Published online Mar 26, 2022. doi: 10.12998/wjcc.v10.i9.2783
Peer-review started: November 5, 2021
First decision: January 11, 2022
Revised: January 26, 2022
Accepted: February 15, 2022
Article in press: February 15, 2022
Published online: March 26, 2022
Processing time: 137 Days and 4.6 Hours
Ubiquilins (UBQLNs) are important factors for cell proteostasis maintenance. UBQLNs are involved in the modulation of the cell cycle, as well as in apoptosis, membrane receptors regulation, DNA repair, epithelial-mesenchymal transition, and miRNA activities. They also affect the selection of double-strand break repair pathways. Abnormal UBQLNs expression can lead to many diseases, including cancer. Studies have found that the expression of Ubiquilin4 (UBQLN4) is associated with the development of several tumor types. However, the association between UBQLN4 and cervical cancer has not been examined yet.
To investigate the expression of UBQLN4 in cervical cancer and to evaluate its correlation with disease prognosis.
Immunohistochemistry was performed to examine the expression of UBQLN4 in 117 cervical cancer tissues and 32 matching pericervical tissues. Paired t-test (two-tailed) was used to compare the differences between groups. We collected patients’ clinical characteristics, including age, histological grade, pathologic type, lymph node metastasis, and FIGO stage (2018) and compared them by chi-square test. All patients were followed for 5.5 to 6.8 years. Kaplan-Meier method and log-rank test were used to compare the differences in the overall survival (OS) and progression-free survival (PFS) among the different groups.
Overexpression of UBQLN4 was observed in 70.9% (83/117) of all cervical cancer tissues and in 15.6% (5/32) of the paired parauterine tissues. The expression of UBQLN4 was associated with lymph node metastasis, poor differentiation, and advanced stage, but the difference was not significant. Kaplan-Meier and log-rank test results suggested the high expression of UBQLN4 was associated with short OS and PFS. Regardless of UBQLN4 expression, the patient age and FIGO stage were also associated with disease prognosis. The statistically significant variables obtained from univariate the Kaplan-Meier analysis were subjected to Cox multivariate survival regression analysis, which showed that, in addition to the FIGO stage and age, UBQLN4 was also an independent prognostic marker for OS and PFS (P = 0.011 and P = 0.024, respectively).
The overexpression of UBQLN4 was associated with poor prognosis in cervical cancer. Our study proposed a novel prognostic factor and improved the existing understanding of the pathogenesis of cervical cancer.
Core Tip: Cervical cancer is the most common malignant tumor in women. At present, the exact molecular mechanism of its occurrence and development mainly focuses on the regulation of gene expression level, while few researches on the regulation of protein level of translation products. Ubiquilin4 (UBQLN4) regulates protein homeostasis in cells through interactions with different products and proteasomal degradation. Studies have found that the expression of UBQLN4 was associated with several tumors. However, the association between UBQLN4 and cervical cancer has not been examined. In this study, we aimed to measure association between UBQLN4 expression and poor prognosis in cervical cancer.
- Citation: Wang LN, Huang KJ, Wang L, Cheng HY. Overexpression of Ubiquilin4 is associated with poor prognosis in patients with cervical cancer. World J Clin Cases 2022; 10(9): 2783-2791
- URL: https://www.wjgnet.com/2307-8960/full/v10/i9/2783.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i9.2783
Cervical cancer is one of the most common malignant tumors in gynecology and the fourth leading cause of cancer death in women worldwide[1,2]. Human genome encodes five major ubiquilin (UBQLN) proteins (UBQLN1, UBQLN2, UBQLN3, UBQLN4, and UBQLNL) belonging to the non-proteasomal ubiquitin-like/ubiquitin-associated (UBL−UBA) family.They contain a domain similar to ubiquitin at the N−terminus and an ubiquitin-associated domain at the C-terminus. UBQLN1, UBQLN2, and UBQLN4 are ubiquitous, whereas UBQLN3 is exclusively expressed in the testes[3].
UBQLNs are important factors for cells proteostasis maintenance; they function as adapters for the delivery of poly-ubiquitinated proteins in the proteasome[4,5]. It is important to note that they participate in autophagosome formation[6,7], as well as in the endoplasmic reticulum-associated protein degradation pathway[8]. UBQLNs are involved in the modulation of important players in the cell cycle, apoptosis, membrane receptors, DNA repair, epithelial-mesenchymal transition, and miRNA activities. Accumulating evidence has shown an association between UBQLNs and neurodegenerative diseases, such as Alzheimer¢s disease or other forms of dementia with locomotor dysfunction, and with other proteinopathies, such as amyotrophic lateral sclerosis[9-11].
UBQLN4 as Bcl-2-associated athanogene 6 (BAG6)-binding factor is necessary for the elimination of incorrectly localized proteins in the cytosolic transmembrane that has failed to reach their exact destination on the endoplasmic reticulum membrane. UBQLN4 recognizes mislocalized transmembrane proteins in the cytosol and targets them, performing their proteasomal degradation. Moreover, a previous study suggested that UBQLN4 was located within the BAG6 complex, but not in other members of the UBQLN family[12].
Apart from protein degradation, UBQLN4 is also associated with DNA double-strand break (DSB) repair. A previous study indicated that the UBQLN4 protein was phosphorylated in an ataxia telangiectasia mutant (ATM)-dependent manner and was recruited to DNA damage sites to shift DSB repair to non-homologous end-ligation by functional inhibition of homologous reprogramming repair, which further promoted genomic instability and oncogenic transformation[13].
Accumulating evidence has shown the involvement of UBQLNs in human cancer. UBQLN4 abnormality has been associated with many diseases, especially malignancies, such as liver, lung, stomach, and ovarian cancers and other tumors[13-15]. However, to the best of our knowledge, the association between UBQLN4 and cervical cancer has not been elucidated.
In this study, we aimed to measure the expression of UBQLN4 in cervical cancer tissues and establish its association with disease clinicopathological features as well as prognosis.
A total number of 117 cervical cancer tissue and 32 matching pericervical tissue were obtained from the cervical cancer cohort (HUTES154SU01 Outdo Biotech, Shanghai, China) from January 2010 to October 2011. All patients were diagnosed with stage IB1-IIA1 according to the FIGO 2009 staging criteria, and were reclassified based on the FIGO 2018 staging criteria. All patients received radical surgery but not radiotherapy and chemotherapy before and after surgery. None of the patients developed serious medical complications. Patients’ clinical characteristics, including age, histological grade, pathologic type, lymph node metastasis, and FIGO stage (2018) were collected and compared.
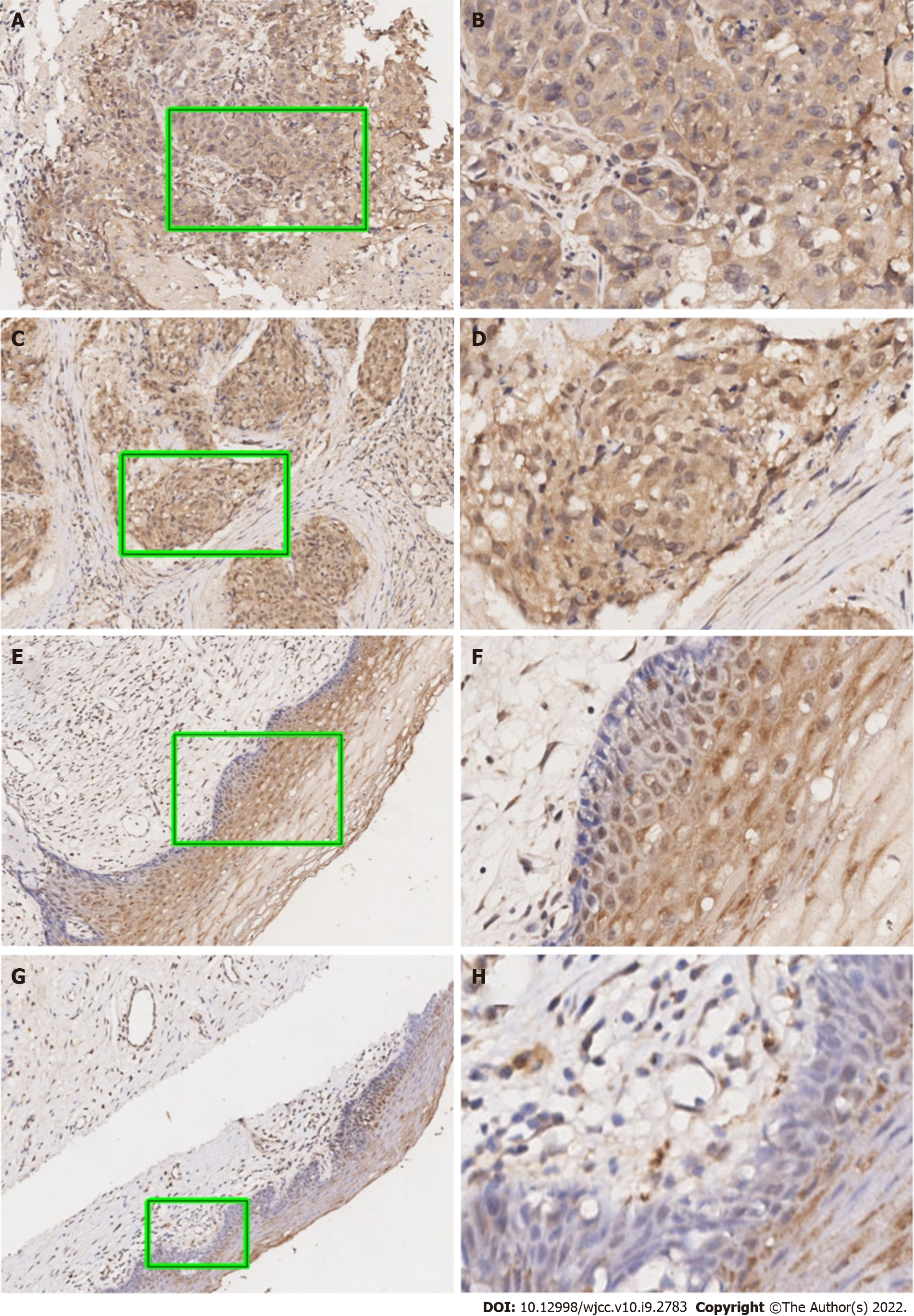
Immunohistochemistry (IHC) analysis was performed as previously described[16-18]. Briefly, tissue sections were deparaffinized, rehydrated, and blocked with goat serum. After incubation with an anti-UBQLN4 antibody (1:500, PAB16793, Abnova) overnight at 4 °C, the slides were rinsed three times with PBS and incubated with an HRP-conjugated secondary antibody (1:300, K8002, Dako) for 30 min at room temperature. Following three 5-min rinses with PBS, the slides were incubated with 3,3′-diaminobenzidine for 10 min and then counterstained with 0.1% hematoxylin.
Immunohistochemical results were evaluated by two independent pathologists without clinical information. The expression of UBQLN4 was determined by the staining intensity and the percentage of positive cells. The intensities were divided into four scores: ‘0’, no brown particles staining; ‘1’, light-brown particles; ‘2’, medium-brown particles; and ‘3’, dark-brown particles. The percentage of the positive cells in UBQLN4 was also divided into four scores: ‘0’ referred to < 10% positive cells, ‘1’ to 10%–40%; ‘2’ to 40%–70%; and ‘3’ to ≥ 70%. The ratio between the ‘staining intensity score’ and the ‘staining positive rate’ was employed for further grouping; a ratio value of 1 was considered the cutting point.
All patients were followed up for a period from 5.5 to 6.8 years. A number of 33 patients died during that period. Progression-free survival (PFS) was defined as the time between the date of surgery and the date of the confirmation of local recurrence or distant metastasis. Overall survival (OS) was defined as the time between the dates of surgery and death due to any reason, or the date of the last contact with the patient.
Chi-square test was applied to compare the clinicopathological features of different groups. Paired t-test (two-tailed) was utilized to compare the differences between groups. Univariate and multivariate Cox regression analysis were employed to determine the independent prognostic factors in patients with cervical cancer. Kaplan-Meier and the log-rank tests were used for comparisons of the differences in the OS among different groups. Data analysis was performed using SPSS V23.0 (IBM, Armonk, NY, United States). All statistical tests were bilateral. P < 0.05 was considered to indicate statistically significant differences.
The statistical methods of this study were reviewed by Huang KJ and Wang L from Harbin Medical University Cancer Hospital (Harbin, China).
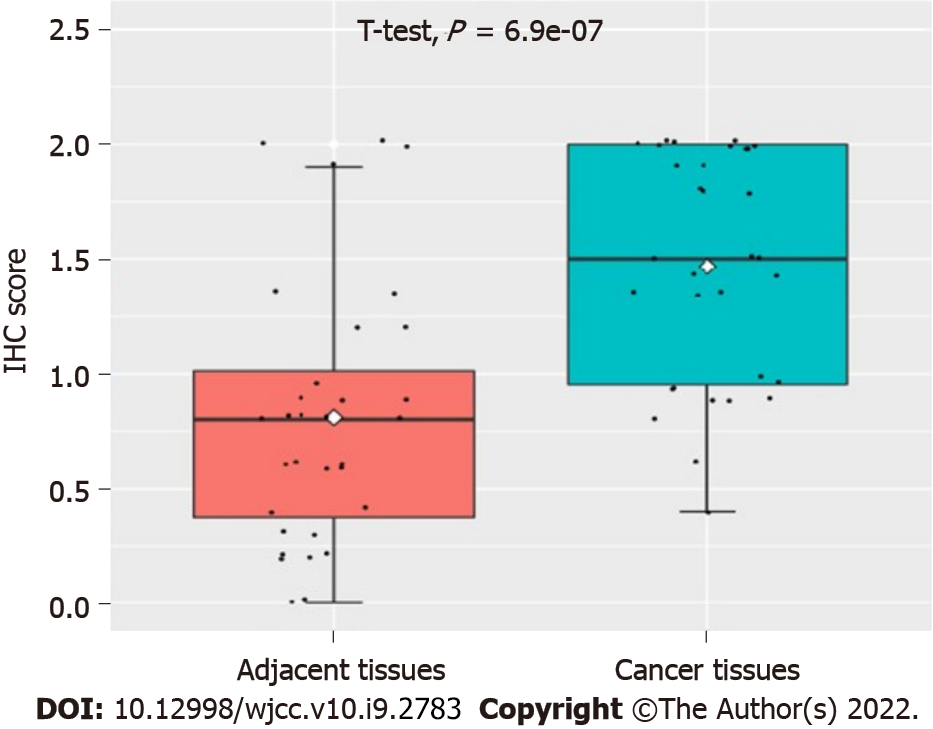
As can be seen in Figure 1, our data suggested that the UBQLN4 expression can be identified in the cytoplasm and nucleus of cells in both cervical cancer and paracancerous tissue. However, the positive rate of UBQLN4 in cervical cancer tissues was significantly higher than in parauterine tissues (70.9% vs 15.6%, P < 0.05) (Figure 2).
We then summarized the associations between UBQLN4 expression and clinicopathological features. As presented in Table 1, the expression of UBQLN4 was associated with lymph node metastasis, poor differentiation and advanced stage, but the difference was not significant.
| Variables | UBQLN4 expression | Total | Χ2 | P value | ||
| Low | High | |||||
| Age (yr) | 0.713 | 0.398 | ||||
| ≤ 45 | 34 | 24 (41.4%) | 58 | |||
| > 45 | 30 | 29 (49.2%) | 59 | |||
| Null | ||||||
| Pathologic type | 0.244 | 0.885 | ||||
| AD | 2 | 1 (33.3%) | 3 | |||
| ASC | 3 | 2 (40.0%) | 5 | |||
| SCC | 59 | 50 (45.9%) | 109 | |||
| Lymph node metastasis | 0.242 | 0.623 | ||||
| No | 53 | 42 (44.2%) | 95 | |||
| Yes | 11 | 11 (50.0%) | 22 | |||
| Histological grade | 1.518 | 0.279 | ||||
| I/II | 16 | 12 (42.9%) | 28 | |||
| III | 39 | 50 (56.2%) | 89 | |||
| FIGO stage (2018) | 0.242 | 0.623 | ||||
| I/II | 53 | 42 (44.2%) | 95 | |||
| III | 11 | 11 (50.0%) | 22 | |||
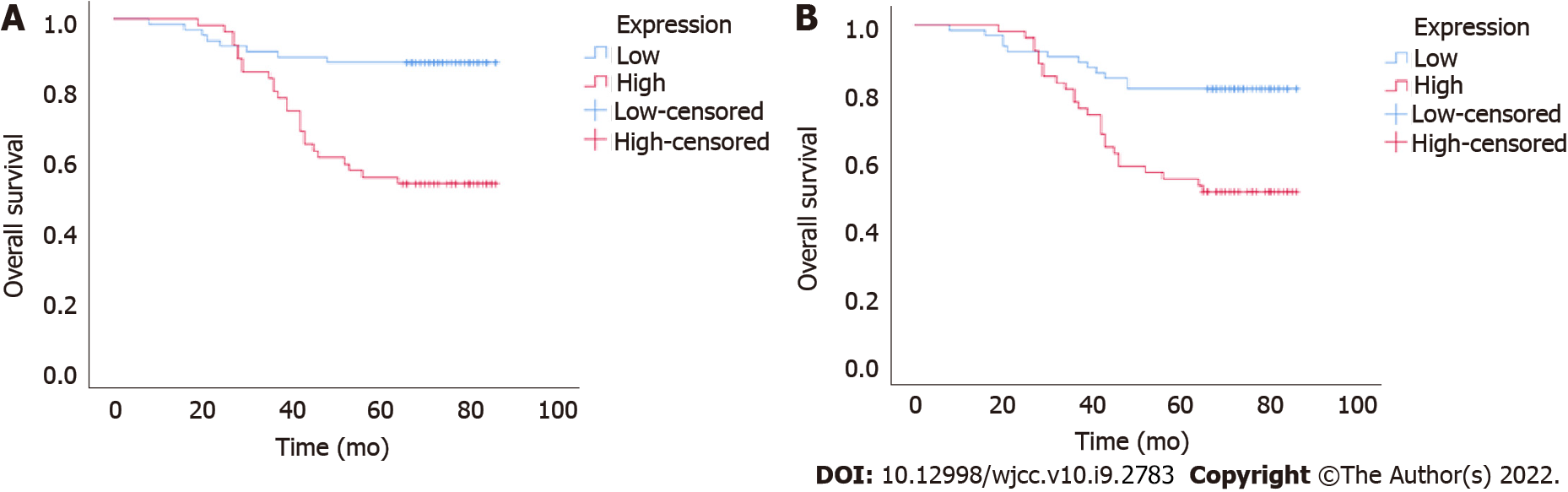
To further investigate the potential clinical value of UBQLN4 expression in cervical cancer, we examined its association with OS and PFS. Kaplan-Meier and log-rank test suggested high expression of UBQLN4 was associated with short OS and PFS (Figure 3). Regardless of UBQLN4 expression, patient age and FIGO stage were also associated with disease prognosis. Statistically significant variables obtained from univariate Kaplan-Meier analysis were included in Cox multivariate survival regression analysis which showed that in addition to FIGO stage and age, UBQLN4 was also an independent prognostic marker for OS and PFS (P = 0.011 and P = 0.024, respectively) (Table 2).
| Clinical variables | Univariate analysis (PFS) | Multivariate analysis (PFS) | Univariate analysis (OS) | Multivariate analysis (OS) | ||||||||
| HR | 95%CI | P value1 | HR | 95%CI | P value1 | HR | 95%CI | P value | HR | 95%CI | P value1 | |
| UBQLN4 (low vs high) | 3.026 | 1.524-6.006 | 0.002 | 2.261 | 1.115-4.583 | 0.024 | 4.357 | 1.962-9.679 | 0 | 2.93 | 1.284-6.683 | 0.011 |
| Age (≤ 45 vs > 45) | 4.944 | 2.261-10.809 | 0 | 4.331 | 1.953-9.605 | 0 | 7.171 | 2.763-18.611 | 0 | 5.927 | 2.241-15.678 | 0 |
| Grade (I/II vs III) | 1.819 | 0.805-4.108 | 0.15 | 2.444 | 0.913-6.542 | 0.075 | ||||||
| FIGO stage (I/II vs III) | 2.869 | 1.983-4.151 | 0 | 2.659 | 1.807-3.911 | 0 | 3.372 | 2.234-5.089 | 0 | 3.048 | 1.968-4.72 | 0 |
UBQLN protein is a critical regulator of protein homeostasis by transferring ubiquilinated proteins to proteasomes for protein degradation[5]. In addition, it also participates in the process of cell proliferation and apoptosis. UBQLN4 is a member of the UBQLN protein family that not only participates in protein degradation, but also promotes the activities of other UBQLN family members. The involvement of UBQLN1 has been identified in a variety of cancers, such as lungs[19] and breast cancers[20]. Additionally, they were found to participate in cell migration, invasion, and epithelial-mesenchymaltransformation[19]. UBQLN1 interacts with microtubule-associated protein light chain (LC3) to assist autophagosome maturation only in the presence of UBQLN4[6,21]. Therefore, UBQLN4 might more regulatory functions than those of the other members of this family.
In this study, we found that UBQLN4 was overexpressed in cervical cancer tissues, which was consistent with previous results obtained for other solid tumors[14]. We established that UBQLN4 was expressed in both the cytoplasm and the nucleus. An earlier investigation revealed that despite the main localization of UBQLN4 activities in the nucleus, it does not contain a nuclear localization sequence and could be detected in the cytoplasm[13]. These authors speculated that UBQLN4 was transferred to the nucleus in an undetermined post-translational modification. We further analyzed the association between UBQLN4 and the clinicopathological features of cervical cancer patients. Our data suggested that the expression of UBQLN4 was associated with lymph node metastasis, poor differentiation, and late stage, with clear but statistically insignificant trends. These inconclusive results might be associated with the small number of the samples analyzed. Additionally, they might suggest that UBQLN4 is has a stronger association with disease development rather than with cancer cell invasion and metastasis. Moreover, UBQLN4 is associated with DNA damage repair, autophagy, and other mechanisms[21], suggesting its involvement in radiotherapy sensitivity and chemotherapy resistance of cervical cancer cells. Future studies focusing on these questions will be carried out with larger sample sizes.
Increasing results have shown that p53, a well-known tumor suppressor, is tightly associated with the progression of cell cycle, and induces G2/M arrest in various types of tumors, including cervical cancer[22,23]. Stabilization of p53 by UBQLN4 overexpression enables transcriptional regulation of p21 by p53. In addition, the acetylated tumor protein p53 activates a cyclin-dependent kinase 1 inhibitor, a gene encoding p21. p21 protein level can be controlled translationally and post-translationally, especially by ubiquitination. Several E ubiquitin ligase complexes, including SCFSkp2, MKRN1, CRL4CDT, APC∕CCDC20, and RNF114 promote p21 ubiquitination and degradation. The overexpression of UBQLN4 induced cell aging and arrest at the G1-S phase of the cell cycle via the p53/p21 axis in a gastric cancer cell line[15]. Thus, the overexpression of UBQLN4 may play a role in cervical cancer via p53–associated activities. The increased expression of UBQLN4 was found to be associated with poor OS in neuroblastoma, ovarian cancer, melanoma, lung cancer, and breast cancer[13].
Our findings also suggest that the overexpression of UBQLN4 is associated with poor prognosis in cervical cancer. Our multivariate COX analysis showed that UBQLN4, AGE, and the FIGO stage were independent prognostic markers, which is consistent with the results of other studies.
In conclusion, the findings of our study indicated that the expression of UBQLN4 was increased in cervical cancer, and was associated with poor prognosis. Here, we not only propose a novel prognostic factor, but also improve the existing understanding of cervical cancer pathogenesis.
Ubiquilins (UBQLNs) are important factors for cell proteostasis maintenance. Abnormal UBQLNs expression can lead to many diseases, including cancer. Studies have found that the expression of Ubiquilin4 (UBQLN4) is associated with the development of several tumor types.
The association between UBQLN4 and cervical cancer has not been examined yet.
To investigate the expression of UBQLN4 in cervical cancer and to evaluate its correlation with disease prognosis.
Immunohistochemistry was performed to examine the expression of UBQLN4 in 117 cervical cancer tissues and 32 matching pericervical tissues. Paired t-test (two-tailed) was used to compare the differences between groups. We collected patients’ clinical characteristics, including age, histological grade, pathologic type, lymph node metastasis, and FIGO stage (2018) and compared them by chi-square test. All patients were followed for 5.5 to 6.8 years. Kaplan-Meier method and log-rank test were used to compare the differences in the overall survival (OS) and progression-free survival (PFS) among the different groups.
Overexpression of UBQLN4 was observed in 70.9% (83/117) of all cervical cancer tissues and in 15.6% (5/32) of the paired parauterine tissues. The expression of UBQLN4 was associated with lymph node metastasis, poor differentiation, and advanced stage, but the difference was not significant. Kaplan-Meier and log-rank test results suggested the high expression of UBQLN4 was associated with short OS and PFS. UBQLN4 was also an independent prognostic marker for OS and PFS (P = 0.011 and P = 0.024, respectively).
The expression of UBQLN4 was increased in cervical cancer, and was associated with poor prognosis.
We not only propose a novel prognostic factor, but also improve the existing understanding of cervical cancer pathogenesis.
The authors express their gratitude to all participants in this study.
| 1. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13326] [Article Influence: 1332.6] [Reference Citation Analysis (4)] |
| 2. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13302] [Article Influence: 1662.8] [Reference Citation Analysis (4)] |
| 3. | Marín I. The ubiquilin gene family: evolutionary patterns and functional insights. BMC EvolBiol. 2014;14:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 4. | Ko HS, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566:110-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 144] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 5. | Lee DY, Brown EJ. Ubiquilins in the crosstalk among proteolytic pathways. BiolChem. 2012;393:441-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 6. | Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, Monteiro MJ. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219-3232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 195] [Article Influence: 12.2] [Reference Citation Analysis (8)] |
| 7. | Şentürk M, Lin G, Zuo Z, Mao D, Watson E, Mikos AG, Bellen HJ. Ubiquilins regulate autophagic flux through mTORsignalling and lysosomal acidification. Nat Cell Biol. 2019;21:384-396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 108] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 8. | Lim PJ, Danner R, Liang J, Doong H, Harman C, Srinivasan D, Rothenberg C, Wang H, Ye Y, Fang S, Monteiro MJ. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol. 2009;187:201-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 122] [Article Influence: 7.2] [Reference Citation Analysis (8)] |
| 9. | El Ayadi A, Stieren ES, Barral JM, Boehning D. Ubiquilin-1 and protein quality control in Alzheimer disease. Prion. 2013;7:164-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 10. | Zhang KY, Yang S, Warraich ST, Blair IP. Ubiquilin 2: a component of the ubiquitin-proteasome system with an emerging role in neurodegeneration. Int J Biochem Cell Biol. 2014;50:123-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 11. | Jantrapirom S, Lo Piccolo L, Yamaguchi M. Non-ProteasomalUbL-UbA Family of Proteins in Neurodegeneration. Int J MolSci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Suzuki R, Kawahara H. UBQLN4 recognizes mislocalizedtransmembrane domain proteins and targets these to proteasomal degradation. EMBO Rep. 2016;17:842-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 61] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 13. | Jachimowicz RD, Beleggia F, Isensee J, Velpula BB, Goergens J, Bustos MA, Doll MA, Shenoy A, Checa-Rodriguez C, Wiederstein JL, Baranes-Bachar K, Bartenhagen C, Hertwig F, Teper N, Nishi T, Schmitt A, Distelmaier F, Lüdecke HJ, Albrecht B, Krüger M, Schumacher B, Geiger T, Hoon DSB, Huertas P, Fischer M, Hucho T, Peifer M, Ziv Y, Reinhardt HC, Wieczorek D, Shiloh Y. UBQLN4 Represses Homologous Recombination and Is Overexpressed in Aggressive Tumors. Cell. 2019;176:505-519.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 14. | Yu Y, Xu P, Cui G, Xu X, Li K, Chen X, Bao J. UBQLN4 promotes progression of HCC via activating wnt-β-catenin pathway and is regulated by miR-370. Cancer Cell Int. 2020;20:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Huang S, Li Y, Yuan X, Zhao M, Wang J, Lin H, Zhang Q, Wang W, Li D, Dong X, Li L, Liu M, Huang W, Huang C. The UbL-UBA Ubiquilin4 protein functions as a tumor suppressor in gastric cancer by p53-dependent and p53-independent regulation of p21. Cell Death Differ. 2019;26:516-530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 16. | Xu S, Li Y, Lu Y, Huang J, Ren J, Zhang S, Yin Z, Huang K, Wu G, Yang K. LZTS2 inhibits PI3K/AKT activation and radioresistance in nasopharyngeal carcinoma by interacting with p85. Cancer Lett. 2018;420:38-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 17. | Xie J, Li Y, Jiang K, Hu K, Zhang S, Dong X, Dai X, Liu L, Zhang T, Yang K, Huang K, Chen J, Shi S, Zhang Y, Wu G, Xu S. CDK16 Phosphorylates and Degrades p53 to Promote Radioresistance and Predicts Prognosis in Lung Cancer. Theranostics. 2018;8:650-662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 18. | Wang Q, Ma J, Lu Y, Zhang S, Huang J, Chen J, Bei JX, Yang K, Wu G, Huang K, Xu S. CDK20 interacts with KEAP1 to activate NRF2 and promotes radiochemoresistance in lung cancer cells. Oncogene. 2017;36:5321-5330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 19. | Shah PP, Lockwood WW, Saurabh K, Kurlawala Z, Shannon SP, Waigel S, Zacharias W, Beverly LJ. Ubiquilin1 represses migration and epithelial-to-mesenchymal transition of human non-small cell lung cancer cells. Oncogene. 2015;34:1709-1717. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 20. | Wang Y, Lu J, Zhao X, Feng Y, Lv S, Mu Y, Wang D, Fu H, Chen Y, Li Y. Prognostic significance of Ubiquilin1 expression in invasive breast cancer. Cancer Biomark. 2015;15:635-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Lee DY, Arnott D, Brown EJ. Ubiquilin4 is an adaptor protein that recruits Ubiquilin1 to the autophagy machinery. EMBO Rep. 2013;14:373-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 22. | Zhang Z, Wang CZ, Du GJ, Qi LW, Calway T, He TC, Du W, Yuan CS. Genistein induces G2/M cell cycle arrest and apoptosis via ATM/p53-dependent pathway in human colon cancer cells. Int J Oncol. 2013;43:289-296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 23. | Yao G, Qi M, Ji X, Fan S, Xu L, Hayashi T, Tashiro S, Onodera S, Ikejima T. ATM-p53 pathway causes G2/M arrest, but represses apoptosis in pseudolaric acid B-treated HeLa cells. Arch BiochemBiophys. 2014;558:51-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Choudhary RK, Šarenac TM S-Editor: Fan JR L-Editor: A P-Editor: Fan JR