Published online Dec 18, 2025. doi: 10.13105/wjma.v13.i4.108504
Revised: May 16, 2025
Accepted: September 18, 2025
Published online: December 18, 2025
Processing time: 246 Days and 16.9 Hours
Periodontitis is a chronic inflammatory disorder influenced by both behavioral and genetic factors. Among epigenetic regulators, the ANRIL gene has been pro
To analyze the association between the rs1333048 genetic polymorphism in the ANRIL gene and periodontitis via meta-analysis.
A literature search was performed for studies published before May 2, 2025. The Review Manager statistical program was used in analyses with calculations of heterogeneity index (I²) and odds ratio (OR) with 95% of confidence intervals (CI). Begg’s test and the Egger’s linear regression test were used for publication bias evaluation using Comprehensive meta-analysis software. P < 0.05 was considered significant.
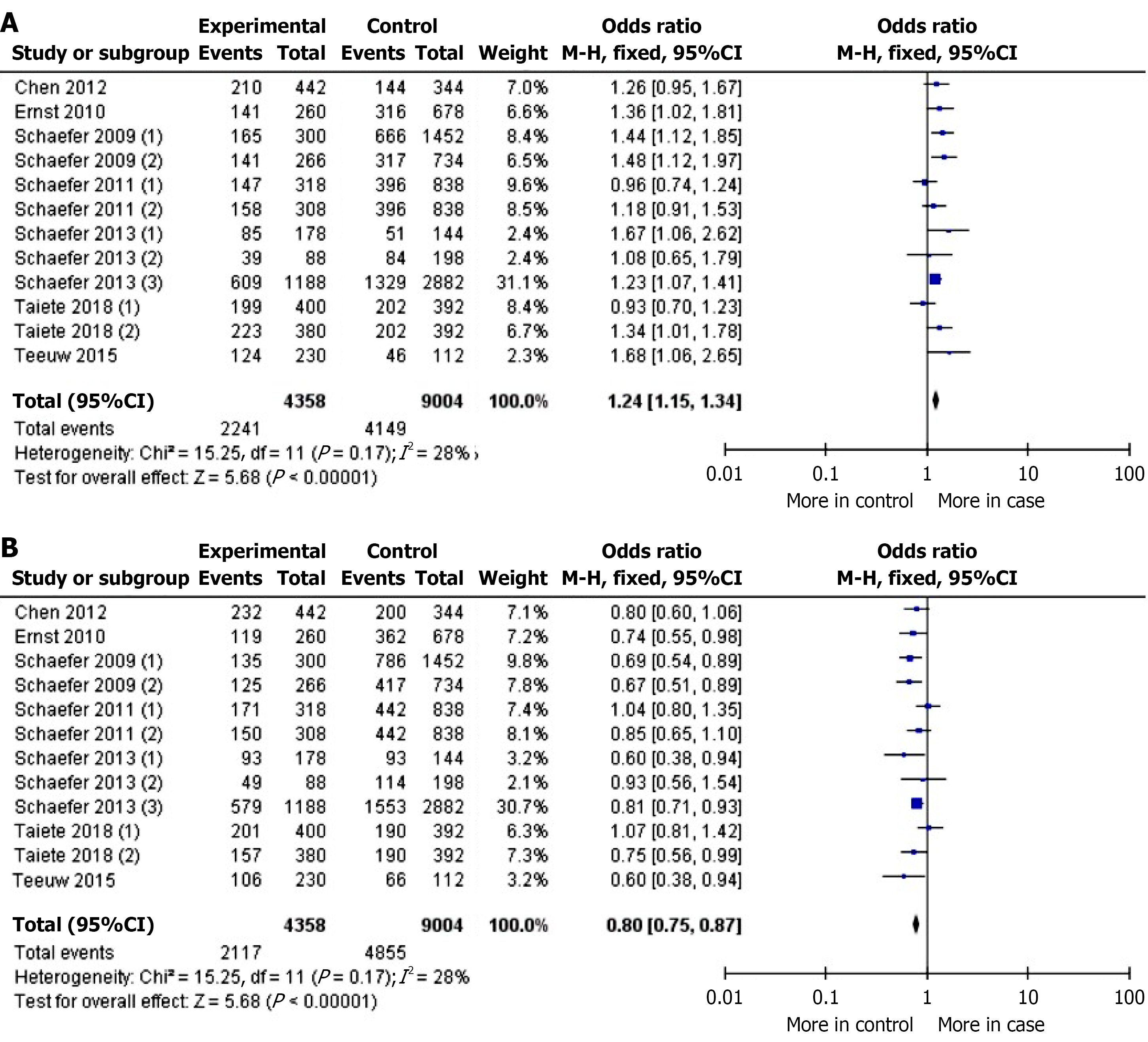
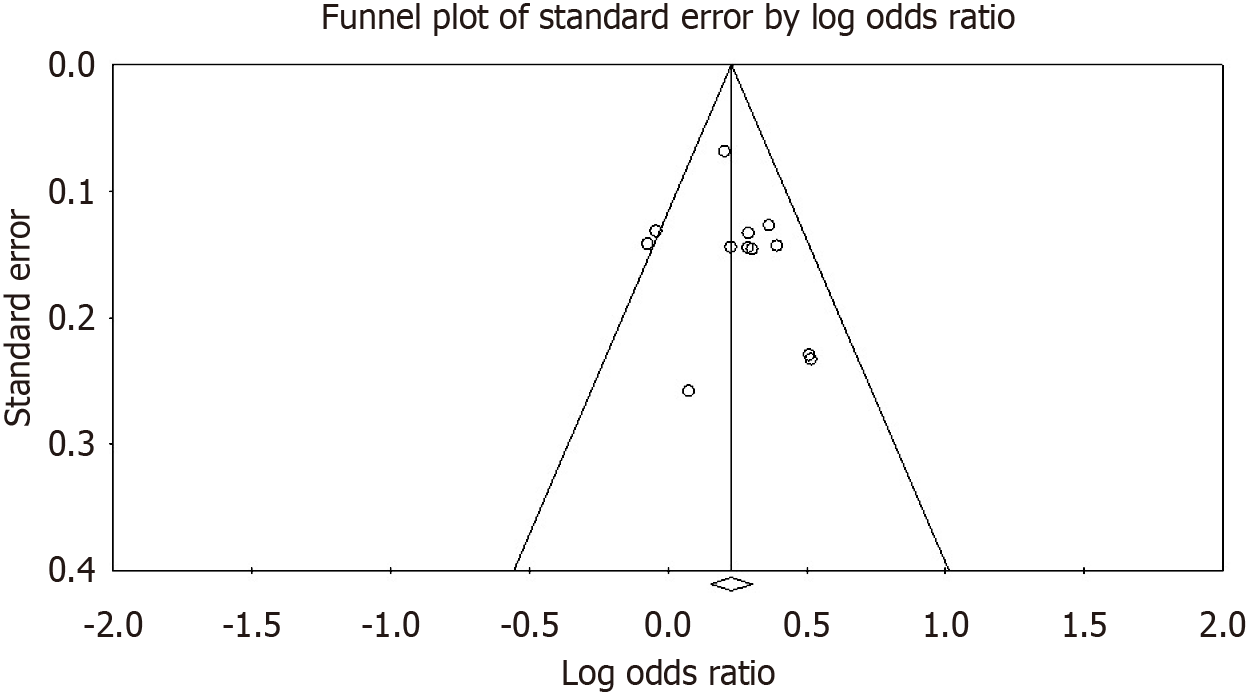
From 12 studies including 5489 participants across multiple ethnic groups, we observed a statistically significant association between ANRIL gene polymorphisms and periodontitis in the allelic contrast model (OR = 1.24 95%CI: 1.15-1.34, P < 0.00001). Conversely, the wild-type allele was significantly associated with the control group (OR = 0.80 95%CI: 0.75-0.87, P < 0.00001). Heterogeneity was low (I² = 28%, Pheterogeneity = 0.17), and no significant risk of publication bias was detected (P > 0.05).
In conclusion, this meta-analysis demonstrated a significant association between the rs1333048 polymorphism and periodontitis in the overall analysis and in stratified analyses of Caucasian populations, but in for mixed-race populations.
Core Tip: Many case-control studies have identified a relationship between the rs1333048 polymorphism and periodontitis. However, the impact of this genetic variation on the clinical aspect of periodontitis is still unknown.
- Citation: da Silva FRP, Leal ALAB, Galeno JG, Monteiro AVO, Araujo FAH, de Oliveira Tavares K, Souza Monteiro JR, de Oliveira ACA, Pereira AL, da Cunha Pereira ACT, Vasconcelos DFP. Association of the rs1333048 polymorphism in the ANRIL gene with different clinical forms of periodontitis: A meta-analysis. World J Meta-Anal 2025; 13(4): 108504
- URL: https://www.wjgnet.com/2308-3840/full/v13/i4/108504.htm
- DOI: https://dx.doi.org/10.13105/wjma.v13.i4.108504
Periodontal diseases arise from dysbiotic shifts in the oral microbiota, with the host immune response serving as the central etiological factor. Hyper-inflammation drives tissue damage in the periodontium, ultimately leading to tooth loss[1]. Periodontal diseases are classified in several ways, with chronic periodontitis (CP) and aggressive periodontitis (AgP) being the most common forms.
The epidemiology of periodontitis is substantial, with recent data indicating a high global prevalence. Severe forms affect approximately 10% of the global population[2]. In the United States, periodontitis has been reported in 42% of adults aged 30 years and older[3].
Multiple factors contribute to the development and progression of periodontitis, including poor oral hygiene, specific subgingival bacterial species[4], and genetic variations in inflammatory mediators[5]. Epigenetic events also play an important role by altering local gene expression linked to inflammation[6].
Epigenetic events are complex but potentially reversible processes that influence gene expression without altering DNA sequence. They involve mechanisms such as DNA methylation, histone modifications and regulation by long non-coding RNAs (lnRNAs)[7]. It has been hypothesized that lnRNAs interact with histone-modifying machinery and thereby contribute to diverse biological conditions, ranging from cancer to inflammatory diseases.
The antisense non-coding RNA in the INK4 locus (ANRIL) is one such lnRNA, identified as a candidate molecule in periodontitis. ANRIL regulates the expression of multiple genes through both cis- and trans-acting mechanisms, inte
Schaefer et al[9] first reported significant associations between two ANRIL polymorphisms (rs1333042 and rs1333048) and generalized AgP. However, a subsequent study in a Han Chinese population did not find a significant association between these polymorphisms and the disease[10].
Other studies have reported on the associations between single nucleotide polymorphisms (SNPs) in the ANRIL gene and periodontitis[11,12]. However, the results remained unclear and are challenging to reconcile. Contradictory findings may be better evaluated through meta-analyses, as demonstrated by many previous studies on polymorphisms and inflammatory conditions[13-15].
Although a previous meta-analysis has attempted to clarify the relationship between the rs1333048 polymorphism and periodontitis[16], the authors’ approach did not allow a complete evaluation on this genetic variation and periodontitis risk, as stratified which analyses by different clinical forms of periodontitis are lacking.
Therefore, this study aimed to perform a meta-analysis of the association between the rs1333048 polymorphism in the ANRIL gene and periodontitis risk, including an evaluation of stratified data.
This meta-analysis was registered in the PROSPERO database with the following assigned number: CRD42018116487.
For the design and conduct of this study, the authors followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines[17] and completed the corresponding PRISMA Checklist (Supplementary Table 1).
Is periodontitis directly influenced by a genetic variation in the ANRIL gene? What is the association between the rs1333048 polymorphism in ANRIL gene and periodontitis? What is the clinical significance of this polymorphism in relation to periodontitis?
The following inclusion criteria were applied in the literature search: (1) Case-control studies or replication genetic studies; (2) Diagnosis of periodontitis (in any of its clinical manifestations) confirmed through rigorous clinical evaluation[18]; (3) Control patients with clinically healthy periodontium; (4) Complete documentation of genotypic frequencies; (5) Studies conducted in human subjects; and (6) Participants (cases and controls) without cardiovascular diseases, autoimmune conditions, or pregnancy.
The literature search strategy was independently conducted by two authors in the Cochrane Library, EMBASE, Google Scholar, MEDLINE, PubMed and Web of Science. Studies published before May 2, 2025, were considered. The following combination of keywords was used: “periodontitis OR periodontal disease OR chronic periodontitis OR aggressive periodontitis” and “polymorphism OR genetic variation” and “ANRIL OR human chromosome region 9p21.3” and “rs1333048 polymorphism”.
No language restrictions were applied, and references from the included studies were evaluated for inclusion.
Three investigators collected the data using a standardized form, which was used to compile the table of study characteristics presented in the Results. The data were extracted by author, year of publication, ethnicity/country of the included participants, study design, clinical diagnosis, subject type evaluated, sample size, mean age of the participants, respect to the Hardy-Weinberg Equilibrium, genotyping method, risk of bias assessment, described below.
To assess methodological quality, the scale proposed by Nibali[19] for improving the quality of meta-analyses in periodontal genetic studies was applied. Studies that received fewer than 10 points on this binary scale were excluded.
The Review Manager software version 5.3 (RevMan, Nordic Cochrane Centre, The Cochrane Collaboration, 2012) for systematic reviews and meta-analyses was used for statistical analysis. Publication bias was evaluated using the Comprehensive Meta-analysis statistical software (Software version 3.3.070 (2014) available free online as a trial.
The presence or absence of true heterogeneity (I²) was calculated by Cochran’s Q test or the χ2Q-based statistical test. I² was analyzed through visualization of the Funnel plot for heterogeneity. When the observed value of I² presented a non-statistical significance and was defined as mild or moderate (I² < 50%, P > 0.05), the authors used the Fixed-effect model for the pooled odds ratio (OR) calculation. When I² presented a statistically significant value and was defined as elevated (I² > 50%, P < 0.05), the Random-effects statistical model was used for the OR calculations. A P value < 0.05 was considered statistically significant.
To quantify the influence of genetic variation on disease risk, six genetic models were calculated, using “M” to denote the major allele and “m” the minor allele. Therefore, the calculations included: (1) Allelic comparisons (M vs m; m vs M); (2) Genotypic comparisons (MM vs mm; mm vs MM); and (3) Combined genotypic models (MM vs mm + Mm; and Mm vs MM + mm). In addition, a sensitivity analysis was performed by omitting one study at a time to assess any significant changes in the OR value. For the assessment of publication bias, Begg’s test and Egger’s linear regression test were used (with P < 0.05). Funnel plot asymmetry for publication bias was also considered to validate Begg’s test and Egger’s test results. All included data in the studies have been dichotomous data expressed as OR with 95% of confidence intervals (CI) to determine the possible association between the genetic variations and periodontitis.
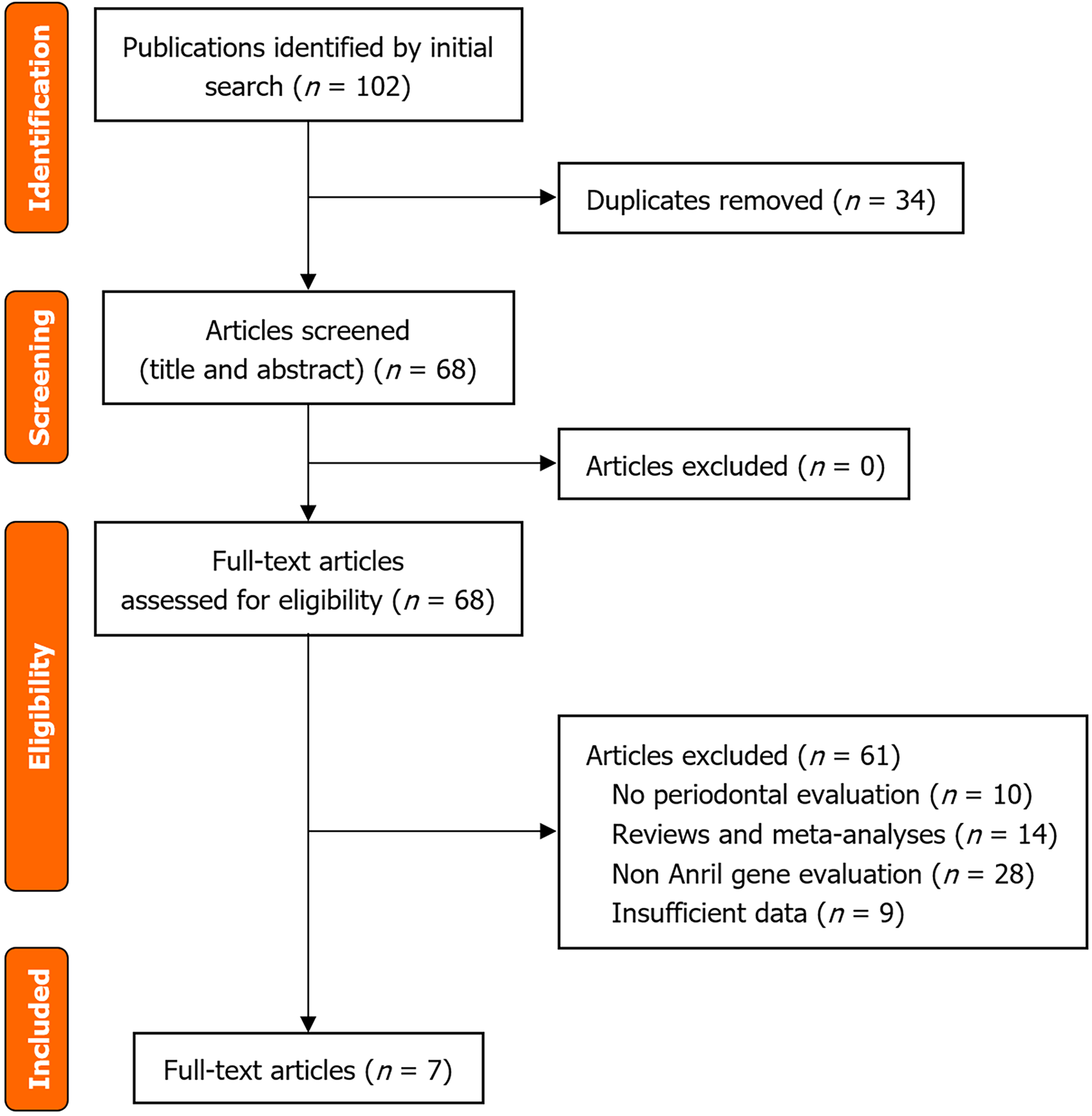
The initial literature search identified 102 records across the selected databases. After screening and evaluation of eligibility, 95 studies were excluded, leaving seven for inclusion in the qualitative synthesis (Figure 1). In total, the ana
| Reference | Ethnicity/country | Study design | Diagnosis (clinical subtypes) | Subject type | Sample size (Cc/Co) | Age (years) (Cc/Co) | HWE | Methods of genotyping | Score |
| Chen et al[10], 2012 | Asian/China | Case/Control | CP | - | 221/172 | 41.6 ± 8.8/40.0 ± 9.2 | Yes | Real time PCR | 14 |
| Ernst et al[11], 2010 | Caucasian/German and Northern Ireland | Case/Control | gAgP | - | 130/339 | 15-35/40-80 | Yes | Real time PCR | 13 |
| Schaefer et al[9], 2009 | Caucasian/Germany | Verification/Replication | gAgP | - | 151/736 | 38 ± 6.6/63 ± 7.4 | Yes | SNPlex/TaqMan | 14 |
| LAgP | - | 137/368 | 37 ± 7.0/37 ± 9.6 | ||||||
| Schaefer et al[12], 2011 | Caucasian/Germany and Netherlands | Case/Control | CP | - | 154/421 | 45.2 ± 9.5/32.3 ± 9.0 | Yes | SNPlex/TaqMan | 15 |
| AgP | 159/421 | 33.8 ± 4.7/32.3 ± 9.0 | |||||||
| Schaefer et al[20], 2013 | Caucasian/Turkey, Italy and Germany | Exploration | AgP | Turkey | 89/72 | - | Yes | Array Immunochip | 18 |
| Italy | 44/99 | Yes | |||||||
| Germany | 594/1441 | Yes | |||||||
| Taiete et al[21], 2018 | Mixed/Brazil | Case/Control | AgP | - | 200/196 | 34.0 ± 4.6/30.5 ± 5.8 | No | PCR-RFLP | 17 |
| CP | - | 190/196 | 50.0 ± 7.2/30.5 ± 5.8 | Yes | 16 | ||||
| Teeuw et al[22], 2015 | Caucasian/Netherlands | Case/Control | Periodontitis | - | 115/56 | 45.5 ± 9.9/43.9 ± 13.2 | Yes | TaqMan |
The included articles, published between 2009 and 2018, encompassed several ethnic groups and comprised seven articles with a total of 12 studies on the ANRIL gene polymorphism[9,10,11,12,20-22]. The studies were performed in Cau
In the included studies, periodontitis was diagnosed as CP[10,12,20,21] generalized AgP[9,11] and localized AgP[9]. All included studies reached more than 10 points on the scale proposed by Nibali[19] as shown in Table 1.
In the overall analysis, a statistically significant association was found between the mutant allele of the ANRIL gene polymorphism and increased risk of periodontitis (OR = 1.24, 95%CI: 1.15-1.34, P < 0.05). Conversely, the wild-type allele was significantly associated with the control group (OR = 0.80, 95%CI: 0.75-0.87, P < 0.05). A non-significant I² value (28%, Pheterogeneity = 0.17) was observed for both evaluations (Figure 2). Similar results were obtained for the mutant homozygous and wild-type homozygous genotypes. Due to the non-significant interference of heterogeneity, the Fixed-effect statistical model was applied. The OR values and data about I² are shown in Table 2. Sub-group evaluation demonstrated a significant association between the rs1333048 polymorphism in ANRIL gene and CP (P = 0.006), AgP (P < 0.05), and the Caucasian population (P < 0.05), but not in the mixed-race population (P = 0.55). The Fixed-effect statistical model was applied due to the decreased I² value. However, the calculations for the mixed-race population were affected by significant heterogeneity.
| Comparison (n) | OR (95%CI) | P value (Z test) | I², % | Pheterogeneity | Statistical model used |
| Overall (n = 12) | |||||
| M vs m | 1.24 (1.15-1.34) | P < 0.05 | 28 | P = 0.17 | F |
| m vs M | 0.80 (0.75-0.87) | P < 0.05 | 28 | P = 0.17 | F |
| MM vs mm | 1.53 (1.32-1.78) | P < 0.05 | 30 | P = 0.16 | F |
| mm vs MM | 0.65 (0.56-0.76) | P < 0.05 | 30 | P = 0.16 | F |
| MM vs Mm/mm | 1.38 (1.22-1.56) | P < 0.05 | 27 | P = 0.18 | F |
| Mm vs MM/mm | 0.97 (0.87-1.08) | P = 0.53 | 25 | P = 0.20 | F |
| CP (n = 3) | |||||
| M vs m | 1.25 (1.07-1.47) | P = 0.006 | 0 | P = 0.81 | F |
| m vs M | 0.80 (0.68-0.94) | P = 0.006 | 0 | P = 0.81 | F |
| MM vs mm | 1.58 (1.14-2.18) | P = 0.005 | 0 | P = 0.77 | F |
| mm vs MM | 0.63 (0.46-0.87) | P = 0.005 | 0 | P = 0.77 | F |
| MM vs Mm/mm | 1.36 (1.05-1.77) | P = 0.02 | 0 | P = 0.80 | F |
| Mm vs MM/mm | 0.98 (0.79-1.23) | P = 0.89 | 0 | P = 0.92 | F |
| AgP (n = 8) | |||||
| M vs m | 1.23 (1.13-1.34) | P < 0.05 | 47 | P = 0.07 | F |
| m vs M | 0.81 (0.75-0.89) | P < 0.05 | 47 | P = 0.07 | F |
| MM vs mm | 1.49 (1.25-1.77) | P < 0.05 | 48 | P = 0.06 | F |
| mm vs MM | 0.67 (0.57-0.80) | P < 0.05 | 48 | P = 0.06 | F |
| MM vs Mm/mm | 1.37 (1.19-1.58) | P < 0.05 | 51 | P = 0.05 | F |
| Mm vs MM/mm | 0.95 (0.84-1.08) | P = 0.44 | 49 | P = 0.06 | F |
| Caucasian (n = 9) | |||||
| M vs m | 1.27 (1.16-1.38) | P < 0.05 | 25 | P = 0.22 | F |
| m vs M | 0.79 (0.72-0.86) | P < 0.05 | 25 | P = 0.22 | F |
| MM vs mm | 1.59 (1.34-1.88) | P < 0.05 | 26 | P = 0.21 | F |
| mm vs MM | 0.63 (0.53-0.74) | P < 0.05 | 26 | P = 0.21 | F |
| MM vs Mm/mm | 1.39 (1.21-1.60) | P < 0.05 | 43 | P = 0.08 | F |
| Mm vs MM/mm | 0.97 (0.87-1.08) | P = 0.94 | 33 | P = 0.15 | F |
| Mixed (n = 2) | |||||
| M vs m | 1.11 (0.91-1.36) | P = 0.55 | 68 | P = 0.08 | R |
| m vs M | 0.90 (0.63-1.28) | P = 0.55 | 68 | P = 0.08 | R |
| MM vs mm | 1.22 (0.83-1.81) | P = 0.55 | 70 | P = 0.07 | R |
| mm vs MM | 0.82 (0.55-1.21) | P = 0.55 | 70 | P = 0.07 | R |
| MM vs Mm/mm | 1.26 (0.93-1.72) | P = 0.14 | 0 | P = 0.49 | F |
| Mm vs MM/mm | 0.84 (0.64-1.11) | P = 0.23 | 30 | P = 0.23 | F |
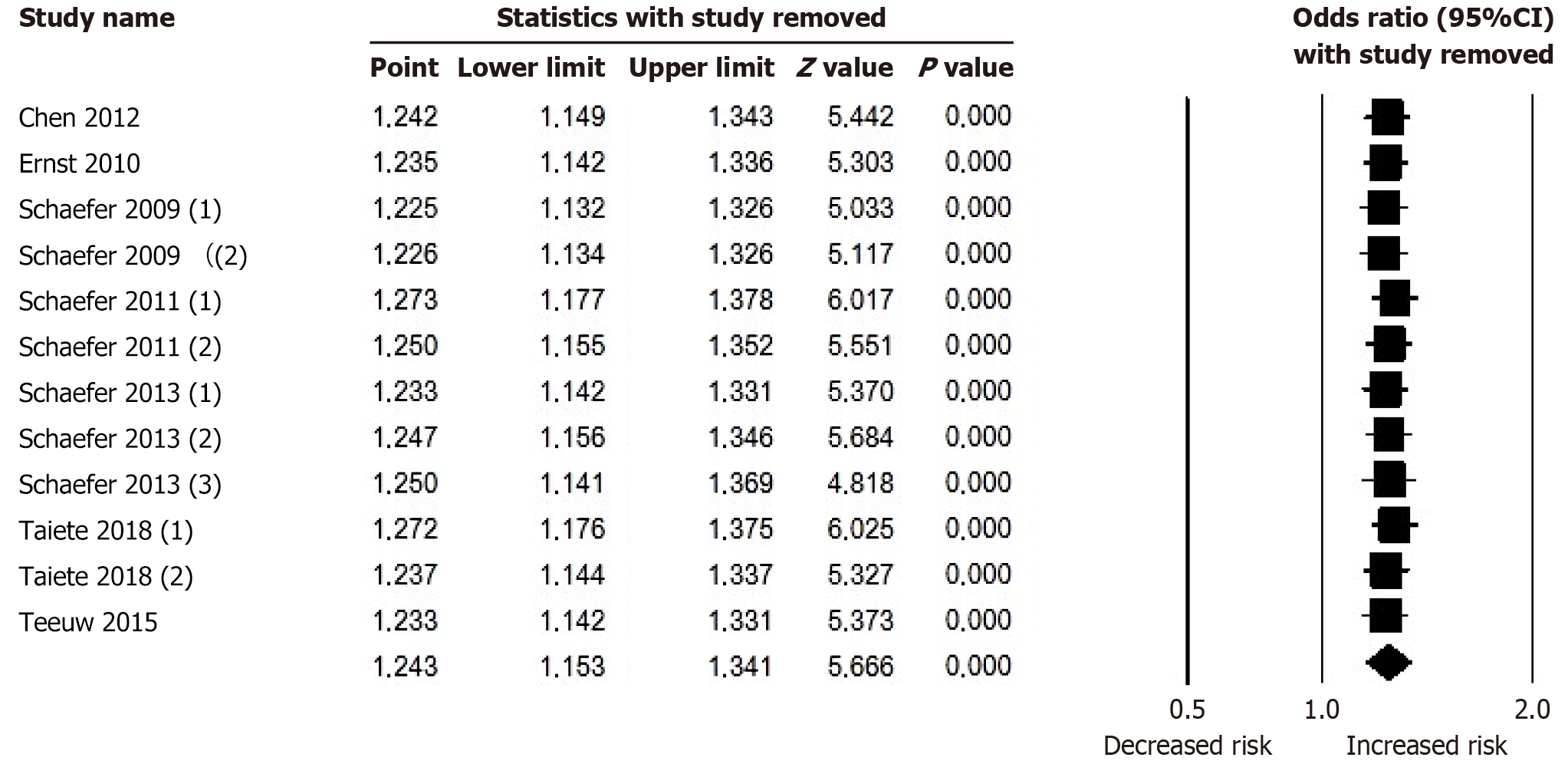
Sensitive analysis showed that no single study significantly altered the pooled OR values, supporting the robustness of the results (Figure 3). Furthermore, no obvious asymmetry was detected in the Funnel plot for the analyzed polymorphi
To the best of our knowledge, this is the first systematic evaluation with meta-analysis study to investigate the association between the rs1333048 polymorphism in the ANRIL gene and periodontitis, with subgroup analyses and a larger number of included studies and participants than previous meta-analyses in this field[16]. The non-significant heterogeneity, results of sensitivity analysis, absence of publication bias, and quality scores of the included studies all support the methodological rigor and robustness of our findings.
The ANRIL gene consists of 19 exons spanning 126.3 kb within the p15/CDKN2B-p16/CDKN2A-p14/ARF gene cluster at the 9p21.3 locus[23]. At the genetic level, our results demonstrated a significant association between the C allele of the rs1333048 polymorphism and periodontitis (P < 0.05), with an increased OR (OR = 1.24). The C allele also showed a notable minor allele frequency (MAF) in the global population (C = 0.442)[24]. These findings corroborate previous reports indicating that the MAF of the C allele was significantly associated with patients diagnosed with periodontitis[22]. Moreover, this mutant allele has also been linked to prostate cancer risk and benign prostatic hyperplasia in Caucasian patients from an Iranian population[25].
Previous findings are inconsistent with these associations. The rs1333048 polymorphism was associated with cancer risk only at the haplotype level, when combined with the mutant allele of rs333045 and the wild-type alleles of rs3977574 and rs10757278[26]. Similar associations have also been reported between other ANRIL gene SNPs and multiple sclerosis[27].
A previous in vitro experiment with vascular smooth muscle cells demonstrated that the ANRIL transcript can affect physiological pathways in a splicing variant-dependent manner, with exon 1 and exon 19 influencing the expression of IL-1A and IL-1RN, respectively[28]. It is well established that IL-1 belongs to the pro-inflammatory cytokine family, and polymorphisms in the IL1A[29] and IL1B[30] genes have been associated with periodontal inflammation. These findings suggest a possible molecular link between ANRIL expression and genetic variations previously implicated in periodontitis.
Bochenek[31] suggested a molecular linkage between ANRIL regulation and periodontitis pathways, identifying that ANRIL transcripts containing exon 13 were correlated with reduced expression of key genes involved in glucose and fatty acid metabolism, with polymorphisms in these genes associated with AgP. This relationship may be mediated by the interaction of ANRIL with polycomb repressive complexes, a group of proteins critical for chromatin remodeling[8] and inflammation[32]. In addition, ANRIL has been shown to interfere with Nuclear Factor kB (NF-kB) pathways[33], where NF-kB functions as a central mediator of cytokine expression and periodontal tissue disruption[34]. Recent findings further demonstrate that ANRIL overexpression can alter the transcription of genes involved in inflammation, NF-κB signaling, type I interferon-mediated signaling, and the innate immune response[35].
The significant association observed in the overall analysis was also evident when evaluating different forms of periodontitis. Although fewer studies focused on CP (n = 3) compared to AgP (n = 8), the results were similar across these clinical forms of the disease (Table 2), both showing a significant association with increased OR values.
The clinical classifications of CP and AgP were dissolved in the current periodontitis classification proposed by Caton et al[36], which unified these two forms under a single category of periodontitis. The current classification could introduce bias in our subgroup analysis evaluating the distinct clinical disease presentations. However, pooled overall analysis resolved this issue.
To further evaluate the influence of confounding factors in the overall analyses, this meta-analysis stratified the studies by participant ethnicity, distinguishing between Caucasian and mixed-race populations. We identified a significant asso
The rs333048 polymorphism was not significantly associated with periodontitis in the mixed-race population (P > 0.05). This may be explained by two important factors: The limited number of included studies (n = 2) and the increased I² value, which interferes with the statistical model applied on the data. Finally, the mixed-race population in the studies is from Brazil, and Brazilians are characterized by an elevated level of genetic admixture[37].
Although the results from this meta-analysis highlight an association between the rs1333048 polymorphism and periodontitis with complete ethnic evaluation, important limitations still exist. First, no studies were available in the li
In conclusion, this systematic meta-analysis composed by 12 studies and 5489 participants demonstrated a significant association between the rs1333048 polymorphism in the ANRIL gene and periodontitis for both primary clinical forms and the Caucasian population.
We thank teacher Abilio Borgi for assistance with English language review.
| 1. | Van Dyke TE, Sima C. Understanding resolution of inflammation in periodontal diseases: Is chronic inflammatory periodontitis a failure to resolve? Periodontol 2000. 2020;82:205-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 2. | Frencken JE, Sharma P, Stenhouse L, Green D, Laverty D, Dietrich T. Global epidemiology of dental caries and severe periodontitis - a comprehensive review. J Clin Periodontol. 2017;44 Suppl 18:S94-S105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 588] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 3. | Eke PI, Thornton-Evans GO, Wei L, Borgnakke WS, Dye BA, Genco RJ. Periodontitis in US Adults: National Health and Nutrition Examination Survey 2009-2014. J Am Dent Assoc. 2018;149:576-588.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 464] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 4. | Di Stefano M, Polizzi A, Santonocito S, Romano A, Lombardi T, Isola G. Impact of Oral Microbiome in Periodontal Health and Periodontitis: A Critical Review on Prevention and Treatment. Int J Mol Sci. 2022;23:5142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 161] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 5. | da Silva FRP, Pessoa LDS, Vasconcelos ACCG, de Aquino Lima W, Alves EHP, Vasconcelos DFP. Polymorphisms in interleukins 17A and 17F genes and periodontitis: results from a meta-analysis. Mol Biol Rep. 2017;44:443-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 6. | Larsson L. Current Concepts of Epigenetics and Its Role in Periodontitis. Curr Oral Health Rep. 2017;4:286-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 7. | Suzuki S, Yamada S. Epigenetics in susceptibility, progression, and diagnosis of periodontitis. Jpn Dent Sci Rev. 2022;58:183-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 8. | Aarabi G, Zeller T, Heydecke G, Munz M, Schäfer A, Seedorf U. Roles of the Chr.9p21.3 ANRIL Locus in Regulating Inflammation and Implications for Anti-Inflammatory Drug Target Identification. Front Cardiovasc Med. 2018;5:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 9. | Schaefer AS, Richter GM, Groessner-Schreiber B, Noack B, Nothnagel M, El Mokhtari NE, Loos BG, Jepsen S, Schreiber S. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009;5:e1000378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 178] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 10. | Chen D, Wei N, Bao XN, Wang LM, Zhou CF, Zhang YL, Zhang J. [Association of variants in chromosome 9p21.3 and chronic periodontitis in the Han Chinese population]. Shanghai Kou Qiang Yi Xue. 2012;21:659-662. [PubMed] |
| 11. | Ernst FD, Uhr K, Teumer A, Fanghänel J, Schulz S, Noack B, Gonzales J, Reichert S, Eickholz P, Holtfreter B, Meisel P, Linden GJ, Homuth G, Kocher T. Replication of the association of chromosomal region 9p21.3 with generalized aggressive periodontitis (gAgP) using an independent case-control cohort. BMC Med Genet. 2010;11:119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Schaefer AS, Richter GM, Dommisch H, Reinartz M, Nothnagel M, Noack B, Laine ML, Folwaczny M, Groessner-Schreiber B, Loos BG, Jepsen S, Schreiber S. CDKN2BAS is associated with periodontitis in different European populations and is activated by bacterial infection. J Med Genet. 2011;48:38-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | da Silva FRP, Galeno JG, Leal ALAB, Koga RS, Batista NY, da Conceição Furtado S, Vasconcelos DFP, Carvalho MD, Barcellos JFM. Non-significant association between - 330 T/G polymorphism in interleukin-2 gene and chronic periodontitis: findings from a meta-analysis. BMC Oral Health. 2020;20:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Silva FRP, Leal ALAB, Nibali L, Shin JI, Carvalho MD, Koga RS, de Andrade Figueira MB, Galeno JG, Toro DM, de Andrade ZG, Batista NY, Barcellos JFM. Lack of association between Mannose Binding Lectin-2 gene polymorphisms and periodontitis: a meta-analysis. Meta Gene. 2020;26:100757. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 15. | da Silva FRP, Leal ALAB, Koga RS, de Lira JASP, da Silva HA, Ayala KNR, Gomes PRC, da Cunha Pereira ACT, Vasconcelos DFP. Relationship between the rs333 polymorphism in the CC chemokine receptor type five (CCR5) gene and immunological disorders: data from a meta-analysis. Int J Stat Med Res. 2021;10:85-96. [DOI] [Full Text] |
| 16. | Öztürk A, Ada AO. The roles of ANRIL polymorphisms in periodontitis: a systematic review and meta-analysis. Clin Oral Investig. 2022;26:1121-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 17. | Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Moher D. Updating guidance for reporting systematic reviews: development of the PRISMA 2020 statement. J Clin Epidemiol. 2021;134:103-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 1384] [Article Influence: 276.8] [Reference Citation Analysis (0)] |
| 18. | Page RC, Eke PI. Case definitions for use in population-based surveillance of periodontitis. J Periodontol. 2007;78:1387-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 940] [Cited by in RCA: 1111] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 19. | Nibali L. Suggested guidelines for systematic reviews of periodontal genetic association studies. J Clin Periodontol. 2013;40:753-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 20. | Schaefer AS, Bochenek G, Manke T, Nothnagel M, Graetz C, Thien A, Jockel-Schneider Y, Harks I, Staufenbiel I, Wijmenga C, Eberhard J, Guzeldemir-Akcakanat E, Cine N, Folwaczny M, Noack B, Meyle J, Eickholz P, Trombelli L, Scapoli C, Nohutcu R, Bruckmann C, Doerfer C, Jepsen S, Loos BG, Schreiber S. Validation of reported genetic risk factors for periodontitis in a large-scale replication study. J Clin Periodontol. 2013;40:563-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 21. | Taiete T, Casati MZ, Stolf CS, Corrêa MG, Santamaria MP, Andere NMRB, Coletta RD, Sallum EA, Nociti Júnior FH, Silvério KG, Casarin RCV. Validation of reported GLT6D1 (rs1537415), IL10 (rs6667202), and ANRIL (rs1333048) single nucleotide polymorphisms for aggressive periodontitis in a Brazilian population. J Periodontol. 2019;90:44-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 22. | Teeuw WJ, Laine ML, Bizzarro S, Loos BG. A Lead ANRIL Polymorphism Is Associated with Elevated CRP Levels in Periodontitis: A Pilot Case-Control Study. PLoS One. 2015;10:e0137335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 23. | Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 361] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 24. | NCBI. National Center for Biotechnology Information. Available from: https://www.ncbi.nlm.nih.gov/snp/rs1333048#frequency_tab. |
| 25. | Taheri M, Pouresmaeili F, Omrani MD, Habibi M, Sarrafzadeh S, Noroozi R, Rakhshan A, Sayad A, Ghafouri-Fard S. Association of ANRIL gene polymorphisms with prostate cancer and benign prostatic hyperplasia in an Iranian population. Biomark Med. 2017;11:413-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 26. | Khorshidi HR, Taheri M, Noroozi R, Sarrafzadeh S, Sayad A, Ghafouri-Fard S. ANRIL Genetic Variants in Iranian Breast Cancer Patients. Cell J. 2017;19:72-78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 27. | Rezazadeh M, Gharesouran J, Moradi M, Noroozi R, Omrani MD, Taheri M, Ghafouri-Fard S. Association Study of ANRIL Genetic Variants and Multiple Sclerosis. J Mol Neurosci. 2018;65:54-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 28. | Congrains A, Kamide K, Katsuya T, Yasuda O, Oguro R, Yamamoto K, Ohishi M, Rakugi H. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem Biophys Res Commun. 2012;419:612-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 29. | da Silva FR, Guimarães-Vasconcelos AC, de-Carvalho-França LF, di-Lenardo D, Rodrigues LS, Barreto-do-Nascimento ML, Vasconcelos DF. Relationship between -889 C/T polymorphism in interleukin-1A gene and risk of chronic periodontitis: Evidence from a meta-analysis with new published findings. Med Oral Patol Oral Cir Bucal. 2017;22:e7-e14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | da Silva FRP, Vasconcelos ACCG, de Carvalho França LF, Di Lenardo D, Nascimento HMS, Vasconcelos DFP. Association between the rs1143634 polymorphism in interleukin-1B and chronic periodontitis: Results from a meta-analysis composed by 54 case/control studies. Gene. 2018;668:97-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 31. | Bochenek G, Häsler R, El Mokhtari NE, König IR, Loos BG, Jepsen S, Rosenstiel P, Schreiber S, Schaefer AS. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet. 2013;22:4516-4527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 32. | Bayarsaihan D. Epigenetic mechanisms in inflammation. J Dent Res. 2011;90:9-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 221] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 33. | Guo F, Tang C, Li Y, Liu Y, Lv P, Wang W, Mu Y. The interplay of LncRNA ANRIL and miR-181b on the inflammation-relevant coronary artery disease through mediating NF-κB signalling pathway. J Cell Mol Med. 2018;22:5062-5075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 34. | Preshaw PM, Taylor JJ. How has research into cytokine interactions and their role in driving immune responses impacted our understanding of periodontitis? J Clin Periodontol. 2011;38 Suppl 11:60-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 279] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 35. | Wufuer A, Luohemanjiang X, Du L, Lei J, Shabier M, Han DF, Ma J. ANRIL overexpression globally induces expression and alternative splicing of genes involved in inflammation in HUVECs. Mol Med Rep. 2023;27:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 36. | Caton JG, Armitage G, Berglundh T, Chapple ILC, Jepsen S, Kornman KS, Mealey BL, Papapanou PN, Sanz M, Tonetti MS. A new classification scheme for periodontal and peri-implant diseases and conditions - Introduction and key changes from the 1999 classification. J Clin Periodontol. 2018;45 Suppl 20:S1-S8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 705] [Article Influence: 88.1] [Reference Citation Analysis (0)] |
| 37. | Kehdy FS, Gouveia MH, Machado M, Magalhães WC, Horimoto AR, Horta BL, Moreira RG, Leal TP, Scliar MO, Soares-Souza GB, Rodrigues-Soares F, Araújo GS, Zamudio R, Sant Anna HP, Santos HC, Duarte NE, Fiaccone RL, Figueiredo CA, Silva TM, Costa GN, Beleza S, Berg DE, Cabrera L, Debortoli G, Duarte D, Ghirotto S, Gilman RH, Gonçalves VF, Marrero AR, Muniz YC, Weissensteiner H, Yeager M, Rodrigues LC, Barreto ML, Lima-Costa MF, Pereira AC, Rodrigues MR, Tarazona-Santos E; Brazilian EPIGEN Project Consortium. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci U S A. 2015;112:8696-8701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 205] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 38. | Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 155] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 39. | Chambrone L, Armitage GC. Commentary: Statistical Significance Versus Clinical Relevance in Periodontal Research: Implications for Clinical Practice. J Periodontol. 2016;87:613-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 40. | Leal ALAB, da Silva FA, Shin JI, Jeong GH, Ferreira GP, Vasconcelos DFP, Monteiro JRS, de Sousa AA, da Silva FRP, da Cunha Pereira ACT. Polymorphisms in immune-mediator genes and the risk of dengue virus infection: Lights from a systematic revaluation by Bayesian approaches. Cytokine. 2022;157:155955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 41. | Penha Mesquita A, Victor Oliveira Monteiro A, Luiz Araújo Bentes Leal A, Dos Santos Pessoa L, de Siqueira Amorim Júnior J, Rogério Souza Monteiro J, Andrade de Sousa A, Fernando Pereira Vasconcelos D, Carolina Alves de Oliveira A, Leão Pereira A, Rodolfo Pereira da Silva F. Gene variations related to the hepatocellular carcinoma: Results from a field synopsis and Bayesian revaluation. Gene. 2023;869:147392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 42. | da Silva FRP, Pessoa LDS, Shin JI, Alves EHP, Koga RS, Smith CV, Vasconcelos DFP, Pereira ACTDC. Polymorphisms in the interleukin genes and chronic periodontitis: A field synopsis and revaluation by Bayesian approaches. Cytokine. 2021;138:155361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/