Published online Mar 26, 2024. doi: 10.4252/wjsc.v16.i3.287
Peer-review started: December 19, 2023
First decision: January 12, 2024
Revised: January 21, 2024
Accepted: February 28, 2024
Article in press: February 28, 2024
Published online: March 26, 2024
Processing time: 96 Days and 18.9 Hours
The self-assembly of solid organs from stem cells has the potential to greatly expand the applicability of regenerative medicine. Stem cells can self-organise into microsized organ units, partially modelling tissue function and regeneration. Dental pulp organoids have been used to recapitulate the processes of tooth development and related diseases. However, the lack of vasculature limits the utility of dental pulp organoids.
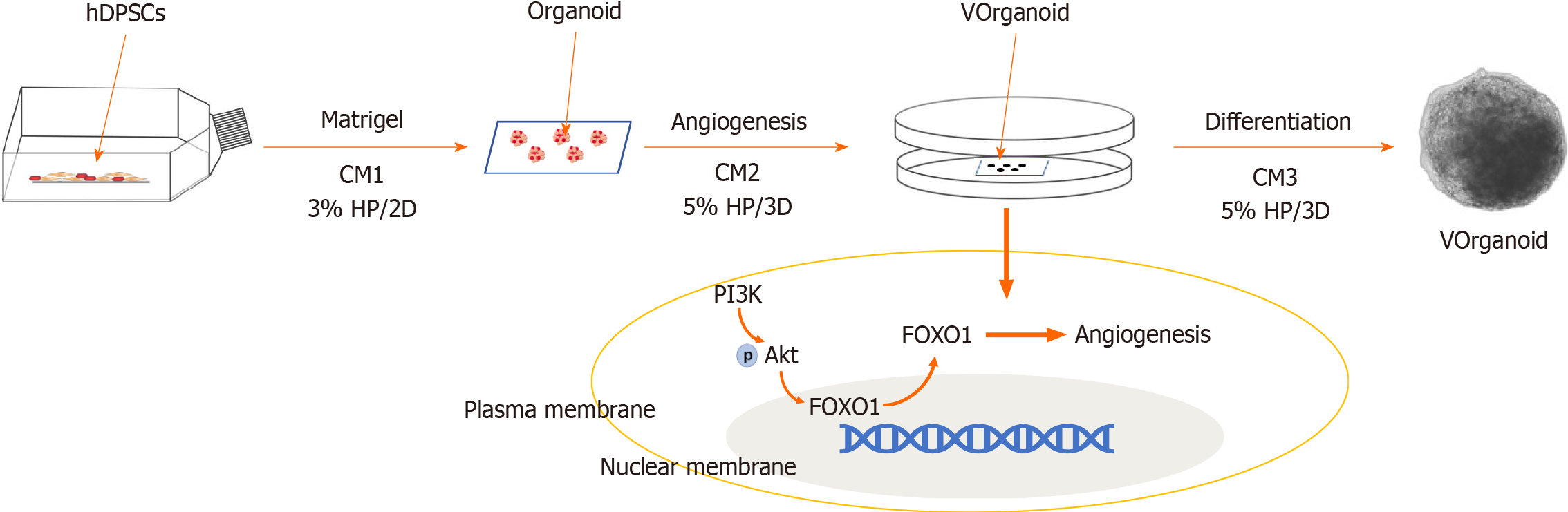
To improve survival and aid in recovery after stem cell transplantation, we demonstrated the three-dimensional (3D) self-assembly of adult stem cell-human dental pulp stem cells (hDPSCs) and endothelial cells (ECs) into a novel type of spheroid-shaped dental pulp organoid in vitro under hypoxia and conditioned medium (CM).
During culture, primary hDPSCs were induced to differentiate into ECs by exposing them to a hypoxic en
The combination of these two agents resulted in prevascularized human dental pulp organoids (Vorganoids) that more closely resembled dental pulp tissue in terms of morphology and function. Single-cell RNA sequencing of dental pulp tissue and RNA sequencing of Vorganoids were integrated to analyse key regulatory pathways associated with angiogenesis. The biomarkers forkhead box protein O1 and fibroblast growth factor 2 were identified to be involved in the regulation of Vorganoids.
In this innovative study, we effectively established an in vitro model of Vorganoids and used it to elucidate new mechanisms of angiogenesis during regeneration, facilitating the development of clinical treatment strategies.
Core Tip: We demonstrated the three-dimensional self-assembly of adult stem cell-human dermal papilla cells and endothelial cells into a novel type of spheroid-shaped dental pulp organoid in vitro under hypoxia and conditioned medium. These organoids have been constructed to be morphologically and functionally closer to dental pulp tissue. Through the integration and analysis of single-cell RNA sequencing and RNA sequencing data, forkhead box protein O1 and fibroblast growth factor 2 were identified as crucial markers involved in the regulation of organoid angiogenesis. In this innovative study, we effectively established an in vitro model of prevascularized dental pulp organoids and used it to elucidate new mechanisms of angiogenesis during regeneration, facilitating the development of clinical treatment strategies.
- Citation: Liu F, Xiao J, Chen LH, Pan YY, Tian JZ, Zhang ZR, Bai XC. Self-assembly of differentiated dental pulp stem cells facilitates spheroid human dental organoid formation and prevascularization. World J Stem Cells 2024; 16(3): 287-304
- URL: https://www.wjgnet.com/1948-0210/full/v16/i3/287.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v16.i3.287
The main experimental tool and goal of tissue regeneration engineering is the implantation of specific stem cells into diseased environments to repair and compensate for the dysfunction of damaged tissues and organs. However, the complexity and variability of the tissue microenvironment at the site of injury, such as hypoxic lethality, the inflammatory response, immune resistance and inadequate blood supply, leads to low survival rates (success rates of approximately 1%-3%) and poor maturation rates of directed differentiation after stem cell transplantation, all of which limit the clinical application and promotion of stem cells[1,2]. Therefore, it is critical for stem cells to form additional microvessels after implantation to quickly connect to the host’s circulatory system and form functional blood vessels to ensure nutrient and oxygen delivery after implantation.
Organoids are constructed in vitro following an in vivo developmental programme that allows cells to grow, migrate, differentiate and function in three-dimensional (3D). A variety of organoids are currently constructed in vitro[3,4]. Compared to traditional 2D culture methods, 3D culture facilitates cell access to bio-factors and reduces intercellular shear force, which promotes the proliferation and differentiation of dental pulp stem cells. With the advent of 3D pulp culture technology, great progress has been made in regenerative endodontic procedures[5-7]. Although the construction of organoid models has outstanding advantages in terms of clinical application, it still faces a major challenge, namely, the lack of model nourishment due to the absence of angiogenesis, which is the main dilemma for the in vitro application of such models. In recent years, numerous studies have investigated the relationship between angiogenesis and pulp regeneration[7,8]. Several researchers have proposed that the addition of high concentrations of nutrients and their controlled and sustained release from scaffolds are important strategies for optimising angiogenesis during pulp regeneration[9]. A hydrogel scaffold was used in combination with conditioned media to release bio-factors in a controlled manner, which subsequently promoted blood vessel and nerve formation and dental tissue repair[10].
Our previous study revealed that hypoxia-activated phosphatidylinositol 3 kinase/protein kinase B (PI3K/Akt) inhibits oxidative stress in human dental pulp stem cells (hDPSCs) by regulating reactive oxygen species[11], thereby maintaining stem cell stemness during in vitro expansion. In addition, a number of signalling pathways associated with angiogenesis are activated, and the expression of regulatory factors associated with angiogenesis is altered. Does using hypoxic hDPSCs in in vitro organoid cultures better induce angiogenesis? How can prevascularized organoids be constructed to address current challenges in stem cell applications? Is angiogenesis regulated by the same mechanisms in dental pulp and prevascularized dental pulp organoids (Vorganoids)?
We induced endothelial cells (ECs) and hDPSCs in culture and subsequently fused the two cell types to obtain Vorganoids, which are morphologically and functionally more similar to dental pulp tissue. Single-cell RNA sequencing (scRNA-seq) of dental pulp tissue and RNA-seq of Vorganoids were subsequently integrated to analyse key regulatory pathways associated with angiogenesis in dental pulp tissue. Vascular endothelial growth factor A (VEGFA), a key signalling pathway regulating the differentiation of vascular ECs in dental pulp tissue, was also significantly enriched in the development of Vorganoids. The biomarkers forkhead box protein O1 (FOXO1) and fibroblast growth factor 2 (FGF2) were identified to be involved in the regulation of Vorganoids. In this innovative study, we effectively established an in vitro model of prevascularized dental pulp organoids and used it to elucidate new mechanisms of angiogenesis during regeneration, facilitating the development of clinical treatment strategies.
We downloaded the GSE161266 dataset from the Gene Expression Omnibus database[12] and performed the following analyses: (1) Quality control and selection of cells for further analysis; (2) Background correction; (3) Selection of high-variability features (identification of high-variability markers in single-cell populations as candidate regulatory genes); (4) Dimensionality reduction (principal component analysis, linear dimensionality reduction, and determination of the appropriate “dimensionality” of the dataset); (5) Clustering of cell populations based on principal component analysis; and (6) Nonlinear dimensionality reduction by the UMAP/tSNE algorithm. Identification of differentially expressed genes (DEGs) in cell subpopulations. Dataset quality control was performed by removing cells with < 200 expressed genes, low-quality/dying cells, and empty droplets (cells with a mitochondrial genome accounting for > 5% of the total genome were selected)[13-15]. Subsequently, global scaling normalisation of overall expression was performed for the cells included in the analyses, and the expression levels were log-transformed. To reduce the noise in the data, Seurat’s nonlinear dimensionality reduction algorithm was used to extract the principal components of the core data regarding the “meta-features” of the clustered dataset. Based on the similarity of the cell expression profiles, cells were grouped into highly related subpopulations and clusters using the KNN algorithm[13,15].
Cell clusters were reannotated by the SingleR and scCATCH algorithms[14]. For the cell clusters with inconsistent annotation results, cell markers were visualised for analysis and determination of the cell subpopulation. Pseudotime-series analysis was performed on the entire cell population and core cell subpopulations. The Monocle algorithm was used to analyse the serial changes in gene expression experienced by each cell during the process of cell-state transition; this algorithm reveals the overall “trajectory” of gene expression changes and defines the appropriate regulatory point of each cell in the trajectory[16]. Based on the Monocle algorithm, cells were arranged in 2D space according to their global expression profiles, and the cell state trajectory was plotted. Subsequently, DEGs with kinetic correlations were identified by differential expression analysis based on the pseudotime values[13,15].
Based on the above analyses, core cell subpopulations were selected, and DEGs with kinetic correlations were subjected to pathway enrichment analysis: (1) The clusterProfiler package was used for Gene Ontology (GO) enrichment analysis (biological process, molecular function, and cellular component)[17]; and (2) GSVA was used to calculate the enrichment scores of pathways[18]: (1) Cell expression profiles were obtained from the preparation of samples for calculation of gene expression; (2) In preparation of the gene set, the core pathway gene list was downloaded from the MSIGDB database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp); and (3) The gene set was introduced into the algorithm, the pathways were scored by the GSVA algorithm for each sample, and the pathway scores were subjected to relative quantification.
The RNA-seq data units corresponding to the FPKM values of the samples were included. Differential gene expression analysis was performed on the samples from the two clusters with the most significant difference in survival. The DESeq algorithm was used to normalise the gene expression profiles and filter out genes with low expression[19]. The criteria for selecting DEGs were as follows: Log2-fold change ≥ 1.5 and Benjamini-Hochberg (B-H) adjusted P value < 0.05. Through unsupervised cluster analysis, the identified DEGs were clustered according to the sample group, and GO enrichment analysis of these genes was subsequently performed to explore their potential biological functions through the ToppGene Suite (https://toppgene.cchmc.org)[20].
Gene set enrichment analysis (GSEA) was performed to identify the core pathways and regulatory genes whose expre
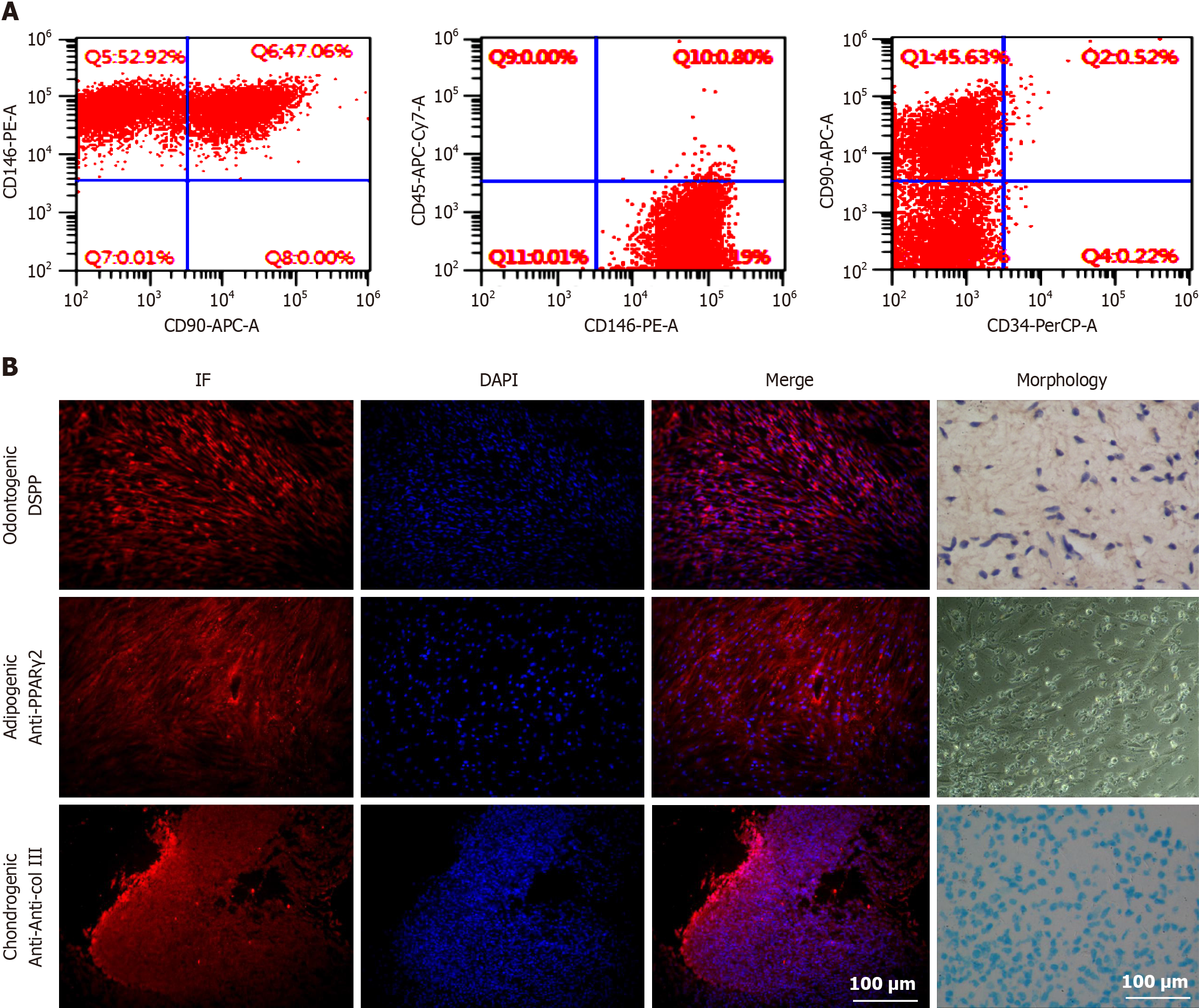
The hDPSCs were collected from pulp tissues of extracted third molars from patients aged 18 to 25 years (12 males and 8 females). Cells from the first to fifth passages were used in this study. All patients were informed, agreed to participate in this study, and signed an informed consent. The study protocol was approved by the Ethics Committee of Guangdong Second Provincial General Hospital. Multiple differentiation assays for hDPSCs were performed according to a pre
The hDPSCs were washed and resuspended in phosphate buffered saline (containing 1% foetal bovine serum) and then labelled with monoclonal anti-human CD146-PE, CD90-APC, CD34-PerCP and CD45-APC (Chemicon, Temecula, CA) for 30 min. The cells were washed twice, resuspended in staining buffer and analysed by flow cytometry.
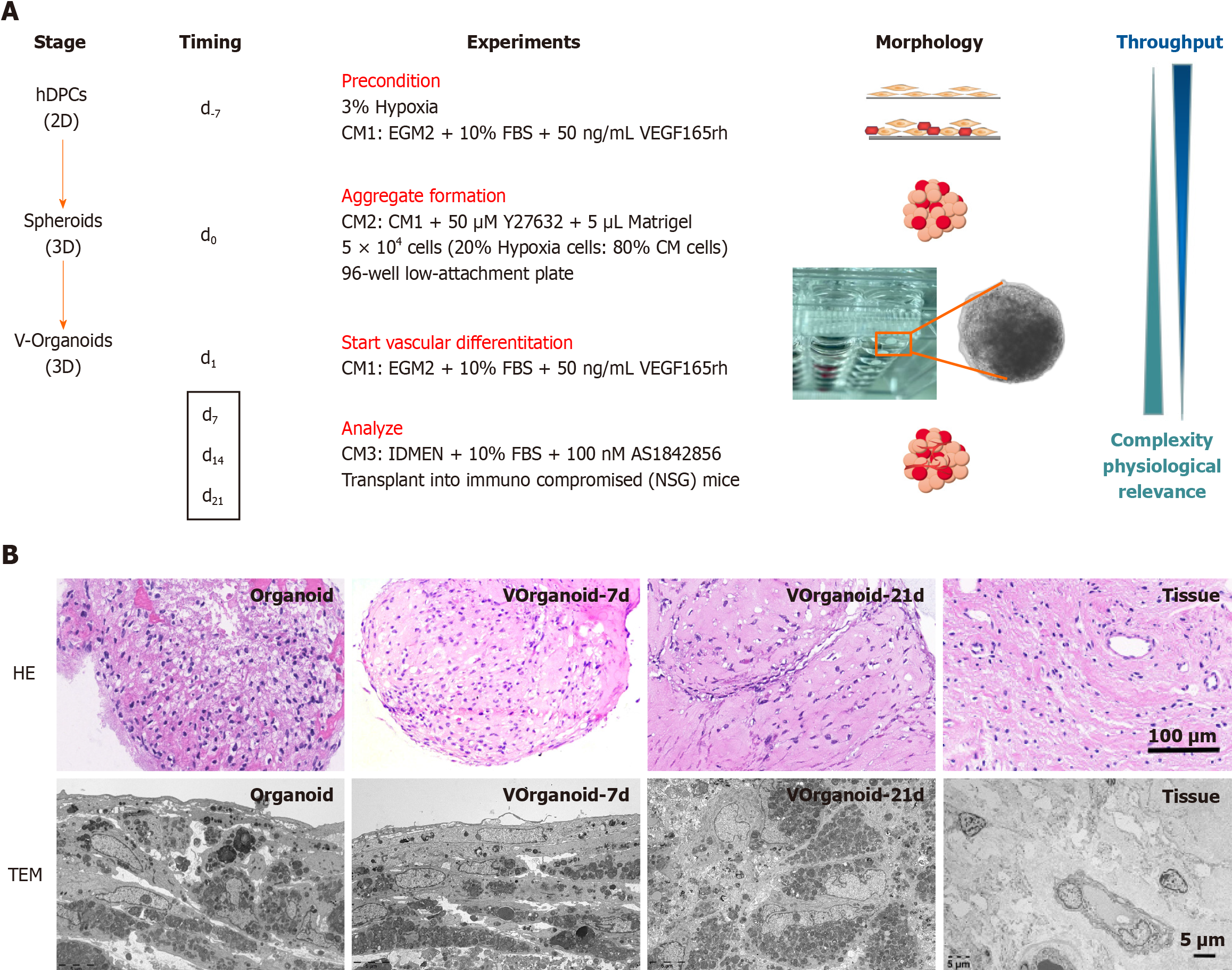
Preconditioning: Primary hDPSC cultures were incubated under 3% hypoxia beginning at passage 1. When the cells had expanded to passage 3, vasculogenic conditioned medium 1 (CM1) was added, and the culture was continued under hypoxic conditions for 7 d. CM1: EGM2 + 10% foetal bovine serum + 50 ng/mL VEGF 165 rh.
Aggregate formation: Hypoxic cells were then mixed with conditionally induced cells at a 2:8 ratio (total 5 × 104 cells) in a 96-well low-attachment plate containing 150 μL of CM2 per well. CM2: CM1 + 50 μM Y27632 + 5 μL Matrigel. After 1 d of 3D culture, the medium was changed to CM1, and the incubation continued for 6 d.
Analysis: The medium was changed to normal medium, and the experimental application phase of the Vorganoids experimental model was started (Figure 1). The standard experimental procedures for[5] identifying the organoids were performed according to previous methods.
Various tissues were placed in 4% paraformaldehyde for histological examination. The tissues were dehydrated, embedded in paraffin, cut into 4 μm thick slices, and baked in an oven at 60 °C for 3 h. Paraffin was removed for hematoxylin-eosin (HE) staining, and the sections were observed and photographed under a microscope (Nikon, Tokyo, Japan).
For morphological analysis, the samples were then fixed with 2.5% glutaraldehyde for 12 h at room temperature. The samples were dehydrated in a graded series of ethanol (50%-100%), permeabilized with propylene oxide, and embedded in a poly/bed 812 kit (Polysciences, Washington, PA, United States). After embedding and polymerising the pure fresh resin in an electron microscope oven (DOSAKA) for 24 h, the initial sections were cut at approximately 50-200 nm, stained with toluidine blue (Sigma), and examined via light microscopy. Sections of approximately 70 nm were double stained with 6% uranyl acetate and lead citrate (Thermo Fisher Scientific, Waltham, MA, United States) for comparison. These sections were cut using a Leica EM UC-7 (Leica Microsystems, Tokyo, Japan) instrument equipped with a diamond knife (Diatome, Hatfield, PA, United States) and then transferred to copper and nickel grids. A transmission electron mic
Samples from each group were fixed with 4% paraformaldehyde for 24 h and subsequently incubated with 0.5% Triton-X for 10 min. Bovine serum albumin (1%) was used to block the cells for 1 h. Samples were dehydrated in sections after paraffin embedding. After blocking with 5% goat serum (Life Technologies, New York, NY, United States) for 1 h at 20 °C, the specimens were then treated with rabbit anti-CD31 (Cell Signaling Technology, Boston, United States), anti-VE-cadherin (Cell Signaling Technology, Boston, United States), FGF2 (Ab208687-40, Abcame, Shanghai, United States), anti-FOXO1 (Cell Signaling Technology, Boston, United States), or anti-p-AKT (Cell Signaling Technology, Boston, United States) antibodies for 12 h at 4 °C. Alexa Fluor 594-conjugated goat anti-rabbit secondary antibody (GB25303; Sevicebio, Wuhan, China) was added, and the cells were incubated for 2 h at room temperature. The nuclei were stained with 4’,6-diamidino-2-phenylindole. CD31, VE-cadherin, FGF2, FOXO1, and p-AKT expression was observed under a fluorescence microscope (Nikon Eclipse, Tokyo, Japan). Statistical analysis was performed using GraphPad Prism 7 Software (San Diego, CA, United States).
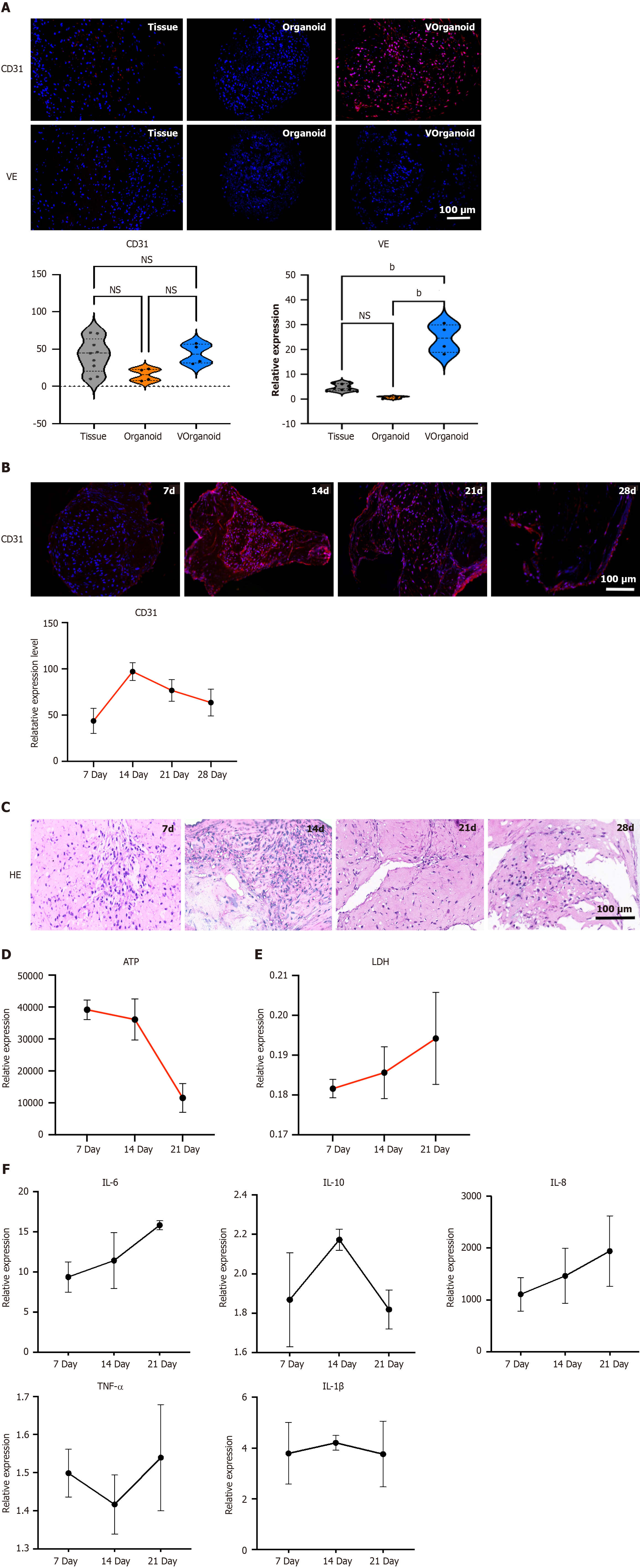
To quantify the metabolic activity of Vorganoids formed at different stages. RIPA buffer was added to lyse the Vorganoids, which were subsequently centrifuged at 4 °C for 5 min at 12000 rpm, after which the supernatant was removed for adenosine triphosphate (ATP) determination. The ATP assay working solution was prepared according to the kit instructions (S0026, Beyotime, China), and an ATP standard curve was constructed. The ATP working solution (100 μL) was added to the test wells and allowed to stand for 3-5 min at room temperature to eliminate background effects. Then, an appropriate amount of sample or standard was added and mixed well. The relative light unit value was measured using a chemiluminescence metre.
To quantify the degree of cell death at Vorganoids that formed at different stages, the culture supernatants were changed to serum-free DMEM 24 h before the culture medium was aspirated from each well, and the sediment was removed by centrifugation at 1000 rpm for 5 min according to the manufacturer’s protocol. A kit was used to measure the lactate dehydrogenase (LDH) concentration in each group of cultures according to the manufacturer’s instructions (C0017, Beyotime, China).
To quantify the expression of inflammatory markers in VOrganoids that formed at different stages, the levels of interleukin (IL)-6, IL-8, IL-10, IL-1β, and tumour necrosis factor-α in culture supernatants were assayed via enzyme-linked immunosorbent assay (BioLegend) according to the manufacturer’s instructions.
The data were analysed using IBM SPSS Statistics 22.0 and Image-Pro Plus 6.0, and the normality and variance of the data distribution were analysed using the Kolmogorov-Smirnov test and Levene’s test, respectively. Means were compared between two groups by t tests, and means were compared between multiple groups by one-way ANOVA with Bon
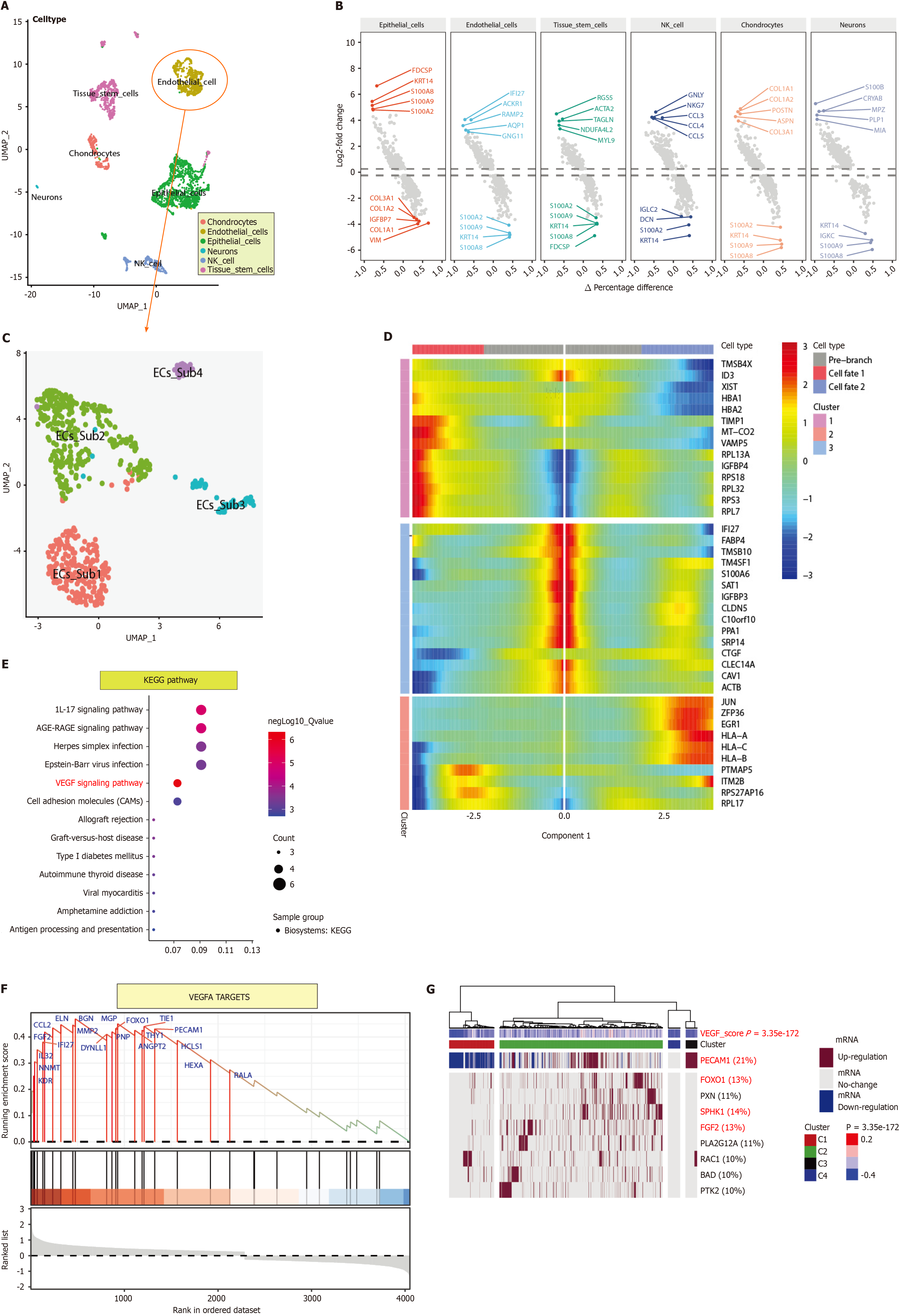
After quality control, we found no differences in the cell cycle distribution between the single-cell subpopulations. Cell clustering by UMAP resulted in 13 cell subpopulations in two spatial dimensions, which were annotated and divided into six classes: Chondrocytes, ECs, epithelial cells, neurons, natural killer cells, and tissue stem cells (Figure 1A). Among the DEGs, FDCSP, KRT14, S100A8, S100A9, and S100A2 were the most common upregulated DEGs, while COL3A1, COL1A2, IGFBP7, COL1A1, and VIM were the most common. For ECs, the upregulated DEGs were IFI27, ACKR1, RAMP2, AQP1, and GNG11, and the downregulated DEGs were S100A2, S100A9, KRT14, and S100A8. For chondrocytes, COL1A1, COL1A2, POSTN, ASPN, and COL3A1 were upregulated DEGs, while S100A2, KRT14, S100A9, and S100A8 were downregulated DEGs (Figure 1B).
ECs were re-extracted and reclustered. The 2D cell distribution determined by UMAP was used to analyse the cell spatial clustering, and four subpopulations were obtained (Figure 1C). Pseudotime-series analysis revealed that the cell po
Pseudotime-series analysis of ECs revealed that CRIP1, IFITM1, and B2M were expressed at high levels in fate2 ECs, while TMSB4X, ID3, and XIST were expressed at low levels. In fate1 cells, MT-ND4, TIMP1, and MT-CO2 were highly expressed, while PTMAP5, ITM2B, and RPS27AP16 were expressed at low levels. In differentiating cells, MT-RNR1, MT-RNR2, and DNASE1L3 were expressed at low levels, while IFI27, FABP4, and TMSB10 were highly expressed (Figure 1D). Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed that the VEGF signalling pathway (B-H adjusted P value = 4.63E-06, gene count = 4), the IL-17 signalling pathway (B-H adjusted P value = 1.18E-05, gene count = 5), and the AGE-RAGE signalling pathway in diabetic complications (B-H adjusted P value= 1.60E-05, gene count = 5) were the predominant pathways enriched (Figure 1E).
GSEA showed that WESTON-VEGFA TARGETS (enrichment score = 0.47, normalised enrichment score = 2.11, and B-H adjusted P value = 0.018) was associated with endodontium differentiation and progression. BGN, ELN, FOXO1, TIE1, and PECAM1, which had relatively high running enrichment scores, were considered key regulators of dental pulp development (Figure 1F). In addition, GSVA and ternary cluster analyses revealed a significant difference in the VEGF signalling pathway among the dental pulp ECs (P = 3.35e-172). PECAM1 (21%), FGF2 (13%), FOXO1 (13%), and SPHK1 (14%) were the core genes with high variability (Figure 1G).
To investigate the multidirectional differentiation potential of hDPSCs, we first established hDPSCs from pulp tissues extracted from third molars (patient age: 15 to 25 years). Cell surface marker identification experiments showed that the hDPSCs were derived from mesenchymal tissue and expressed mesenchymal-specific surface markers (Figure 2A). Multidirectional differentiation experiments revealed that hDPSCs could differentiate into osteogenic cells, cartilage cells, and adipocytes and express their corresponding specific markers (Figure 2B). Collectively, these data proved that hDPSCs exhibit multidirectional differentiation potential.
The cells began to aggregate into clusters at approximately 6 h and formed a single spherical morphology after approximately 1 d. The final diameter of the Vorganoid was approximately 400-600 μm. After the addition of 0.3% Triton X to permeabilize the Vorganoids for 30 min, the cells were unevenly distributed within the Vorganoids, and flocculent and irregularly distributed within the Vorganoids were observed. Long-term cultures of Vorganoids showed irregular cell proliferation and hyaline stroma at the edges, and Vorganoids were cultured continuously for more than 42 d in vitro (Figure 3A). HE staining and transmission electron microscopy revealed that the cells at the edge of the Vorganoids were arranged in a spindle row complex with normal intracellular organelles and fewer necrotic cells. In contrast, lysosome-like vesicles, which are polygonal in shape and tend to be compressed, appear in central cells. Compared to those in organoid culture, Vorganoids in culture had a greater proportion of Matrigel, a looser cell density and internal structure, fewer necrotic cells, and a greater distribution density and morphology similar to those of dental pulp tissue (Figure 3B).
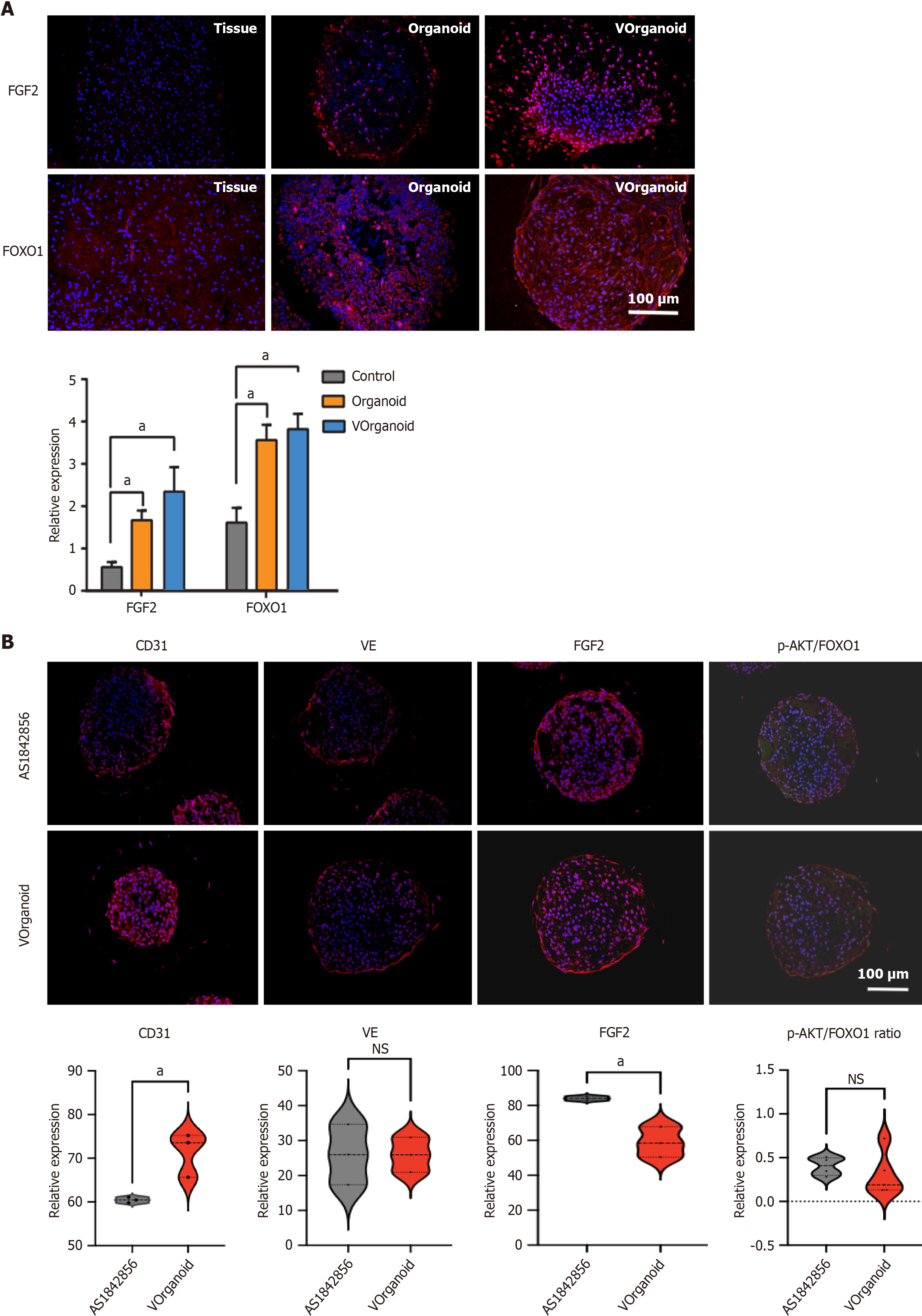
Differences in the expression of the angiogenic markers CD31 and VE were compared in three different tissues. CD31 was weakly expressed in pulp tissue, and VE was less expressed; CD31 was weakly expressed in organoids, while VE was not expressed; and the fluorescence intensities of CD31 and VE were greater in Vorganoid than in pulp tissue and organoids (Figure 4A). CD31 expression increased in the early stages of culture, reaching a peak at 14 d, with a sub
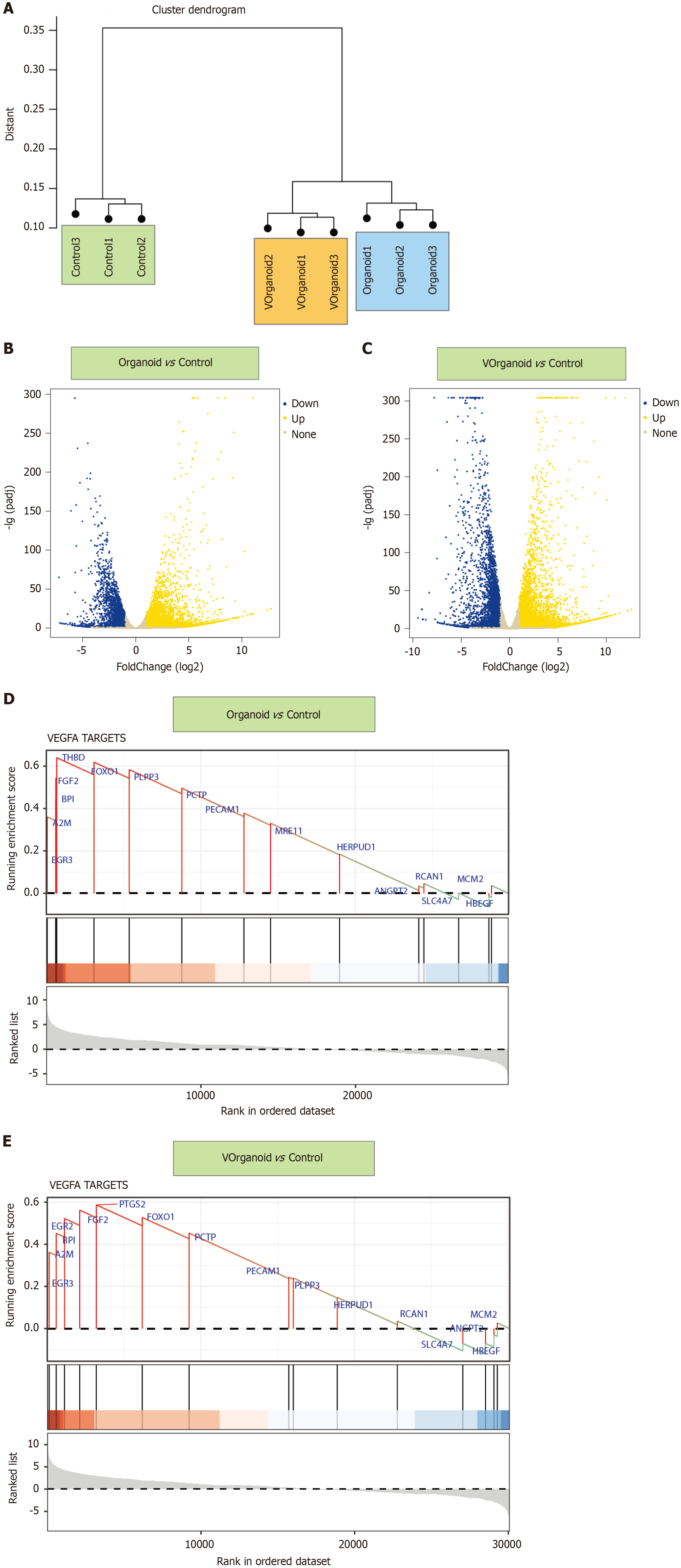
By sample clustering analysis, we found that the pulp cells were well differentiated between the 2D/3D culture and normal groups (Figure 5A). Compared with control, organoid showed 4171 and 2598 up- and downregulated genes, respectively, and Vorganoid showed 5227 and 3355 up- and downregulated genes, respectively (Figure 5B and C). GSEA showed that the DEGs in the VEGFA pathway were significantly enriched in the organoid and Vorganoid groups relative to the control group (organoid vs control: Enrichment score = 0.61, NES = 1.22, P.adjust = 0.0082; Vorganoid vs control: Enrichment Score = 0.64, NES = 1.63, P.adjust = 0.0058) (Figure 5D and E).
The above analyses revealed that the VEGFA TARGETS pathway may be the major pathway associated with the pro
Immunofluorescence staining revealed that the increase in FOXO1 and FGF2 expression was significantly greater in the Vorganoids group (P < 0.05) (Figure 6A). After 7 d of addition of the FOXO1 inhibitor AS1842856 to the Vorganoid medium, a decrease in CD31 expression and an increase in FGF2 expression were observed, as was a decrease in the ratio of p-AKT/FOXO1 expression, with no significant change in VE expression (Figure 6B).
Revascularization plays an important role in tissue engineering. In this study, first, prevascularized human pulp or
To more thoroughly analyse the mechanisms involved in pulp tissue angiogenesis, we extracted single-cell sequencing data from public databases, further analysed the key regulatory pathways linking EC subpopulations in the tissue to angiogenesis, and performed a temporal analysis of key genes at different temporal and spatial nodes[27]. ECs were dominant in Vorganoid culture, and the proportion of ECs in the cell population was quite high in Vorganoid culture. Combined with the RNA-seq results, these findings revealed that the pathways associated with VEGFA targets were significantly enriched in the development of Vorganoids. Among the regulatory factors, the FOXO1 and FGF2 biomarkers were found to be involved in the regulation of Vorganoids.
FOXO1 is a key transcription factor involved in a wide range of biological functions. It is widely expressed in vascular ECs, and vascular cell adhesion molecule 1, intercellular adhesion molecule 1 and E-selectin have been reported to be downstream regulators of vascular ECs[28,29]. Systematic knockout of the FOXO1 gene impaired angiogenesis and killed embryos, but specific knockout of the FOXO1 gene in the adult mouse myocardium did not affect cardiac function[30,31]. Previous experiments showed that FOXO1 transcript expression was reduced in hypoxic culture but increased when the culture environment was changed to conditional induction culture or 3D organoid formation (Figure 7). Thus, FOXO1 may regulate different gene transcripts that act at different stages of angiogenesis, particularly during embryonic deve
The ability of cellular grafts to repair damaged tissues is limited, and the introduction of vascularized grafts brings them closer to the function and maturation of the corresponding tissues. This technique has the potential to overcome the limitations of many other models, such as maintaining in vitro accessibility and scalability, but significant improvements are still needed. In conclusion, the finding of this study suggested that this new model could be applied in the field, which may pave the way for future dental regeneration prospects. In addition, vascularized organoids are more biologically similar to normal tissue. In addition to tissue regeneration and repair, Vorganoids can also be used for disease modelling, toxicity testing and drug screening. The use of human 3D organoids, along with other advances in single-cell technology, has revealed unprecedented insights into human biology and disease mechanisms.
Current organoid models do not fully replicate all cell types, levels of cell maturation, and physiological functions of their respective organs. They only exhibit some of the organ’s functions. In this study, we effectively established an in vitro model of prevascularized dental pulp organoids and used it to elucidate new mechanisms of angiogenesis. Our results suggest that the biomarkers FOXO1 and FGF2 confirm the angiogenic regulatory role of Vorganoids. However, to understand the mechanisms by which organoids interact between structure and function, further investigation is required. Additionally, the use of organoids in simulating inflammation in clinically relevant diseases and the immunogenicity of dental materials should be studied.
Stem cells can self-organise into microsized organ units, which can partially model tissue function and regeneration. Dental pulp organoids have been used to replicate the processes of tooth development and related diseases. However, the lack of vasculature limits the usefulness of dental pulp organo.
The survival of stem cell transplants should be promoted, thereby improving the repair ability of the cells.
Three-dimensional (3D) self-assembly of a novel vascularised dental pulp-like organoid in vitro by hypoxia and con
Human dental pulp stem cells were induced from endothelial cells (ECs) through exposure to a hypoxic environment and conditioned medium. The resulting cells were then mixed with ECs at specific ratios and conditioned in a 3D environment to produce Vorganoids. The biological characteristics of the Vorganoids were analysed, and the regulatory pathways associated with angiogenesis were studied.
Vorganoids are similar in morphology and function to dental pulp tissue. Single-cell RNA sequencing of dental pulp tissue and RNA sequencing of Vorganoids were performed to identify the involvement of the biomarkers forkhead box protein O1 (FOXO1) and fibroblast growth factor 2 (FGF2) in key regulatory pathways associated with Vorganoid angiogenesis.
In this study, we effectively established an in vitro model of prevascularized dental pulp organoids and used it to elucidate novel mechanisms of angiogenesis during dental regeneration. The biomarkers FOXO1 and FGF2 confirmed the angiogenesis-regulating role of angiopoietins.
This innovative study has effectively established an in vitro model of prevascularized dental pulp organoids and used it to elucidate new mechanisms of angiogenesis during regeneration, facilitating the development of clinical treatment strategies.
| 1. | Roche ET, Hastings CL, Lewin SA, Shvartsman D, Brudno Y, Vasilyev NV, O'Brien FJ, Walsh CJ, Duffy GP, Mooney DJ. Comparison of biomaterial delivery vehicles for improving acute retention of stem cells in the infarcted heart. Biomaterials. 2014;35:6850-6858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 122] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 2. | Karp JM, Leng Teo GS. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell. 2009;4:206-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1012] [Cited by in RCA: 1107] [Article Influence: 65.1] [Reference Citation Analysis (0)] |
| 3. | Corsini NS, Knoblich JA. Human organoids: New strategies and methods for analyzing human development and disease. Cell. 2022;185:2756-2769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 109] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 4. | Wimmer RA, Leopoldi A, Aichinger M, Kerjaschki D, Penninger JM. Generation of blood vessel organoids from human pluripotent stem cells. Nat Protoc. 2019;14:3082-3100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 204] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 5. | Jeong SY, Lee S, Choi WH, Jee JH, Kim HR, Yoo J. Fabrication of Dentin-Pulp-Like Organoids Using Dental-Pulp Stem Cells. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 6. | Hemeryck L, Hermans F, Chappell J, Kobayashi H, Lambrechts D, Lambrichts I, Bronckaers A, Vankelecom H. Organoids from human tooth showing epithelial stemness phenotype and differentiation potential. Cell Mol Life Sci. 2022;79:153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 7. | Katata C, Sasaki JI, Li A, Abe GL, Nör JE, Hayashi M, Imazato S. Fabrication of Vascularized DPSC Constructs for Efficient Pulp Regeneration. J Dent Res. 2021;100:1351-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 8. | Stegen S, van Gastel N, Eelen G, Ghesquière B, D'Anna F, Thienpont B, Goveia J, Torrekens S, Van Looveren R, Luyten FP, Maxwell PH, Wielockx B, Lambrechts D, Fendt SM, Carmeliet P, Carmeliet G. HIF-1α Promotes Glutamine-Mediated Redox Homeostasis and Glycogen-Dependent Bioenergetics to Support Postimplantation Bone Cell Survival. Cell Metab. 2016;23:265-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 156] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 9. | Bertassoni LE. Progress and Challenges in Microengineering the Dental Pulp Vascular Microenvironment. J Endod. 2020;46:S90-S100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 10. | Carvalho GL, Sarra G, Schröter GT, Silva LSRG, Ariga SKK, Gonçalves F, Caballero-Flores HV, Moreira MS. Pro-angiogenic potential of a functionalized hydrogel scaffold as a secretome delivery platform: An innovative strategy for cell homing-based dental pulp tissue engineering. J Tissue Eng Regen Med. 2022;16:472-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 11. | Liu F, Huang X, Luo Z, He J, Haider F, Song C, Peng L, Chen T, Wu B. Hypoxia-Activated PI3K/Akt Inhibits Oxidative Stress via the Regulation of Reactive Oxygen Species in Human Dental Pulp Cells. Oxid Med Cell Longev. 2019;2019:6595189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 12. | Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol. 2016;1418:93-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 869] [Cited by in RCA: 1544] [Article Influence: 154.4] [Reference Citation Analysis (0)] |
| 13. | Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2049] [Cited by in RCA: 2984] [Article Influence: 426.3] [Reference Citation Analysis (1)] |
| 14. | Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7652] [Cited by in RCA: 8714] [Article Influence: 1089.3] [Reference Citation Analysis (0)] |
| 15. | Sinha D, Sinha P, Saha R, Bandyopadhyay S, Sengupta D. Improved dropClust R package with integrative analysis support for scRNA-seq data. Bioinformatics. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Slovin S, Carissimo A, Panariello F, Grimaldi A, Bouché V, Gambardella G, Cacchiarelli D. Single-Cell RNA Sequencing Analysis: A Step-by-Step Overview. Methods Mol Biol. 2021;2284:343-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 146] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 17. | Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 5860] [Article Influence: 1172.0] [Reference Citation Analysis (0)] |
| 18. | Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7222] [Cited by in RCA: 10417] [Article Influence: 801.3] [Reference Citation Analysis (3)] |
| 19. | Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12015] [Cited by in RCA: 11748] [Article Influence: 734.3] [Reference Citation Analysis (8)] |
| 20. | Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305-W311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1911] [Cited by in RCA: 2319] [Article Influence: 136.4] [Reference Citation Analysis (0)] |
| 21. | Gandia C, Armiñan A, García-Verdugo JM, Lledó E, Ruiz A, Miñana MD, Sanchez-Torrijos J, Payá R, Mirabet V, Carbonell-Uberos F, Llop M, Montero JA, Sepúlveda P. Human dental pulp stem cells improve left ventricular function, induce angiogenesis, and reduce infarct size in rats with acute myocardial infarction. Stem Cells. 2008;26:638-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 274] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 22. | Iohara K, Imabayashi K, Ishizaka R, Watanabe A, Nabekura J, Ito M, Matsushita K, Nakamura H, Nakashima M. Complete pulp regeneration after pulpectomy by transplantation of CD105+ stem cells with stromal cell-derived factor-1. Tissue Eng Part A. 2011;17:1911-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 235] [Article Influence: 15.7] [Reference Citation Analysis (1)] |
| 23. | Dissanayaka WL, Zhan X, Zhang C, Hargreaves KM, Jin L, Tong EH. Coculture of dental pulp stem cells with endothelial cells enhances osteo-/odontogenic and angiogenic potential in vitro. J Endod. 2012;38:454-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 24. | Dissanayaka WL, Hargreaves KM, Jin L, Samaranayake LP, Zhang C. The interplay of dental pulp stem cells and endothelial cells in an injectable peptide hydrogel on angiogenesis and pulp regeneration in vivo. Tissue Eng Part A. 2015;21:550-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 148] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 25. | Xu X, Li Z, Ai X, Tang Y, Yang D, Dou L. Human three-dimensional dental pulp organoid model for toxicity screening of dental materials on dental pulp cells and tissue. Int Endod J. 2022;55:79-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Wörsdörfer P, Ergün S. The Impact of Oxygen Availability and Multilineage Communication on Organoid Maturation. Antioxid Redox Signal. 2021;35:217-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 27. | Vargas-Valderrama A, Messina A, Mitjavila-Garcia MT, Guenou H. The endothelium, a key actor in organ development and hPSC-derived organoid vascularization. J Biomed Sci. 2020;27:67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 28. | Wang S, Shi M, Li J, Zhang Y, Wang W, Xu P, Li Y. Endothelial cell-derived exosomal circHIPK3 promotes the proliferation of vascular smooth muscle cells induced by high glucose via the miR-106a-5p/Foxo1/Vcam1 pathway. Aging (Albany NY). 2021;13:25241-25255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Zhuang T, Liu J, Chen X, Zhang L, Pi J, Sun H, Li L, Bauer R, Wang H, Yu Z, Zhang Q, Tomlinson B, Chan P, Zheng X, Morrisey E, Liu Z, Reilly M, Zhang Y. Endothelial Foxp1 Suppresses Atherosclerosis via Modulation of Nlrp3 Inflammasome Activation. Circ Res. 2019;125:590-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 146] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 30. | Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ Res. 2010;107:1470-1482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 589] [Cited by in RCA: 576] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 31. | Qi Y, Zhu Q, Zhang K, Thomas C, Wu Y, Kumar R, Baker KM, Xu Z, Chen S, Guo S. Activation of Foxo1 by insulin resistance promotes cardiac dysfunction and β-myosin heavy chain gene expression. Circ Heart Fail. 2015;8:198-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cordelier P, France; Li SC, United States S-Editor: Wang JJ L-Editor: A P-Editor: Zhao YQ