Published online Apr 6, 2021. doi: 10.12998/wjcc.v9.i10.2228
Peer-review started: January 12, 2021
First decision: January 24, 2021
Revised: January 31, 2021
Accepted: February 24, 2021
Article in press: February 24, 2021
Published online: April 6, 2021
Processing time: 77 Days and 6.5 Hours
In this study, recent trends in the distribution and drug resistance of pathogenic bacteria isolated from patients treated at a burn ward between 2006 and 2019 were investigated.
To develop more effective clinical strategies and techniques for the prevention and treatment of bacterial infections in burn patients.
Clinical samples with positive bacteria were collected from patients at the burn ward in Beijing Jishuitan Hospital in China between January 2006 and December 2019. The samples were retrospectively analyzed, the distribution of pathogenic bacteria was determined, and the trends and changes in bacterial drug resistance during different period were assessed. Drug resistance in several main pathogenic bacteria from 2006 to 2011 and from 2012 to 2019 was comparatively summarized and analyzed.
Samples from 17119 patients were collected and analyzed from 2006 to 2019. Surprisingly, a total of 7960 strains of different pathogenic bacteria were isolated at this hospital. Among these bacteria, 87.98% (7003/7960) of the strains were isolated from burn wounds, and only 1.34% (107/7960) were isolated from the blood of patients. In addition, 49.70% (3956/7960) were identified as Gram-positive bacteria, 48.13% (3831/7960) were Gram-negative bacteria, and the remaining 2.17% (173/7960) were classified as fungi or other pathogens. Importantly, Staphylococcus aureus (21.68%), Pseudomonas aeruginosa (14.23%), and Staphylococcus epidermidis (9.61%) were the top three pathogens most frequently isolated from patients.
In patients treated at the burn ward in this hospital from 2006 to 2019, Staphylococcus aureus and Pseudomonas aeruginosa were the predominant clinical pathogens responsible for bacterial infections. The circumstantial detection and detailed monitoring of the intensity and growth of different pathogenic bacteria in clinical patients as well as tests of drug sensitivity during burn recovery are particularly important to provide guidelines for the application of antibiotics and other related drugs. Careful collection and correct, standard culture of bacterial specimens are also crucial to improve the efficiency of bacterial infection detection. Effective monitoring and timely clinical treatment in patients may help reduce the possibility and rate of infection as well as alleviate the effects of drug resistance among patients in burn centers.
Core Tip: This retrospective study analyzed the pathogenic bacteria isolated from clinical samples from patients on burn wards at our hospital between 2006 and 2019 to determine the distribution of different bacterial species and the long-term trends in bacterial drug resistance. Our findings may direct effective strategies, based on scientific evidence, for the prevention, control, and treatment of infections in burn patients.
- Citation: Chen H, Yang L, Cheng L, Hu XH, Shen YM. Distribution and drug resistance of pathogens in burn patients in China from 2006 to 2019. World J Clin Cases 2021; 9(10): 2228-2237
- URL: https://www.wjgnet.com/2307-8960/full/v9/i10/2228.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i10.2228
Bacterial infection is one of the major causes of death of burn patients[1]. Highly effective therapeutic treatment against bacterial infection is very important for the prevention and control of infection. Antibiosis is frequently observed in clinical patients. The condition leads to dramatic changes in the bacterial distribution and resistance to drugs used for the treatment of pathogenic bacterial infections, resulting in clinical difficulties throughout the treatment of infectious diseases. Moreover, in burn patients, there are always bacterial evolutions and changes in spatial and regional sites as well as diverse temporal phases during infection. For hospitalized patients with burns, the distribution of pathogens can directly affect the treatment options and therapeutic effectiveness. It has been shown in both domestic and international studies that there are some differences in the distribution of pathogens in burn wounds in different locations and striking differences in drug resistance also exist[2-4].
There are several major clinical strains responsible for severe infections in burn centers or units in hospitals. For example, Staphylococcus aureus (S. aureus) is the predominant species responsible for burn infections. This organism is easily transmitted via close contact, especially to wounds that require long-term treatment. This organism is easy to detect but difficult to eliminate, especially during progression to systemic infection[5]. The rate of occurrence of methicillin-resistant S. aureus (MRSA) has gradually declined according to recent reports. This decline of drug resistance in S. aureus is related to the initiation of antimicrobial rectification measures in 2011 and the limitation of unreasonable applications of third-generation cephalosporins and third-generation quinolone antibiotics.
Pseudomonas aeruginosa (P. aeruginosa) is one of the most serious causes of life-threatening infections in patients with heat injury[6]. This organism usually colonizes the skin and medical devices. Long lasting colonization of burn wounds can result in prolonged hospitalization, the long-term use of broad-spectrum antibiotics, delayed wound recovery, severe P. aeruginosa infection, and a risk of drug resistance[7].
Another pathogen, called Staphylococcus epidermidis (S. epidermidis), is a constituent of the normal flora of the skin and mucous membranes[8] and is the most commonly detected strain among the coagulase-negative staphylococci. Patients with this organism with low virulence are generally asymptomatic. However, in patients with immunosuppressive conditions (such as thermal injury), coagulase-negative staphylococci are considered to be as pathogenic as S. aureus and often transmit genetic resistance to susceptible S. aureus, converting them into multidrug-resistant S. aureus[5].
Impaired immunity and decreased susceptibility to S. epidermidis have been found in patients who receive various invasive treatments and extensive administration of hormones and immunosuppressive agents[4] This may lead to prosthetic valve endocarditis, venous catheter, peritonitis, blood vessel and artificial joint infections, and other complications.
S. epidermidis has become an important opportunistic pathogen in patients with impaired immune function. For burn patients, S. epidermidis is a common colonizer. Coagulase-negative staphylococci mainly colonize the skin and mucous membranes and the detection rate of this organism has clearly risen in recent years. Strict disinfection procedures to avoid nosocomial infections should be employed. Microbiological sampling procedures also need to be improved and optimized to reduce the potential for bacterial contamination.
Based on previous studies, it is important to monitor the distribution of pathogens, the characteristics and profiles of pathogenic bacteria and drug resistance in pathogenic bacteria in burn patients for the rapid and accurate selection of antibacterial regimens in clinical practice[2,9]. This retrospective analysis of batches of pathogenic bacteria isolated from clinical samples from patients treated at the burn ward in our hospital between 2006 and 2019 provides a new perspective regarding the distribution of bacterial species and drug resistance over a long-term period. Our findings provide useful and valuable scientific evidence and information for the development of new and effective strategies for the prevention, control, and treatment of bacterial infections in burn patients.
All bacteria in this study were isolated from the wound secretions, blood, catheters, and sputum of patients in the burn ward at Beijing Jishuitan Hospital (a Third-Class A hospital) between January 2006 and December 2019. The burn severity in patients was classified according to the classification method adopted at the 1970 National Burn Conference, namely: (1) Mild burn; (2) Moderate burn; (3) Severe burn; and (4) Extremely severe burn.
The VITEK-60 automatic bacterial analysis system (bioMérieux, France) was used to detect common bacteria. The standard reference strains used for quality control were Escherichia coli (E. coli) ATCC25922, S. aureus ATCC25923, P. aeruginosa ATCC27853, and Klebsiella pneumoniae (K. pneumoniae) ATCC700603, which were obtained from the Health Planning Commission Inspection Center.
The sensitivity to drugs was tested with GNS-121 and GPS-107 detection cards (bioMérieux). Microdilution (bioMérieux) and the K-B paper diffusion method were used to test for drug susceptibility. Antibiotic papers were used for drug resistance tests according to the Clinical and Laboratory Standards Institute criteria[1,4].
Categorical data were presented as frequencies and percentages. Categorical variables were analyzed using Pearson’s 2 test. The Fisher exact probability method was used when the theoretical frequency of cells was < 5. The Cochran-Armitage trend test was used to analyze the detection rate of pathogenic bacteria and the rate of drug resistance. The statistical analysis was performed with SPSS 19 software. P values < 0.05 were considered statistically significant.
A total of 17119 patients with an average age of 36.43 ± 17.07 years (95% confidence interval: 1–98) were treated in the burn ward at our hospital from 2006 to 2019, of which 12899 (75.35%) were male and 4220 (24.65%) were female. In our study, patients with mild burns accounted for 70.41% of cases, those with moderate burns accounted for 17.03%, those with severe burns accounted for 5.19%, and those with extremely severe burns accounted for 7.37%.
A total of 7960 strains of pathogenic bacteria were detected among clinical specimens from patients treated at the burn ward in our hospital between January 2006 and December 2019. The pathogenic bacteria were mainly isolated from burn wounds (87.98%, 7003/7960). Only 1.34% (107/7960) isolated from the blood of patients. Gram-positive bacteria accounted for 49.70% (3956/7960) of the isolates, Gram-negative bacteria accounted for 48.13% (3831/7960) and fungi and other pathogens accounted for 2.17% (173/7960).
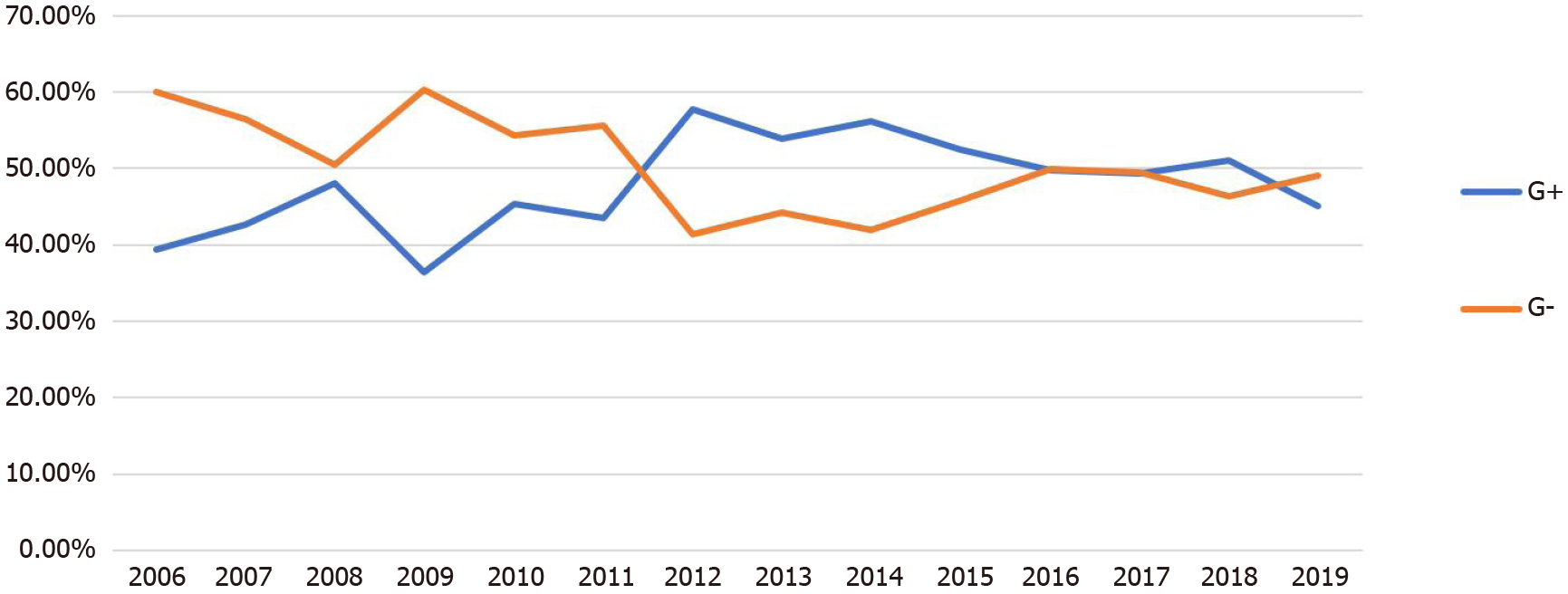
During 2006–2011, the average number of positive samples was 235 per year, compared with 819 per year in 2012–2019 (P < 0.05; Table 1). The proportion of Gram-negative bacteria was higher than that of Gram-positive bacteria during the first 6 years (2006–2011). However, the proportion of Gram-negative bacteria detected in 2012–2015 was lower than that of Gram-positive bacteria. In 2016–2019, the proportions Gram-negative bacteria and Gram-positive bacteria were not significantly different (P > 0.05; Figure 1 and Table 1).
| Year | Strains identified | Gram-positive bacteria, n (%) | Gram-negative bacteria, n (%) | Fungi, n (%) |
| 2006 | 170 | 67 (39.41) | 102 (60.00) | 1 (0.59) |
| 2007 | 232 | 99 (42.67) | 131 (56.46) | 2 (0.86) |
| 2008 | 129 | 62 (48.07) | 65 (50.39) | 2 (1.55) |
| 2009 | 239 | 87 (36.4) | 144 (60.25) | 8 (3.35) |
| 2010 | 293 | 133 (45.39) | 159 (54.26) | 1 (0.34) |
| 2011 | 349 | 152 (43.56) | 194 (55.59) | 3 (0.86) |
| 2012 | 621 | 358 (57.65) | 257 (41.38) | 6 (0.97) |
| 2013 | 606 | 326 (53.8) | 268 (44.22) | 12 (1.98) |
| 2014 | 785 | 441 (56.18) | 329 (41.91) | 15 (1.91) |
| 2015 | 678 | 356 (52.51) | 310 (45.72) | 12 (1.77) |
| 2016 | 785 | 390 (49.68) | 392 (49.94) | 3 (0.38) |
| 2017 | 810 | 400 (49.38) | 401 (49.51) | 9 (1.11) |
| 2018 | 1098 | 560 (51.00) | 508 (46.27) | 30 (2.73) |
| 2019 | 1165 | 525 (45.06) | 571 (49.01) | 69 (5.92) |
| Total | 7960 | 3956 (49.70) | 3831 (48.13) | 173 (2.17) |
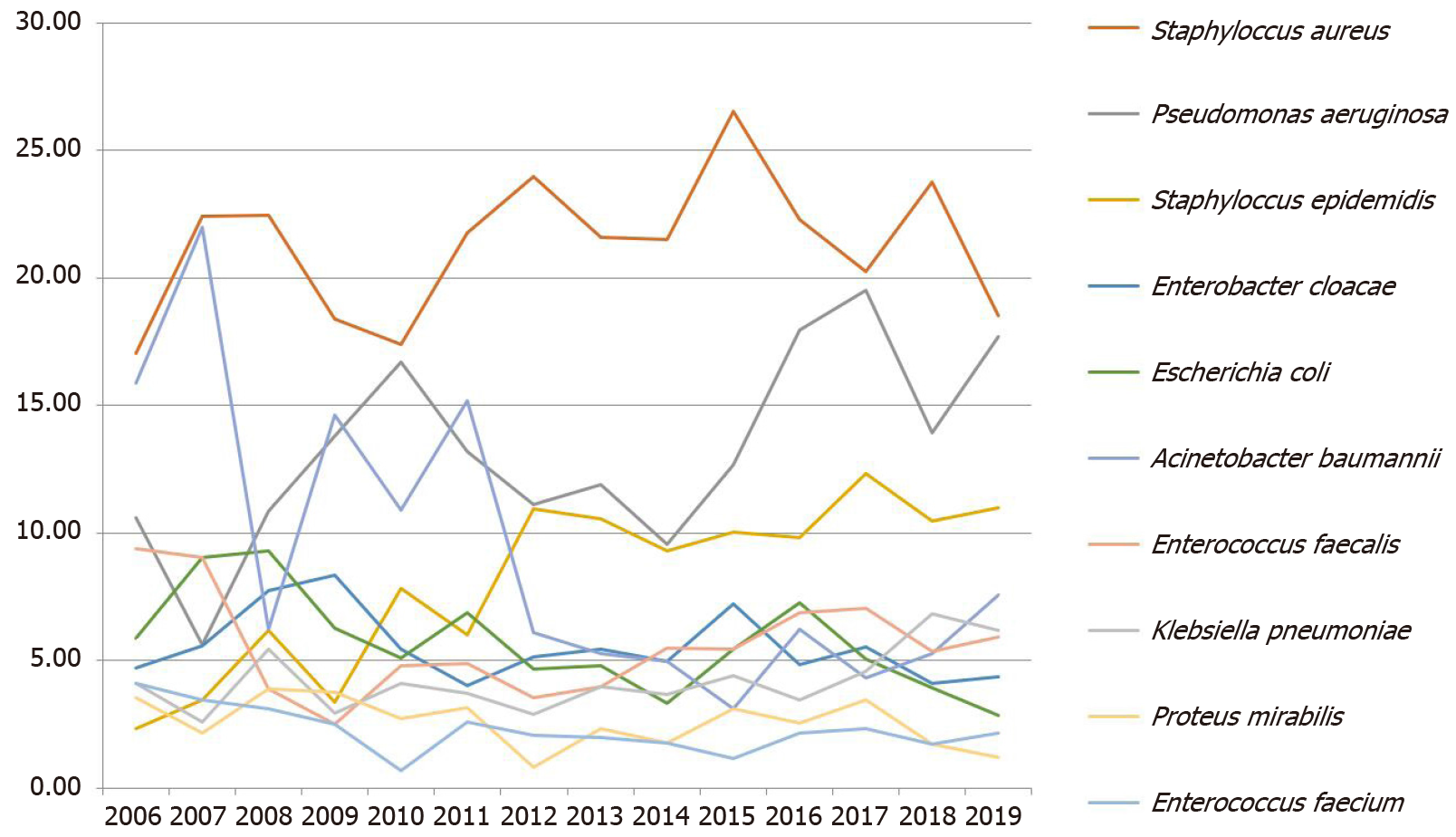
The eight most frequently detected pathogens in this study were S. aureus (21.68%, 1726/7960), P. aeruginosa (14.23%, 1133/7960), S. epidermidis (9.61%, 765/7960), Acinetobacter baumannii (A. baumannii) (7.11%, 566/7960), Enterococcus faecalis (E. faecalis) (5.58%, 444/7960), Enterobacter cloacae (5.19%, 413/7960), E. coli (4.92, 392/7960) and K. pneumoniae (4.57%, 364/7960) (Figure 2 and Table 2).
| 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | Total1 | |
| Staphylococcus aureus | 29 (17.06) | 52 (22.41) | 29 (22.48) | 44 (18.41) | 51 (17.41) | 76 (21.78) | 149 (23.99) | 131 (21.62) | 169 (21.53) | 180 (26.55) | 175 (22.29) | 164 (20.25) | 261 (23.77) | 216 (18.54) | 1726 (21.68) |
| Pseudomonas aeruginosa | 18 (10.59) | 13 (5.6) | 14 (10.85) | 33 (13.81) | 49 (16.72) | 46 (13.18) | 69 (11.11) | 72 (11.88) | 75 (9.55) | 86 (12.68) | 141 (17.96) | 158 (19.51) | 153 (13.93) | 206 (17.68) | 1133 (14.23) |
| Staphylococcus epidermidis | 4 (2.35) | 8 (3.45) | 8 (6.2) | 8 (3.35) | 23 (7.85) | 21 (6.02) | 68 (10.95) | 64 (10.56) | 73 (9.3) | 68 (10.03) | 77 (9.81) | 100 (12.35) | 115 (10.47) | 128 (10.99) | 765 (9.61) |
| Acinetobacter baumannii | 27 (15.88) | 51 (21.98) | 8 (6.2) | 35 (14.64) | 32 (10.92) | 53 (15.19) | 38 (6.12) | 32 (5.28) | 39 (4.97) | 21 (3.1) | 49 (6.24) | 35 (4.32) | 58 (5.28) | 88 (7.55) | 566 (7.11) |
| Enterococcus faecalis | 16 (9.41) | 21 (9.05) | 5 (3.88) | 6 (2.51) | 14 (4.78) | 17 (4.87) | 22 (3.54) | 24 (3.96) | 43 (5.48) | 37 (5.46) | 54 (6.88) | 57 (7.04) | 59 (5.37) | 69 (5.92) | 444 (5.58) |
| Enterobacter cloacae | 8 (4.71) | 13 (5.6) | 10 (7.75) | 20 (8.37) | 16 (5.46) | 14 (4.01) | 32 (5.15) | 33 (5.45) | 39 (4.97) | 49 (7.23) | 38 (4.84) | 45 (5.56) | 45 (4.1) | 51 (4.38) | 413 (5.19) |
| Escherichia coli | 10 (5.88) | 21 (9.05) | 12 (9.3) | 15 (6.28) | 15 (5.12) | 24 (6.88) | 29 (4.67) | 29 (4.79) | 26 (3.31) | 37 (5.46) | 57 (7.26) | 41 (5.06) | 43 (3.92) | 33 (2.83) | 392 (4.92) |
| Klebsiella pneumoniae | 7 (4.12) | 6 (2.59) | 7 (5.43) | 7 (2.93) | 12 (4.1) | 13 (3.72) | 18 (2.9) | 24 (3.96) | 29 (3.69) | 30 (4.42) | 27 (3.44) | 37 (4.57) | 75 (6.83) | 72 (6.18) | 364 (4.57) |
| Proteus mirabilis | 6 (3.53) | 5 (2.16) | 5 (3.88) | 9 (3.77) | 8 (2.73) | 11 (3.15) | 5 (0.81) | 14 (2.31) | 14 (1.78) | 21 (3.1) | 20 (2.55) | 28 (3.46) | 19 (1.73) | 14 (1.2) | 179 (2.25) |
| Enterococcus faecium | 7 (4.12) | 8 (3.45) | 4 (3.1) | 6 (2.51) | 2 (0.68) | 9 (2.58) | 13 (2.09) | 12 (1.98) | 14 (1.78) | 8 (1.18) | 17 (2.17) | 19 (2.35) | 19 (1.73) | 25 (2.15) | 163 (2.05) |
| Sum | 170 | 232 | 129 | 239 | 293 | 349 | 621 | 606 | 785 | 678 | 785 | 810 | 1098 | 1165 | 7960 (100.00) |
Over the past 14 years, S. aureus has been the most frequently isolated pathogen (17.06%–26.55%) in the burn ward, with no obvious upward or downward trend in the detection rate (χ2 = 0.027, P = 0.870). The second most common pathogen isolated from the burn patients was P. aeruginosa. As the predominant Gram-negative bacterium, its detection rate increased from 10.59% in 2006 to 17.68% in 2019 with statistical significance (χ2 = 32.351, P ≤ 0.0001). S. epidermidis showed a similar trend, with its isolation rate increasing significantly starting from 2011. From 2012 to 2019, S. epidermidis was the third most frequently isolated strain in burn patients, with the detection rate increasing from 2.35% in 2006 to 10.99% in 2019. This increasing trend was statistically significant (χ2 = 32.793, P ≤ 0.0001). In contrast, the detection rate of A. baumannii decreased from 15.88% in 2006 to 7.55% in 2019 (χ2= 79.007, P ≤ 0.0001). Significant trends were not observed for any of the other common bacterial species (P > 0.05).
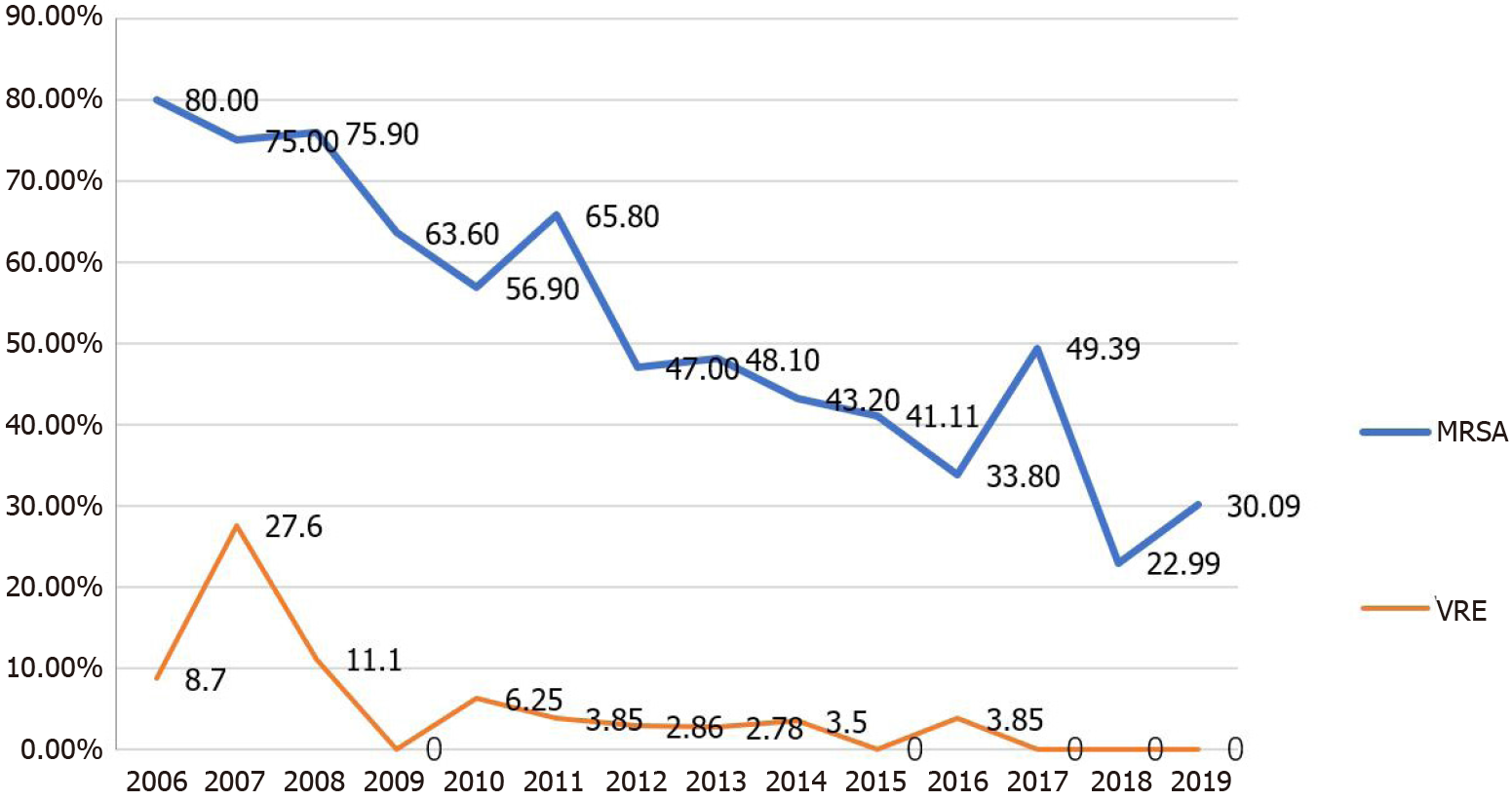
In this study, bacterial drug resistance was evaluated. Four commonly used antimicrobial agents, imipenem, ceftazidime, cefepime, and levofloxacin, were selected for the evaluation of bacterial resistance. Compared with the results from the 2006-2011 period, the resistance of S. aureus to oxacillin decreased significantly over the following 6 years (2012–2019; χ2= 88.0448, P < 0.001). The resistance of Enterococcus to vancomycin also decreased in 2012–2019, and the difference was statistically significant (χ2 = 31.265, P < 0.001; Figure 3 and Table 3). The resistance of P. aeruginosa to all four common antimicrobial agents showed a downward trend in 2012–2019 compared with the trend during the first 6 years (2006–2011), particularly the resistance to cefepime (χ2 = 36.2767, P < 0.001), ceftazidime (χ2 = 6.7138, P = 0.001) and levofloxacin (χ2 = 13.0306, P < 0.001). Similarly, the resistance of A. baumannii to cefepime (χ2 = 21.3591, P < 0.001), ceftazidime (χ2 = 14.7418, P < 0.001) and levofloxacin (χ2 = 71.5236, P < 0.001) decreased significantly over the latter 6 years, whereas the resistance to imipenem remained at a very high level, evidenced by no statistical difference between the two time periods (Table 3).
| 2006-2011 | 2012-2019 | χ2 | P value | ||||
| Strain | Resistant strains | Strain | Resistant strains | ||||
| Staphylococcus aureus | Oxacillin | 281 | 191 (67.97) | 1445 | 545 (37.72) | 88.0448 | < 0.001 |
| Vancomycin | 281 | 0 (0.00) | 1445 | 0 (0.00) | NA | NA | |
| Enterococcus | Vancomycin | 115 | 13 (11.30) | 492 | 6 (1.22) | 31.265 | < 0.001 |
| Pseudomonas aeruginosa | Cefepime | 173 | 109 (63.01) | 960 | 369 (38.44) | 36.2767 | < 0.001 |
| Ceftazidime | 173 | 78 (45.09) | 960 | 334 (34.79) | 6.7138 | 0.001 | |
| Imipenem | 173 | 79 (45.67) | 960 | 390 (40.63) | 1.5347 | 0.215 | |
| Levofloxacin | 173 | 88 (50.87) | 960 | 349 (36.35) | 13.0306 | < 0.001 | |
| Acinetobacter baumannii | Cefepime | 203 | 181 (89.16) | 360 | 261 (72.50) | 21.3591 | < 0.001 |
| Ceftazidime | 203 | 173 (85.22) | 360 | 255 (70.83) | 14.7418 | < 0.001 | |
| Imipenem | 203 | 135 (66.50) | 360 | 233 (64.72) | 0.1817 | 0.67 | |
| Levofloxacin | 203 | 177 (87.19) | 360 | 186 (51.67) | 71.5236 | < 0.001 |
This retrospective analysis was conducted with 7960 potential microbial pathogens found in clinical specimens from burn patients treated at our hospital between January 2006 and December 2019. The study participants consisted of patients from 36 provinces in China; 84% of the patients originated from north China and 11 patients were foreigners. Our findings revealed that the distribution of both Gram-positive and Gram-negative bacteria changed dynamically over time, consistent with previous studies showing that the distribution of bacterial species differed depending on the location of the hospital and the time period investigated[10-12]. The average number of bacteria-positive specimens increased from 235 cases per year in 2006–2011 to 819 cases per year in the subsequent 8 years (P < 0.001). This may reflect the fact that more samples were subjected to etiological diagnosis in our hospital. Our results, which were largely based on culture data, revealed that antibiotic resistance among Gram-positive (S. aureus, E. faecalis) and Gram-negative (P. aeruginosa, A. baumannii) bacteria decreased after antibiotics were used (Figure 3 and Table 3). Taken together, these findings indicate that more clinical samples should be collected for diagnosis to guide the effective utilization of antibiotics. That would help reduce the level of antibiotic resistance and antibiotic misuse.
This study showed that S. aureus was the primary pathogen in burn wards over the 12-year period from 2006 to 2019, accounting for 25.0% of the total positive isolates and consistent with most burn centers in domestic and international hospitals[2,4]. A significant downward trend was observed in the rate of MRSA detection in burn specimens, decreasing from 80% in 2006 to 30.09% in 2019.
The detection rate of P. aeruginosa in the burn wounds of patients increased from 2006 to 2019. From 2012 to 2019, P. aeruginosa remained the second most commonly isolated pathogen and the predominant Gram-negative bacteria. Another study also found that the detection rate of P. aeruginosa was on the rise in samples from the burn ward at Ruijin Hospital in China from 2007 to 2014[3]. A 20-year retrospective study in Germany also showed an increase in the detection rate of P. aeruginosa[2]. In our study, P. aeruginosa resistance to cefepime, ceftazidime, and levofloxacin showed a significant downward trend in the years before and after 2011. It has been suggested that the widespread use of antibacterial drugs has led to gradual bacterial resistance[13]. We speculated that the observed decline in resistance rates may be due to the increasing rational use of antimicrobials in China since 2011. The changes in treatment regimens have undoubtedly contributed to a reduction in the transmission and prevalence of P. aeruginosa-resistant strains in burn wards.
Our study showed that the detection rate of S. epidermidis increased significantly after 2011. In addition, the detection rate of A. baumannii decreased dramatically from 2006 to 2019. As a conditional pathogen, A. baumannii is widely distributed in natural environments and is universally found in patients with impaired immunity, which can lead to nosocomial infections[13]. In recent years, owing to the implementation of control measures for nosocomial infection, the infection rates in hospitals have decreased significantly, and not surprisingly, the detection rate of A. baumannii has also declined.
Our results suggest that the sensitivity of Gram-negative bacteria to third and fourth generation cephalosporins has recovered remarkably, whereas the sensitivity to imipenem has not increased. One possible explanation for this finding is that imipenem is still used as the main treatment for Gram-negative bacilli[14]. High usage of carbapenems leads to the emergence of high levels of drug resistance, and the treatment of carbapenem-resistant P. aeruginosa may become a huge challenge for infections in the future.
There were also some limitations in our study. First, we only collected data on bacteria and drug resistance from burn ward patients and did not record the clinical characteristics of the patients or their demographic information, which may affect bacterial resistance. Second, more samples were collected starting in 2011. A greater number of samples can increase the false-positive rate. Third, bacterial specimens were not subjected to more detailed in-depth molecular identification and homology analysis. In addition, this study was a retrospective analysis and therefore could not prospectively focus on the dynamic changes in pathogen resistance over different treatment periods. These limitations should be considered when interpreting our data, and further clarification and identification are required in subsequent studies.
In conclusion, we analyzed the distribution of pathogens and the long-term trends of bacterial resistance in patients in our burn ward during a 14-year period. After a series of antibacterial drug remediation activities in 2011, the number of positive cases detected increased significantly and resistance to drugs among the major Gram-positive and Gram-negative bacteria showed a downward trend. The monitoring of changes in pathogens and drug susceptibility is especially important to guide the rational use of antibacterial drugs.
To improve the detection rate of pathogens in burn wounds, all processes for specimen collection, inspection, and testing should be standardized. Clinical treatment should be based on the identification of pathogens and profiles of drug resistance to facilitate effective personalized treatment with antibiotics. A series of tailored antibacterial drugs may be administered to combat bacterial infections and to alleviate drug resistance while avoiding the damaging side effects of non-specific antibacterial drug usage.
Highly effective therapeutic treatment against bacterial infection is very important for the prevention and control of infection in burn patients. It is important to monitor the distribution of pathogens, the characteristics and profiles of pathogenic bacteria, and drug resistance in pathogenic bacteria in burn patients for the rapid and accurate selection of antibacterial regimens in clinical practice.
To provide useful and valuable scientific evidence and information for the development of new and effective strategies for the prevention, control, and treatment of bacterial infections in burn patients.
To develop more effective clinical strategies and techniques for the prevention and treatment of bacterial infections in burn patients.
Clinical samples with positive bacteria were collected from patients at the burn ward in Beijing Jishuitan Hospital in China between January 2006 and December 2019. The samples were retrospectively analyzed, the distribution of pathogenic bacteria was determined, and the trends and changes in bacterial drug resistance during different periods were assessed. Drug resistance in several main pathogenic bacteria from 2006 to 2011 and from 2012 to 2019 was comparatively summarized and analyzed.
We analyzed data obtained from samples collected between 2006 and 2019 from 17119 patients. Staphylococcus aureus (21.68%), Pseudomonas aeruginosa (14.23%), Staphylococcus epidermidis (9.61%) were the top three pathogens most frequently isolated from patients.
The circumstantial detection and detailed monitoring of the intensity and growth of different pathogenic bacteria in clinical patients as well as tests of drug sensitivity during burn recovery are particularly important to provide guidelines for the use of antibiotics and other related drugs.
Bacterial specimens should be subjected to more detailed in-depth molecular identification and homology analysis.
| 1. | Kwei J, Halstead FD, Dretzke J, Oppenheim BA, Moiemen NS. Protocol for a systematic review of quantitative burn wound microbiology in the management of burns patients. Syst Rev. 2015;4:150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 2. | Guggenheim M, Zbinden R, Handschin AE, Gohritz A, Altintas MA, Giovanoli P. Changes in bacterial isolates from burn wounds and their antibiograms: a 20-year study (1986-2005). Burns. 2009;35:553-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 88] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 3. | Dou Y, Huan J, Guo F, Zhou Z, Shi Y. Pseudomonas aeruginosa prevalence, antibiotic resistance and antimicrobial use in Chinese burn wards from 2007 to 2014. J Int Med Res. 2017;45:1124-1137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 4. | Cen H, Wu Z, Wang F, Han C. Pathogen distribution and drug resistance in a burn ward: a three-year retrospective analysis of a single center in China. Int J Clin Exp Med. 2015;8:19188-19199. [PubMed] |
| 5. | Miller LG, Eells SJ, David MZ, Ortiz N, Taylor AR, Kumar N, Cruz D, Boyle-Vavra S, Daum RS. Staphylococcus aureus skin infection recurrences among household members: an examination of host, behavioral, and pathogen-level predictors. Clin Infect Dis. 2015;60:753-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 6. | Tredget EE, Shankowsky HA, Rennie R, Burrell RE, Logsetty S. Pseudomonas infections in the thermally injured patient. Burns. 2004;30:3-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 123] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 7. | Armour AD, Shankowsky HA, Swanson T, Lee J, Tredget EE. The impact of nosocomially-acquired resistant Pseudomonas aeruginosa infection in a burn unit. J Trauma. 2007;63:164-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 8. | Azimi T, Mirzadeh M, Sabour S, Nasser A, Fallah F, Pourmand MR. Coagulase-negative staphylococci (CoNS) meningitis: a narrative review of the literature from 2000 to 2020. New Microbes New Infect. 2020;37:100755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Keen EF 3rd, Robinson BJ, Hospenthal DR, Aldous WK, Wolf SE, Chung KK, Murray CK. Prevalence of multidrug-resistant organisms recovered at a military burn center. Burns. 2010;36:819-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 108] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 10. | Yali G, Jing C, Chunjiang L, Cheng Z, Xiaoqiang L, Yizhi P. Comparison of pathogens and antibiotic resistance of burn patients in the burn ICU or in the common burn ward. Burns. 2014;40:402-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Karimi H, Montevalian A, Motabar AR, Safari R, Parvas MS, Vasigh M. Epidemiology of paediatric burns in Iran. Ann Burns Fire Disasters. 2012;25:115-120. [PubMed] |
| 12. | Alrawi M, Crowley TP, Pape SA. Bacterial colonisation of the burn wound: a UK experience. J Wound Care. 2014;23:274-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Vernaz N, Huttner B, Muscionico D, Salomon JL, Bonnabry P, López-Lozano JM, Beyaert A, Schrenzel J, Harbarth S. Modelling the impact of antibiotic use on antibiotic-resistant Escherichia coli using population-based data from a large hospital and its surrounding community. J Antimicrob Chemother. 2011;66:928-935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 70] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 14. | Bayram Y, Parlak M, Aypak C, Bayram I. Three-year review of bacteriological profile and antibiogram of burn wound isolates in Van, Turkey. Int J Med Sci. 2013;10:19-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
Open-Access: This is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Charco R S-Editor: Gao CC L-Editor: Filipodia P-Editor: Liu JH