©The Author(s) 2025.
World J Gastroenterol. Dec 14, 2025; 31(46): 112664
Published online Dec 14, 2025. doi: 10.3748/wjg.v31.i46.112664
Published online Dec 14, 2025. doi: 10.3748/wjg.v31.i46.112664
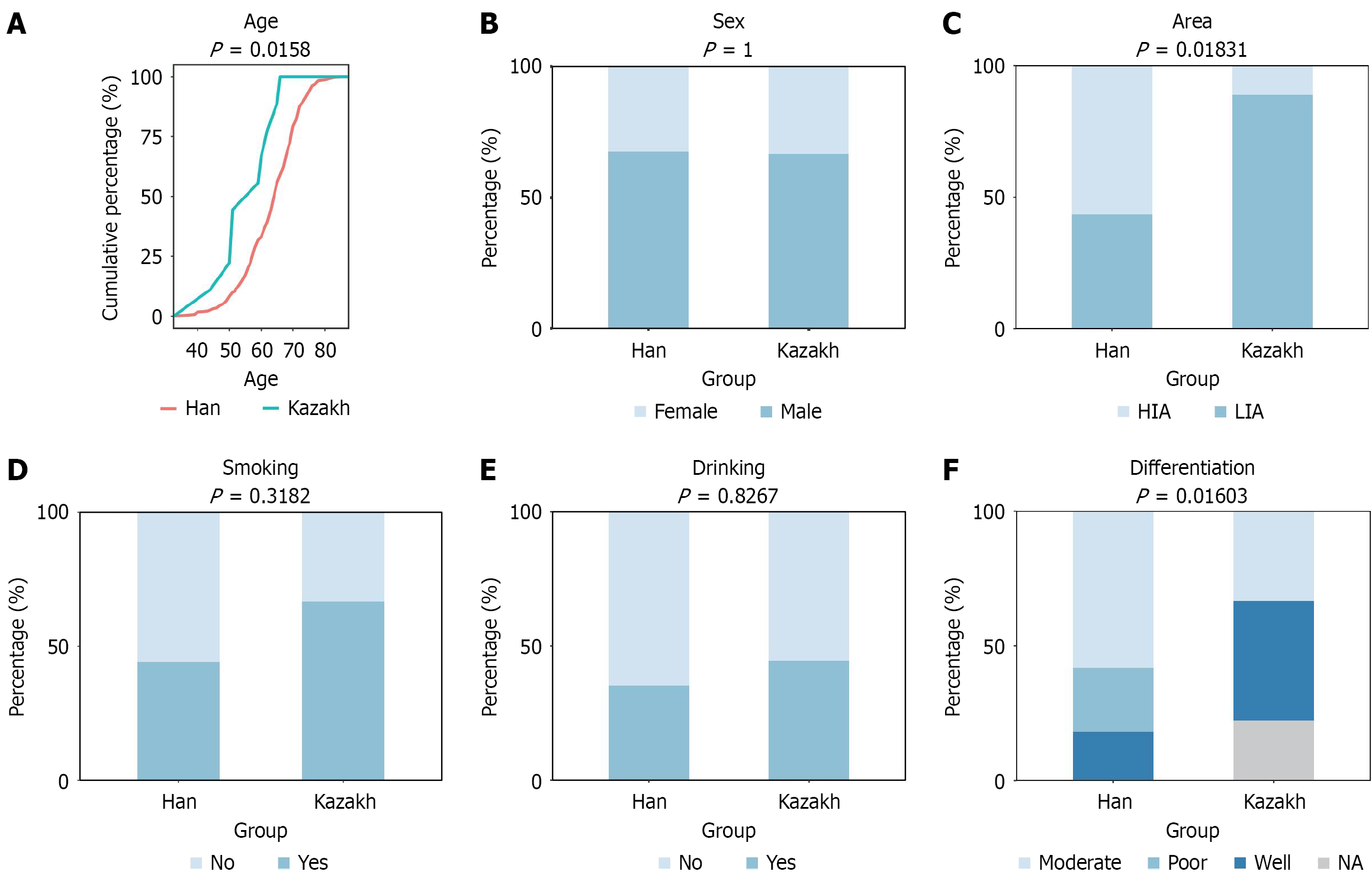
Figure 1 Distribution of five biological factors in two patient groups, justifying a need to balance these confounders.
A: Diagnosis; B: Sex; C: Area; D: Smoking; E: Alcohol drinking; F: Differentiation. Analysis of variance test was used to calculate the P value for age at diagnosis, and χ2 test was used for the other factors. HIA: High incidence area; LIA: Low incidence area.
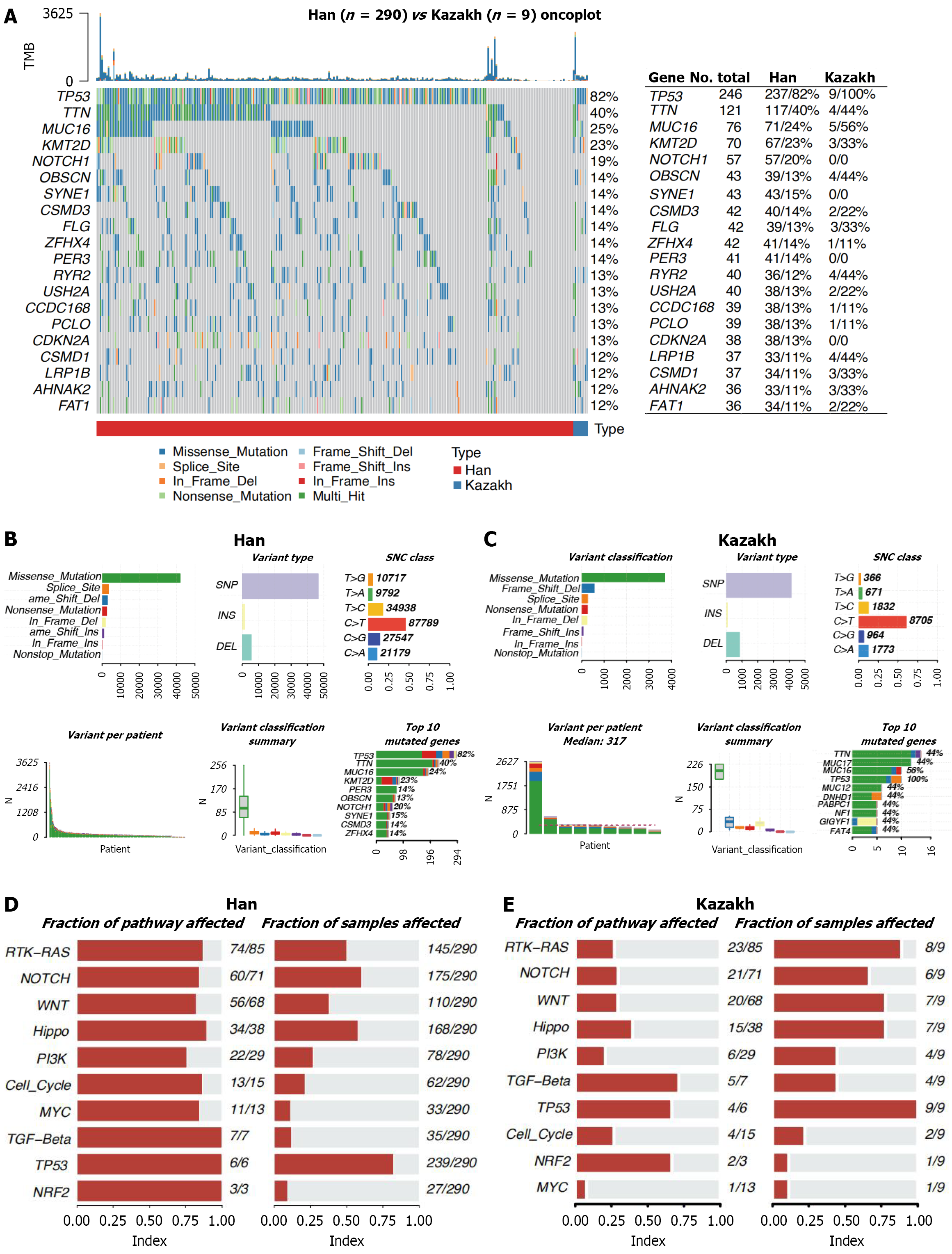
Figure 2 Significantly mutated genes in esophageal squamous cell carcinoma.
A: Significantly mutated genes (SMGs) identified by MutSigCV on a combined cohort of Han and Kazakh ethnics groups, Chinese whole-exome sequence samples. Each column denotes an esophageal squamous cell carcinoma patient, and each row is a gene. On top is the number of somatic mutations per sample. On the right are the mutation frequencies of each SMG. The bar plot on the top shows the composition of mutations in the gene. Genes are ordered by their mutation frequencies. The table on the right shows the number of mutation population of total, Han and Kazakh; B and C: Visualizing mutation summary. This maftools plot shows a summary of the multiple alignment format file. Highlighting the most mutated genes, single-nucleotide variant class, and variant classification distributions within a tumor type for Han and Kazakh patients; D and E: Fraction of pathway and sample affected for Han and Kazakh. SNV: Single nucleotide variant; SNP: Single nucleotide polymorphism; INS: Insertion; DEL: Deletion; PI3K: Phosphatidylinositol 3-kinase; TGF: Transforming growth factor; NRF2: Nuclear factor erythroid 2-related factor 2.
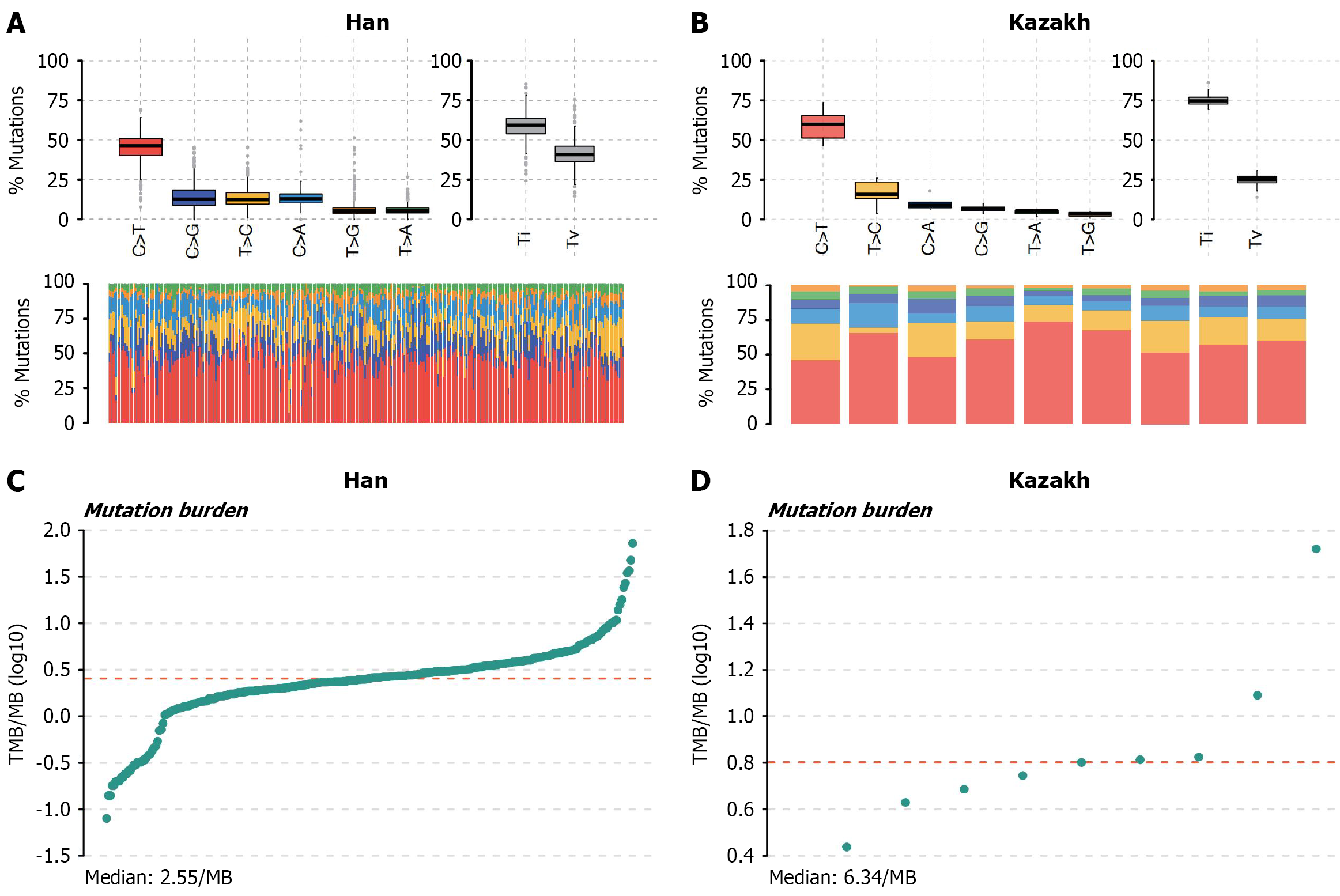
Figure 3 Variant calling and mutation burden.
A and B: The transition and transversion plot illustrates the distribution of single-nucleotide variants (above) in esophageal squamous cell carcinoma, categorized into six transition and transversion events. The core purpose is to describe the distribution characteristics (such as central tendency, dispersion degree, outliers, etc.) of a single set of data (for example, a certain transformation/transposition event). The stacked bar chart (bottom) displays the mutational spectrum distribution of each sample in the multiple alignment format file. Present the “proportion distribution of categorical data” in a visual way to assist in understanding the heterogeneity of the mutation spectrum among different samples; C and D: Mutation burden. The core is to describe the distribution characteristics of tumor mutational burden among the samples, including the central tendency (such as the median) and the degree of dispersion (such as the range of variation). TMB: Tumor mutation burden; MB: Mega base.
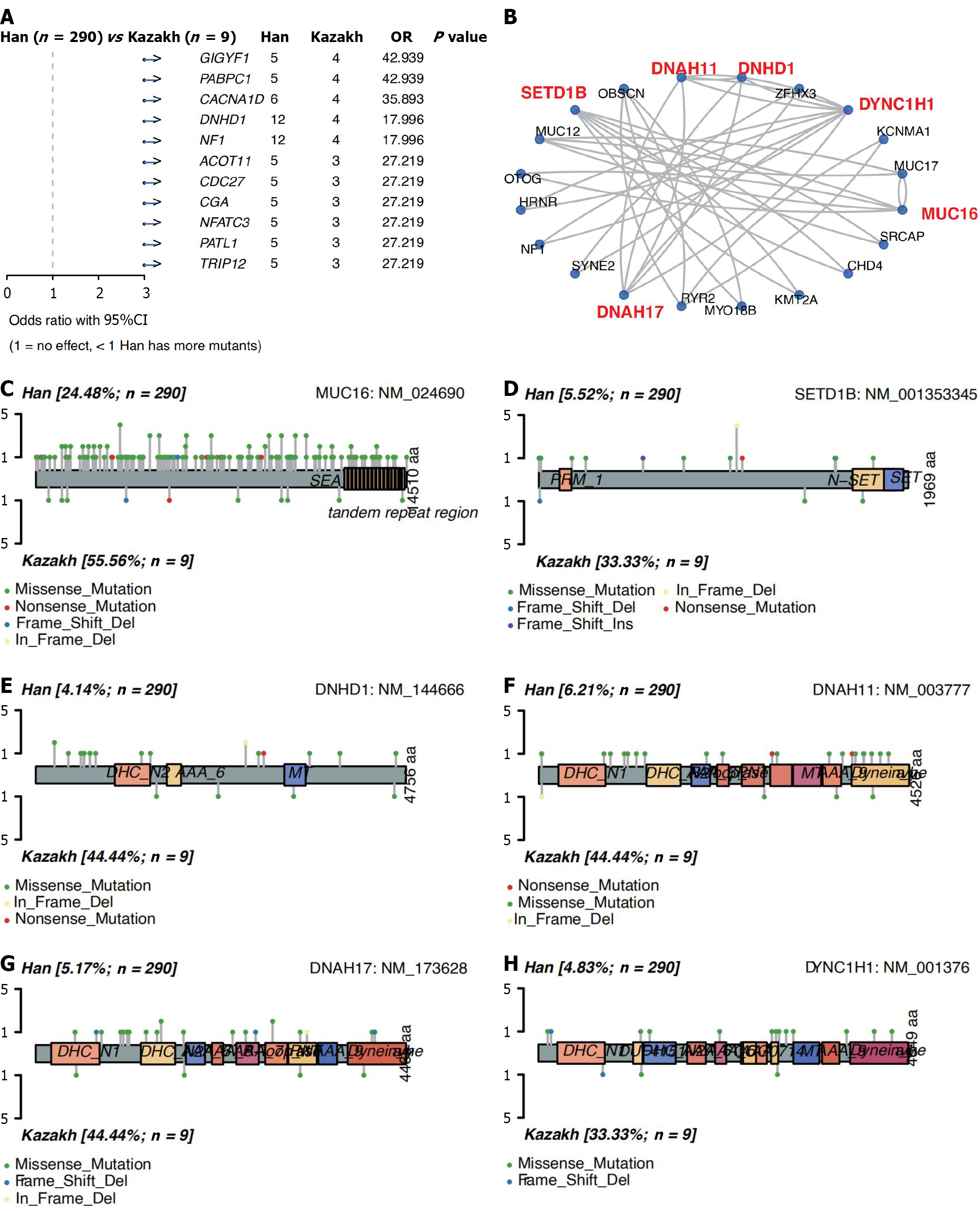
Figure 4 Difference genes and protein domain.
A: Difference gene between the two ethnics groups (P < 0.001); B: Gene-gene interaction network between difference and significant mutation genes (P < 0.05, and the mutation frequency ≥ 5%); C-H: Schematics of protein changes in ethnics-biased gene products. Scattered distribution of somatic mutations in MUC16, SETD1B, DNHD1, DNAH11, DNAH17, DYNC1H1. OR: Odds ratio; CI: Confidence interval.
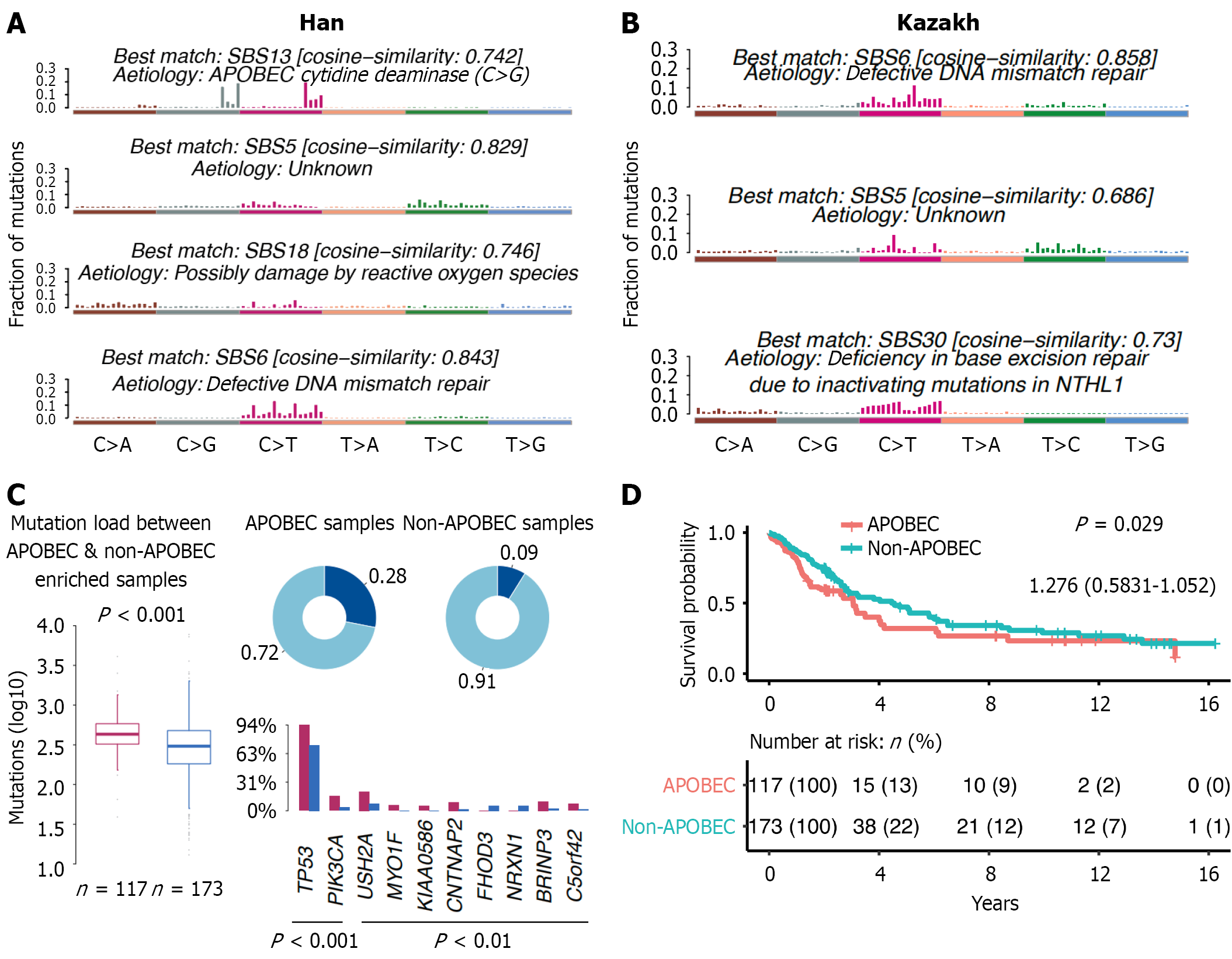
Figure 5 Signature plot.
A: Han; B: Kazakh patients. Each row presents a specific single base substitutions (SBS) signature (Catalogue of Somatic Mutations in Cancer reference signatures with known etiologies) and its best match with known signatures. The distribution of each signature shows the relative frequencies of the six single-base substitution types. Different SBS signatures are associated with different pathogenic mechanisms, such as apolipoprotein B messenger RNA-editing enzyme catalytic polypeptide (APOBEC) cytidine deaminase, unknown mechanisms, possible reactive oxygen species damage, and DNA mismatch repair deficiency, etc.; C: Differences between APOBEC enriched and non-enriched patients for Han patients (none of the samples are enriched for APOBEC in Kazakh patients); D: Survival plot of Han patients. SBS: Single base substitution; APOBEC: Apolipoprotein B messenger RNA-editing enzyme catalytic polypeptide.
- Citation: Wei MX, Lei LL, Xu RH, Liu YX, Wang R, Han WL, Fan ZM, Xiao FK, Sheyhidin I, Ma L, Ku JW, Yin MZ, Ji AF, Bao QD, Gao SG, Han XN, Li XM, Chen PN, Zhao XK, Song X, Wang LD. Ethnic genomic differences in esophageal squamous cell carcinoma: Whole-exome sequencing of Han and Kazakh populations in China. World J Gastroenterol 2025; 31(46): 112664
- URL: https://www.wjgnet.com/1007-9327/full/v31/i46/112664.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i46.112664