©The Author(s) 2025.
World J Gastrointest Oncol. Dec 15, 2025; 17(12): 113289
Published online Dec 15, 2025. doi: 10.4251/wjgo.v17.i12.113289
Published online Dec 15, 2025. doi: 10.4251/wjgo.v17.i12.113289
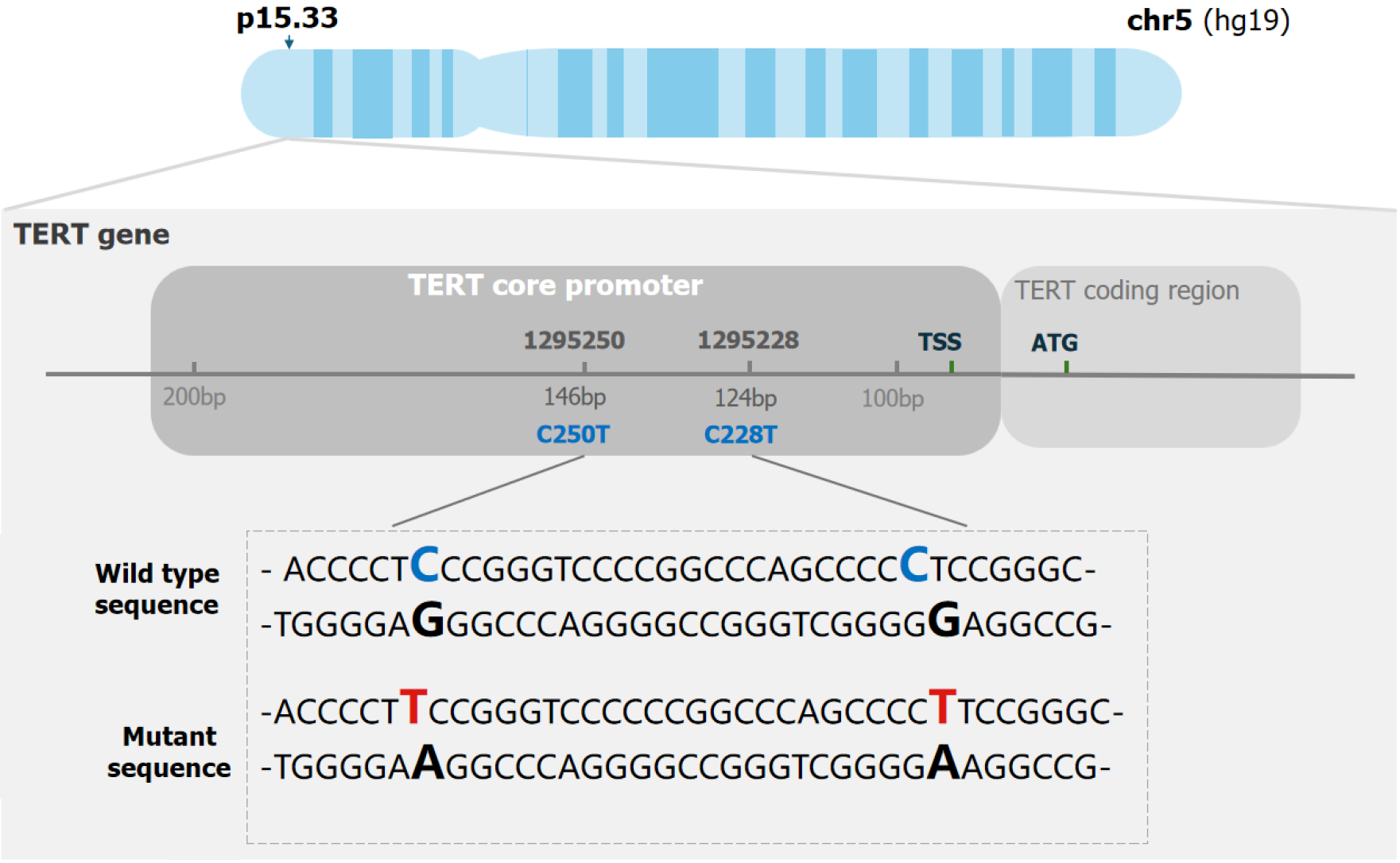
Figure 1 Schematic representation of the commonly mutated nucleotides C228T (124 bp) and C250T (146 bp) in relation to the telomerase reverse transcriptase promoter positions and the genomic positions within chromosomal region 5 p15.
33. TSS: Transcriptional start site; ATG: Adenine-thymine-guanine; TERT: Telomerase reverse transcriptase.
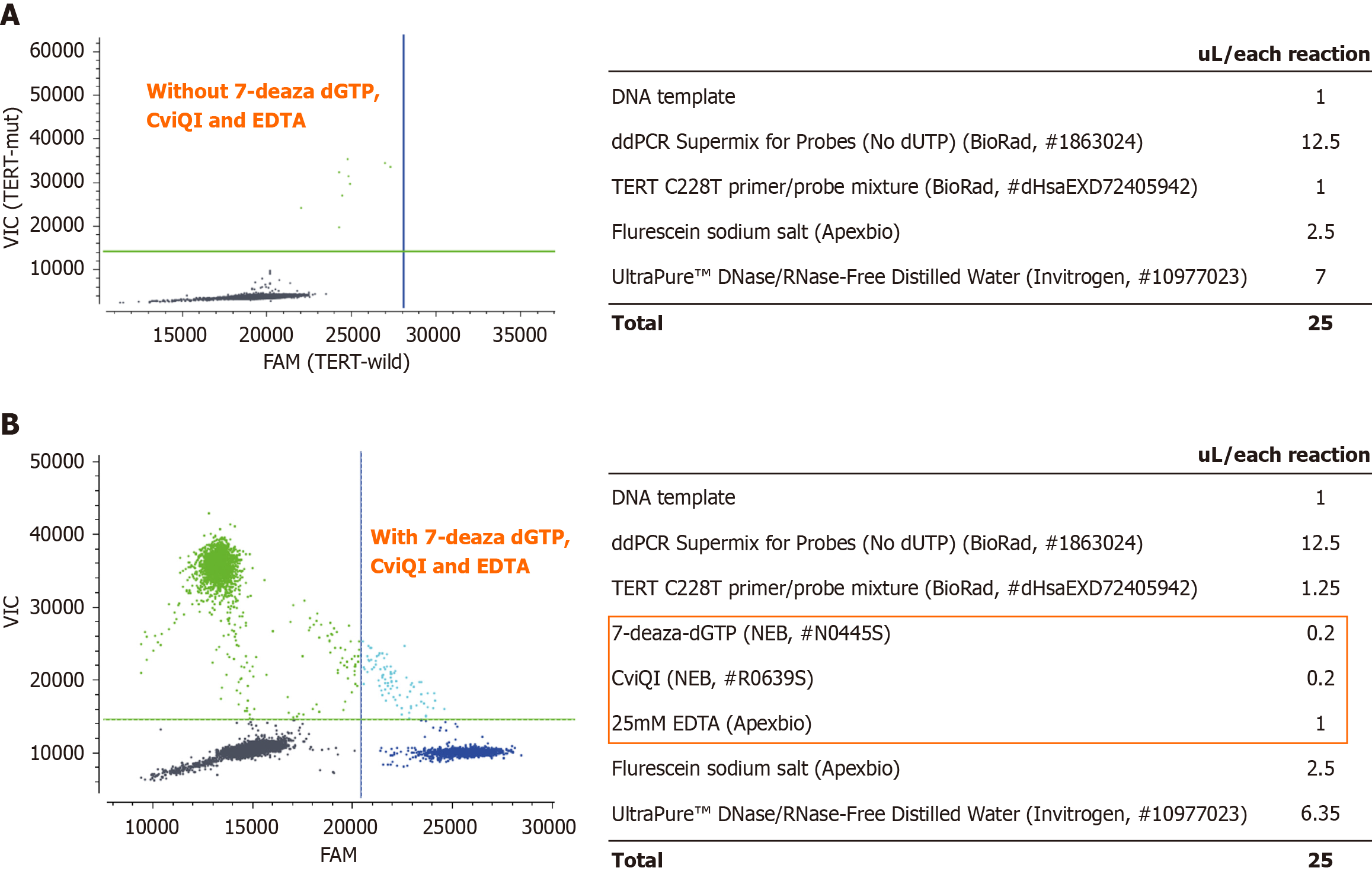
Figure 2 Two-dimensional digital polymerase chain reaction plots showing the telomerase reverse transcriptase C228T promoter assay with and without 7-deaza-dGTP, CviQI and EDTA reagents in patient HC2873.
A: Preliminary results of amplification and detection of the C228T fragment using standard supermixtures and assays (left); the detailed amount of each reagent (right); B: The optimized detection conditions, including 7-deaza-DGTP, CviQI and EDTA reagents, and the experimental results of generating distinct droplet clusters for a C228T fragment (left). The optimized detailed dosage of each reagent (right). Color of line: Green, VIC probe telomerase reverse transcriptase (TERT) C228T wild-type; dark blue, 6-carboxyfluorescein probe for TERT C228T mutant type; light blue, droplets with TERT C228T mutant and wild-type allele; dark grey, droplets negative for template DNA. The green and blue lines are manually set thresholds for a 6-carboxyfluorescein mutant probe and VIC wild-type probe for digital polymerase chain reaction mutation detection, respectively.
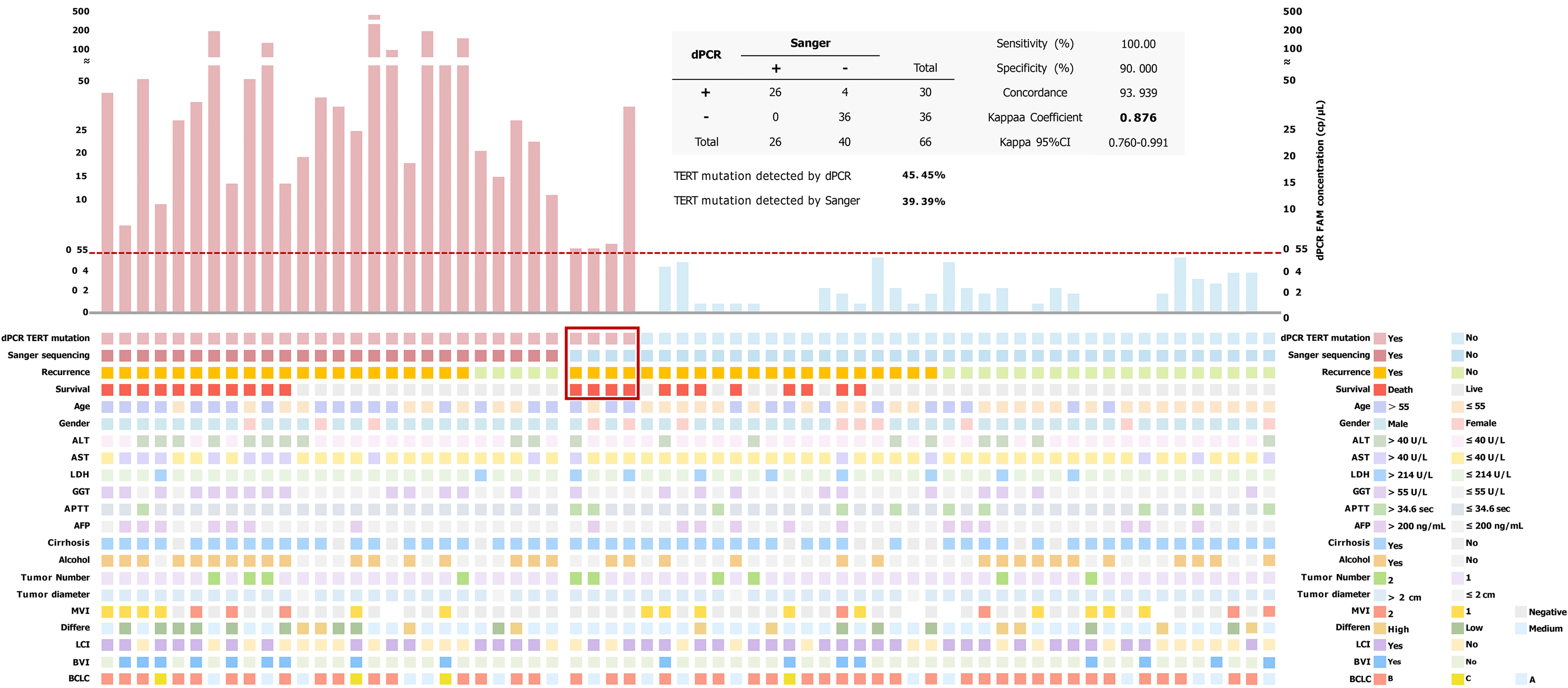
Figure 3 Altered landscape of hepatocellular carcinoma cohort samples, combined with clinical variables (the lower half) and a bar graph of 6-carboxyfluorescein concentration (the upper half) of telomerase reverse transcriptase C228T mutation.
A minimum concentration of 0.55 cp/μL (limit of detection) was evaluated to be positive in digital polymerase chain reaction. Comparison of telomerase reverse transcriptase status detected by Sanger sequencing and digital polymerase chain reaction in our hepatocellular carcinoma cohort (the table at the top of this picture). +: Telomerase reverse transcriptase C228T mutated; -: Telomerase reverse transcriptase C228T non-mutated; dPCR: Digital polymerase chain reaction; TERT: Telomerase reverse transcriptase; ALT: Alanine aminotransferase; AST: Aspartate aminotransferase; LDH: Lactate dehydrogenase; GGT: Gamma-glutamyl transferase; APTT: Activated partial thromboplastin time; AFP: Alpha fetoprotein; MVI: Microvascular invasion; LCI: Liver capsular invasion; BVI: Blood vessel invasion; BCLC: Barcelona Clinic Liver Cancer; CI: Confidence interval.
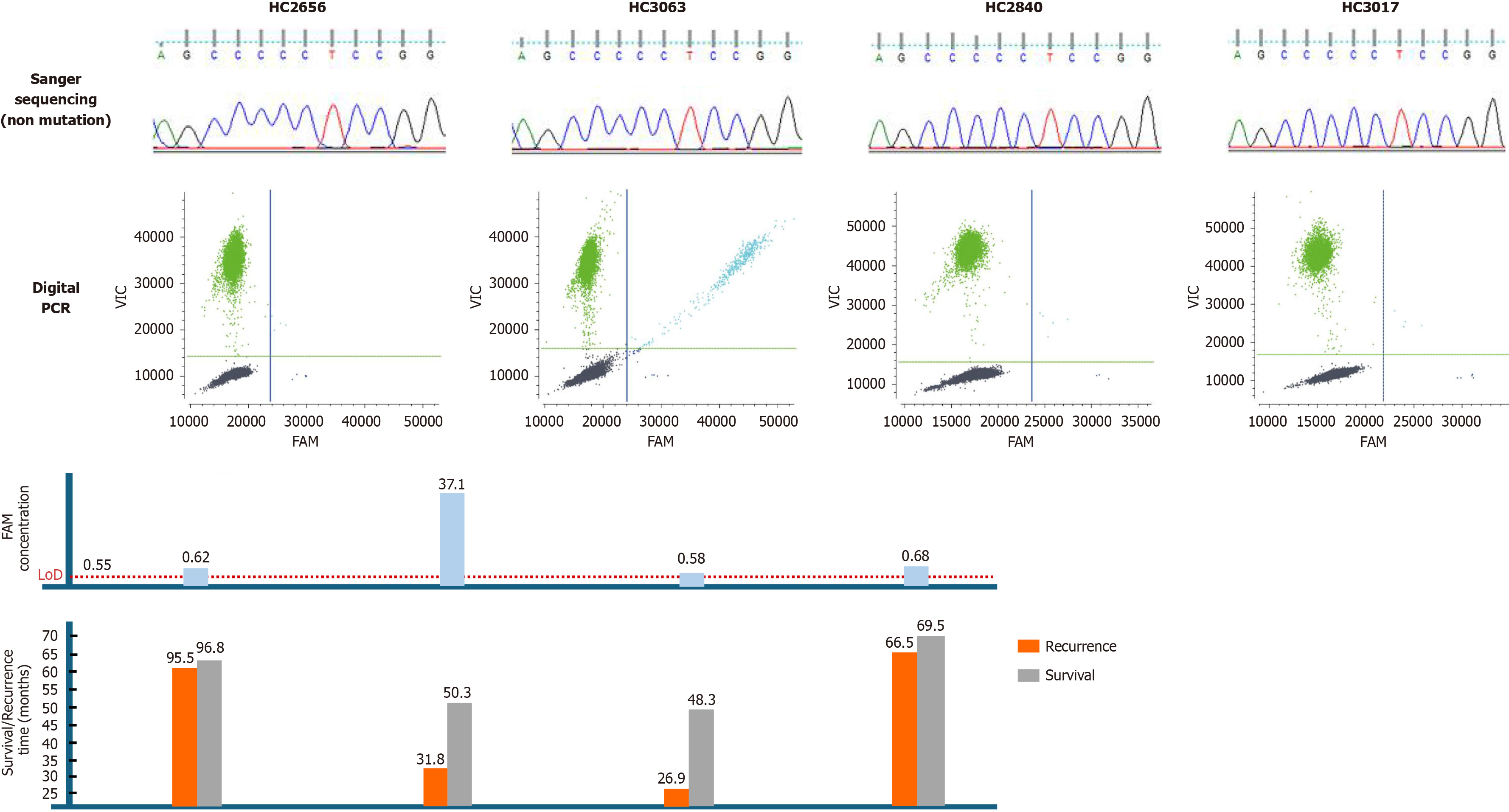
Figure 4 Illustration of sequencing results and follow-up data in four samples, which were positive for telomerase reverse transcriptase C228T by digital polymerase chain reaction assay, but were all negative by Sanger sequencing.
Each column represents a sample of one patient with hepatocellular carcinoma after surgical resection. At the top of the figure, Sanger sequencing results (all were non mutated); In the middle of the figure, the two-dimensional digital polymerase chain reaction plot (the color of line: Green, wild-type; dark blue, mutant type; light blue, droplets with mutant and wild-type allele; dark grey, negative droplets) and the concentration (cp/μL) of the telomerase reverse transcriptase C228T mutant allele targeted by the 6-carboxyfluorescein dye labeled probes. 6-carboxyfluorescein concentration higher than LoD was positive. Clinical follow-up data collected from each patient are shown at the bottom of the figure. The orange and grey bars represent the time from surgery to tumor recurrence and death, respectively. PCR: Polymerase chain reaction.
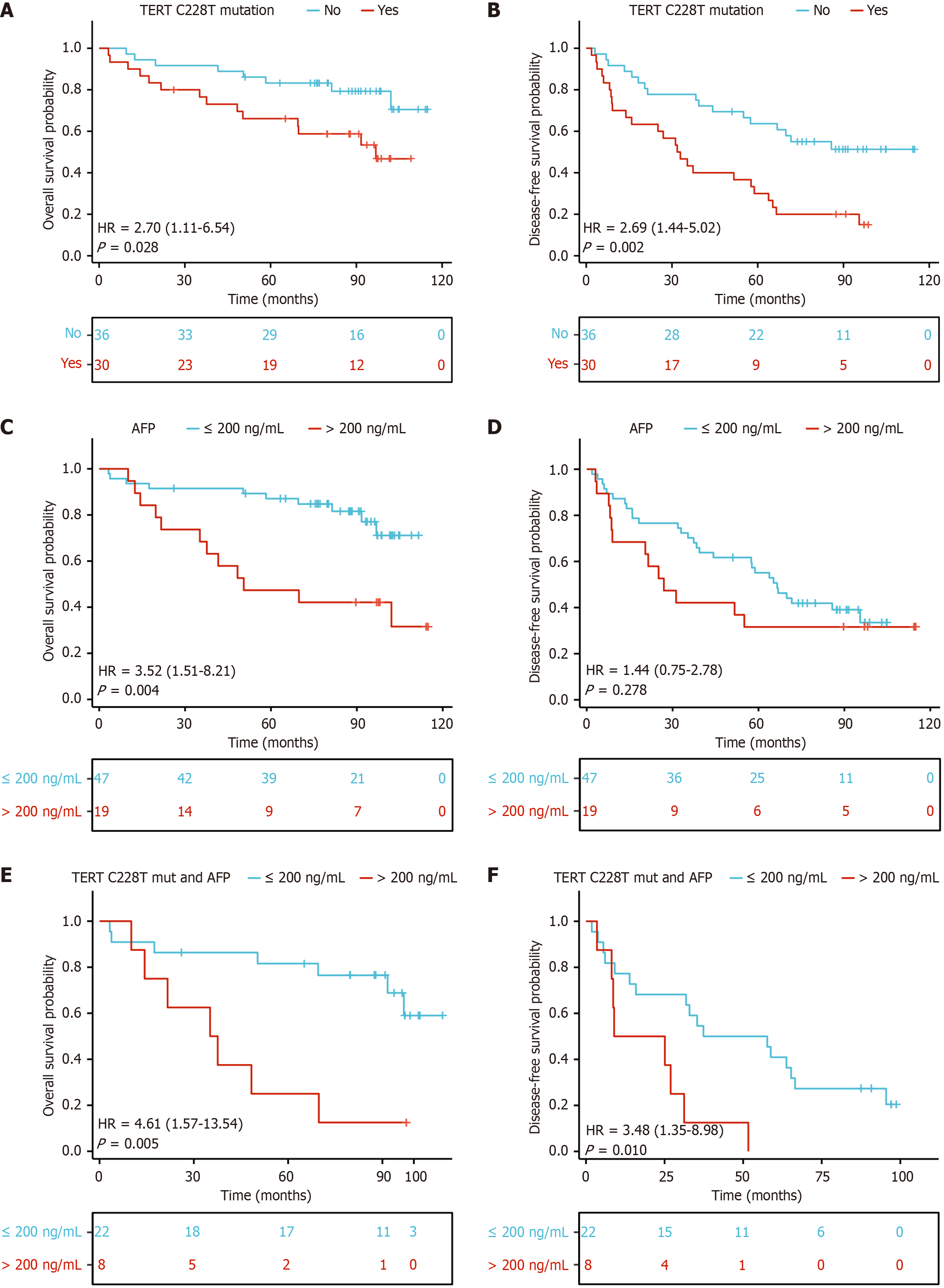
Figure 5 Kaplan-Meier curves based on telomerase reverse transcriptase C228T mutation status and alpha fetoprotein level.
A and B: Kaplan-Meier curve of overall survival (OS; P = 0.028) and disease-free survival (DFS; P = 0.002) in telomerase reverse transcriptase C228T mutated patients; C and D: Kaplan-Meier curve of OS (P = 0.004) and DFS (P = 0.278) stratified by alpha fetoprotein; E and F: The telomerase reverse transcriptase C228T mutated patient’s OS (P = 0.005) and DFS (P = 0.01) stratified by alpha fetoprotein. TERT: Telomerase reverse transcriptase; HR: Hazard ratio; AFP: Alpha fetoprotein.
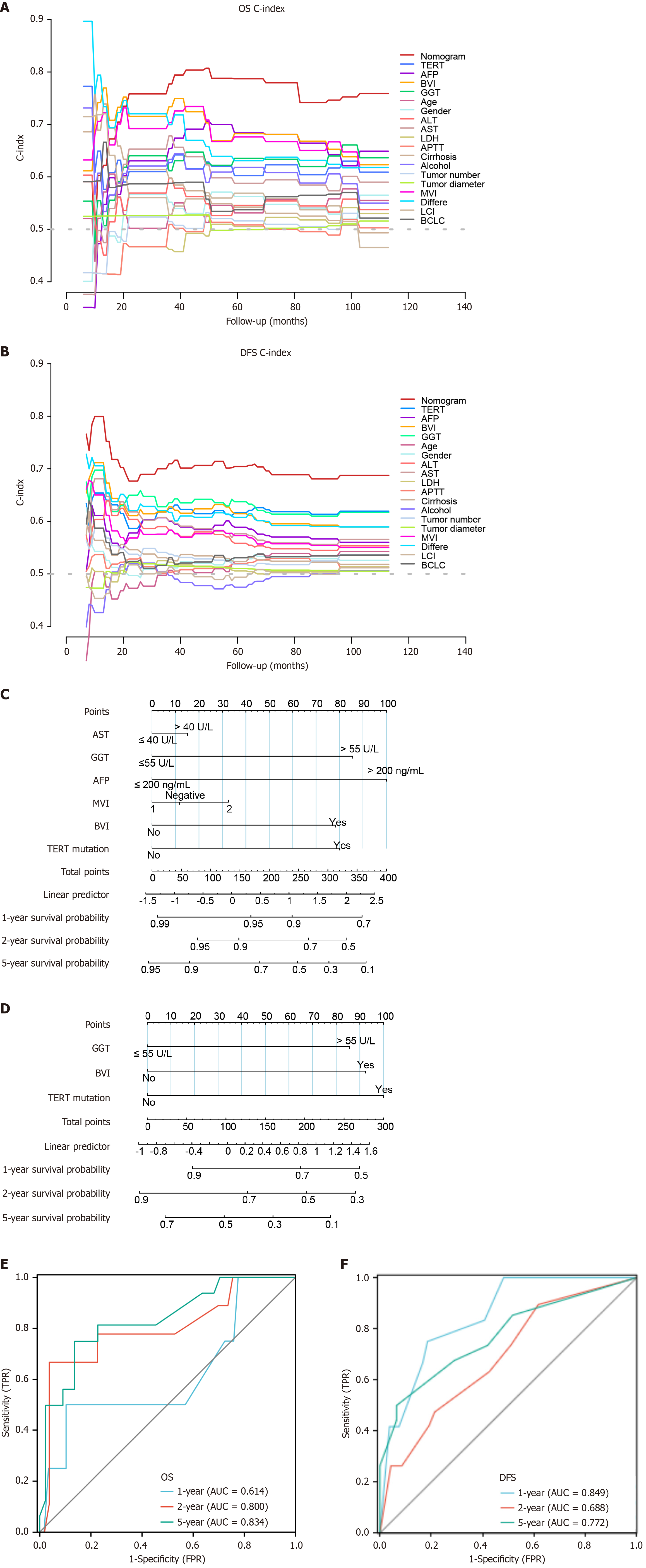
Figure 6 Construction and validation of overall survival and disease-free survival nomograms.
A and B: The prognostic performance between the telomerase reverse transcriptase mutation status and different conventional clinical characteristics was compared by calculating the overall survival (OS) and disease-free survival (DFS) C-index. The maximum C-index of the OS and DFS nomogram model was 0.7651 and 0.6899, respectively. The maximum C-index of telomerase reverse transcriptase (TERT) mutation in OS and DFS was 0.6224 and 0.6228, respectively; C: OS-nomogram was used to predict aspartate aminotransferase, gamma-glutamyl transferase, microvascular invasion, blood vessel invasion and TERT mutation status; D: DFS-nomogram was used to predict gamma-glutamyl transferase, blood vessel invasion and TERT mutation status; E and F: Receiver operating characteristic curves showed the 1-, 2-, and 5-year predictive efficiency of the OS and DFS nomogram model. OS: Overall survival; DFS: Disease-free survival; AST: Aspartate aminotransferase; GGT: Gamma-glutamyl transferase; AFP: Alpha fetoprotein; MVI: Microvascular invasion; BVI: Blood vessel invasion; TERT: Telomerase reverse transcriptase; AUC: Areas under the curve; TPR: True positive rate; FPR: False positive rate.
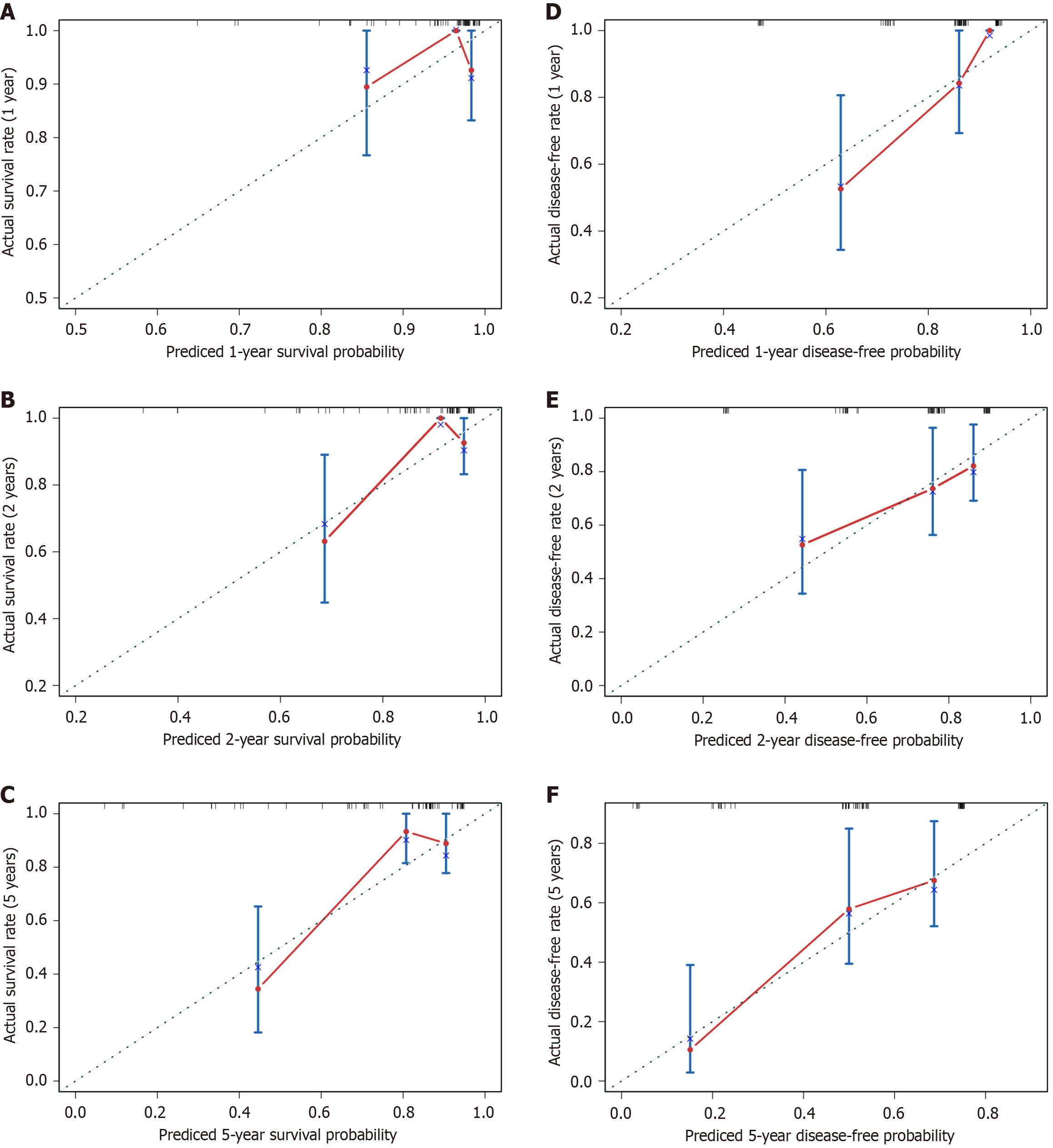
Figure 7 Validation of the overall survival and disease-free survival nomogram.
A-C: Overall survival; D-F: Disease-free survival. Calibration plots were used to predict the 1-, 2-, and 5-year overall survival and disease-free survival in our hepatitis B virus-related hepatocellular carcinoma cohort, respectively.
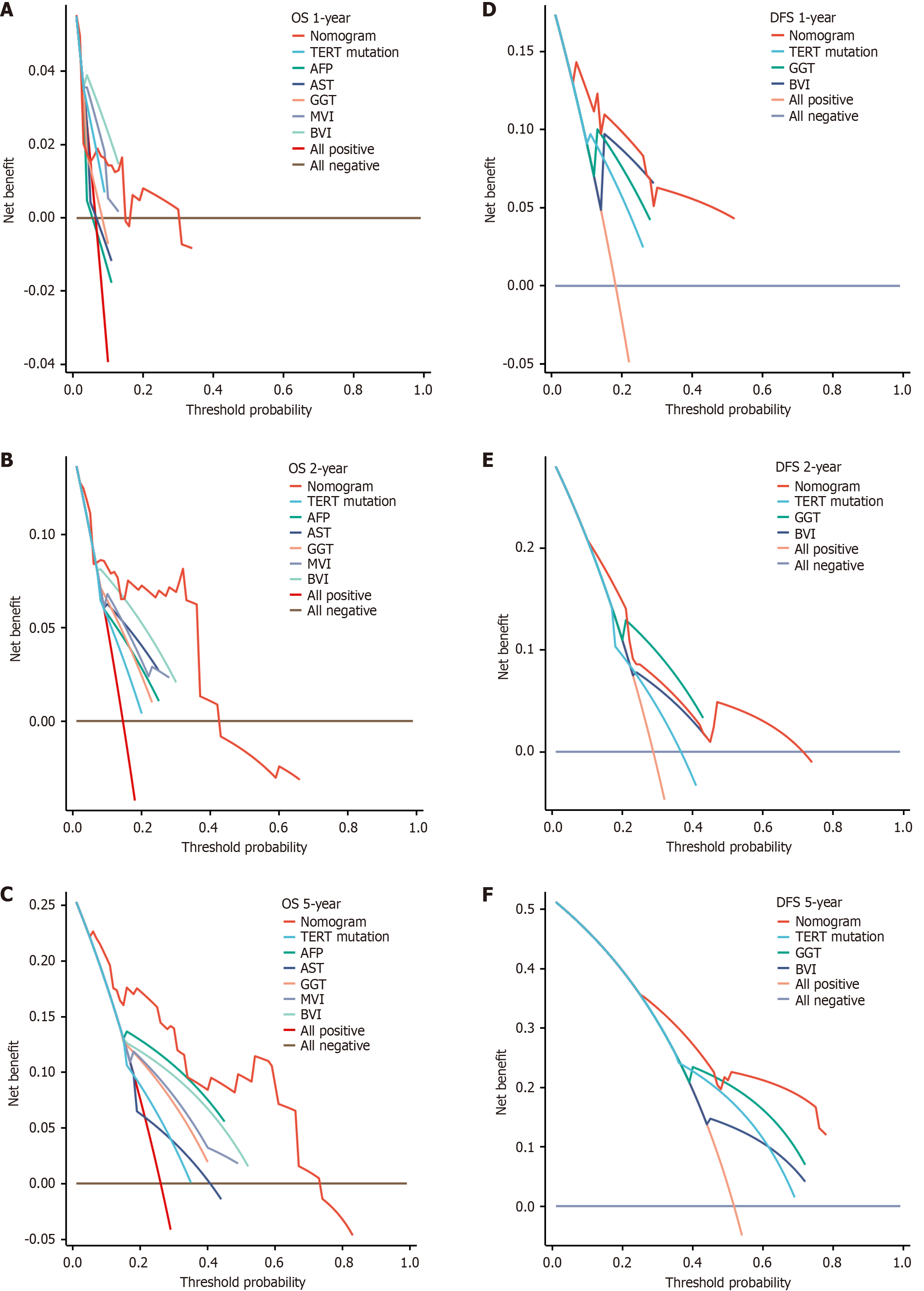
Figure 8 Decision curve analysis.
A-C: Overall survival; D-F: Disease-free survival. Decision curve analysis showed the 1-, 2-, and 5-year predictive efficiency of the overall survival and disease-free survival nomogram model. OS: Overall survival; DFS: Disease-free survival; TERT: Telomerase reverse transcriptase; AFP: Alpha fetoprotein; AST: Aspartate aminotransferase; GGT: Gamma-glutamyl transferase; MVI: Microvascular invasion; BVI: Blood vessel invasion.
- Citation: Aizimuaji Z, Hu N, Li HY, Wang XJ, Ma S, Wang YR, Zheng RQ, Li Z, Zhao H, Rong WQ, Xiao T. Optimized digital polymerase chain reaction enables detection of telomerase reverse transcriptase C228T mutation for prognostic assessment in hepatocellular carcinoma. World J Gastrointest Oncol 2025; 17(12): 113289
- URL: https://www.wjgnet.com/1948-5204/full/v17/i12/113289.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i12.113289