 drive tumor cell proliferation and foster mesenchymal/neuronal cell differentiation in HNF4α-negative PC. HNF4α regulates PC proliferation and molecular subtypes by directly binding to the gene promoter regions of SIX1 and SIX4, suppressing their expression[106]. Further research has found that SIX4 and SIX1 physically interact with their enzymatic cofactors, such as SIX4 with the histone demethylase UTX and SIX1 depending on the Eya family protein tyrosine phosphatases, which are potential drug targets[106]. Moreover, RNA sequencing analysis revealed a gene network coregulated by SIX4 and SIX1, which likely involves multiple cell fate determinants that influence the characteristics of PC cells[106].
SIX6 has been identified as one of the differentially expressed genes consistently associated with PC and its radioresistance. Genome-wide methylation analysis revealed a set of genes whose expression was altered in radioresistant cell lines, with a particular enrichment of genes related to cholesterol biosynthesis pathways. SIX6 is involved in the cholesterol biosynthesis pathway and is speculated to be associated with the radioresistance of PC cells[107]. However, comprehensive research is still lacking to elucidate the specific mechanism of action of SIX6 in PC.
IMAGING IN HEALTHCARE
Although the role of the SIX gene family in GIC has been extensively studied, relying on a single gene is insufficient to fully elucidate the complex mechanisms underlying cancer development. Cancer is a multifactorial disease driven by interactions between gene expression, epigenetic modifications, and environmental influences[108]. To better understand the onset and progression of cancer, technologies that can accurately capture these multiple factors are essential.
Traditional diagnostic methods face limitations in early detection and real-time imaging, particularly for GIC, which are often located deep within internal organs and are not visible to the naked eye or easily detected through non-invasive techniques[109]. However, advances in imaging technologies, especially hyperspectral imaging (HSI) and multispectral imaging, have enabled scientists to capture the spectral characteristics of tissues, leading to a better understanding of gene expression, mutations, and epigenetic changes (Figure 2). Integrating these imaging techniques with artificial intelligence can enhance early cancer detection and provide new perspectives for targeting the SIX gene family[110].
Figure 2 Hyperspectral applications in healthcare: A multifunctional diagnostic tool.
HSI: Hyperspectral.
For example, in the early diagnosis of esophageal cancer, the combination of HSI with artificial intelligence has been shown to differentiate between normal and cancerous tissues through spectral analysis without direct tissue contact[111]. Studies have demonstrated that HSI outperforms traditional endoscopic techniques in sensitivity and specificity for early esophageal cancer detection, reducing reliance on biopsies[112,113]. Furthermore, multispectral imaging can be used not only for diagnosis but also as a surgical navigation tool, allowing real-time evaluation of tissue states and boundaries during surgery[114,115].
This presents new opportunities for exploring the application of HSI in GIC. Combining this technology with research on the SIX gene family offers a novel approach for early GIC detection and can deepen our understanding of the role of SIX genes in cancer initiation and progression. By precisely capturing the spectral characteristics of tumor tissues, HSI can support targeted gene therapy, providing strong backing for personalized cancer treatment[116]. This innovative integration of new imaging technologies and gene-targeted therapies offers a fresh perspective for developing SIX gene family-based therapies to address abnormalities in complex signaling pathways.
SUMMARY AND PROSPECT
At the mechanistic level, the diverse roles of the SIX gene family provide researchers with key insights into its functions in GIC, opening new avenues for therapeutic interventions. Precision-targeted therapies, particularly those designed for specific genetic targets, offer significantly improved efficacy and reduced side effects compared to traditional treatments[117]. For instance, ginsenoside Rh4 targets SIX1, thereby blocking the TGF-β/Smad2/3 signaling pathway. Compared to common chemotherapy agents like oxaliplatin, ginsenoside Rh4 demonstrates lower systemic toxicity and side effects while inhibiting tumor growth and metastasis[61].
Lowering SIX1 expression with siRNA can increase the sensitivity of HepG2 cells to the chemotherapeutic drug paclitaxel, suggesting that SIX1 siRNA may help overcome resistance to chemotherapy in certain cancers[91]. Moreover, to address challenges in target identification, it is crucial to deeply explore relevant ligands and signaling pathways. In GIC contexts, SIX genes often activate the Wnt/β-catenin signaling pathway, promoting tumor cell proliferation and migration. Developing small molecule inhibitors targeting SIX or inhibitors of the Wnt signaling pathway could potentially suppress malignant progression in the GIC tract[73].
Additionally, abnormal regulation of SIX proteins and their cofactors may lead to tumor development and progression. Developing inhibitors targeting these cofactors could be an effective therapeutic strategy. Interactions between SIX1 and SIX4 with their cofactors, such as UTX and the Eya family proteins, are critical for their activity. Using inhibitors like benzbromarone (inhibits Eya family proteins) has suppressed SIX1 and SIX4 expression, which may impact the proliferation of tumor cells dependent on these transcription factors[106].
Although targeting the SIX gene family has demonstrated potential advantages in the treatment of GIC, current research faces several limitations. Most inhibitors are still in the preclinical stage, lacking large-scale clinical validation, and many studies focus on single targets, limiting the coverage of relevant signaling pathways. Consequently, these limitations hinder the clinical application of existing findings. Further research is required to explore the mechanisms of the SIX gene family in different cancer types and to develop broader spectrum small-molecule inhibitors to enhance efficacy and expand applicability. However, due to the limited number and scope of current studies, the widespread clinical use of these inhibitors remains constrained. Future research must expand to cover more targeted pathways and undergo large-scale clinical trials to verify the effectiveness and safety of these therapies across various cancer types.
Additionally, future research directions should include extending the findings on the SIX gene family to other types of cancer. Although this study primarily focused on GIC, other studies have shown that the SIX gene family plays an important role in breast cancer, lung cancer, and other malignancies[118,119]. Therefore, future targeted therapies should explore the application of these genes in other types of tumors and develop inhibitors with broader applicability[120].