©The Author(s) 2024.
World J Clin Cases. Dec 16, 2024; 12(35): 6826-6833
Published online Dec 16, 2024. doi: 10.12998/wjcc.v12.i35.6826
Published online Dec 16, 2024. doi: 10.12998/wjcc.v12.i35.6826
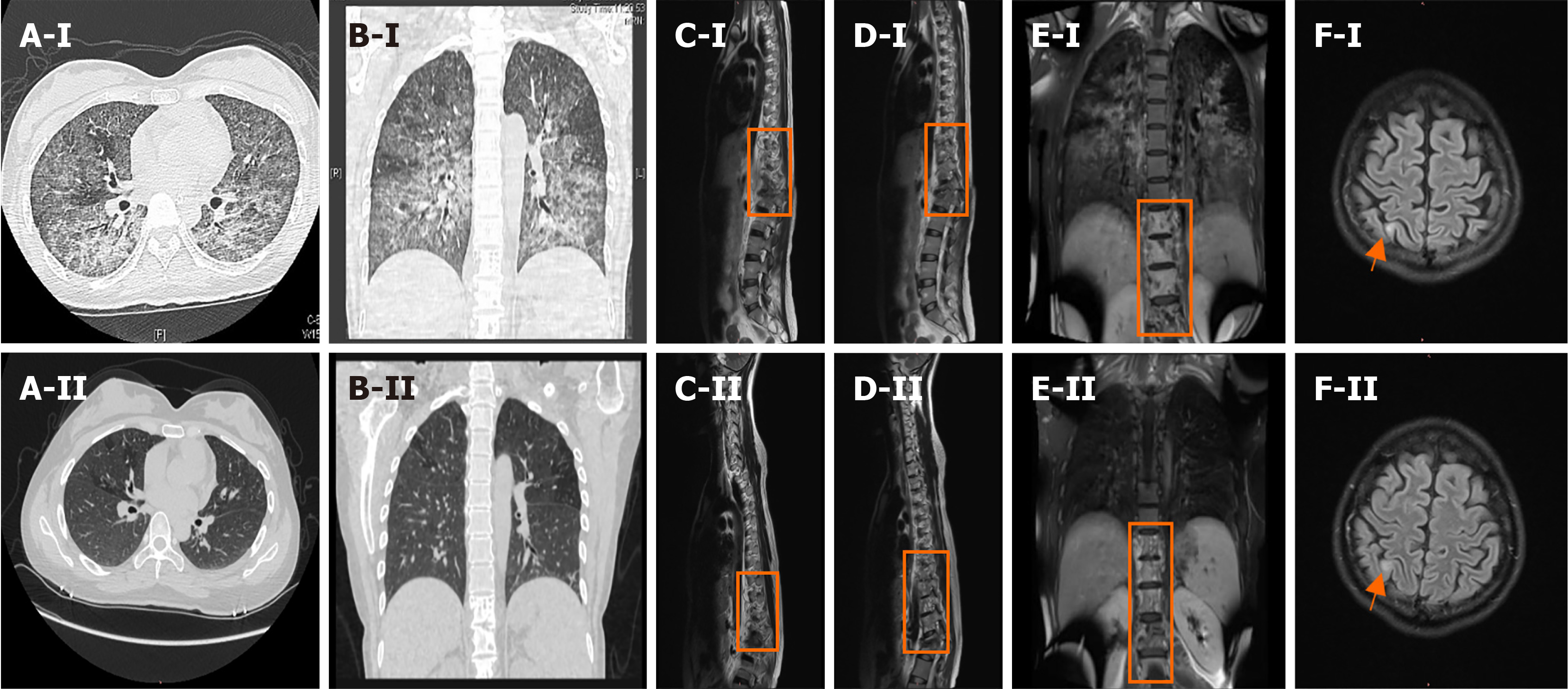
Figure 1 Chest computed tomography and enhanced magnetic resonance imaging of the thoracolumbar spine and brain.
A-I and B-I: Chest computed tomography scans showing diffuse lesions in both lungs on admission; C-I, D-I, and E-I: Enhanced magnetic resonance imaging (MRI) of the thoracolumbar spine performed on day 13, showing abnormal signal shadows from the ninth thoracic vertebral body to the first lumbar vertebral body and the surrounding soft tissues; F-I: Enhanced MRI of the brain performed on day 12, showing an abnormal right parietal lobe signal. A-II and B-II: The chest computed tomography scan performed 6 ½ months after admission, showing an improvement in the diffuse lesions in both lungs. C-II, D-II, and E-II: Enhanced MRI of the thoracic spine performed 6 ½ months after admission, showing improved signal shadows from the ninth thoracic vertebral body to the first lumbar vertebral body and the surrounding soft tissues; F-II: Enhanced MRI of the brain performed 6 ½ months after admission, showing similar signal intensities in the right parietal lobe and the frontal lobe.
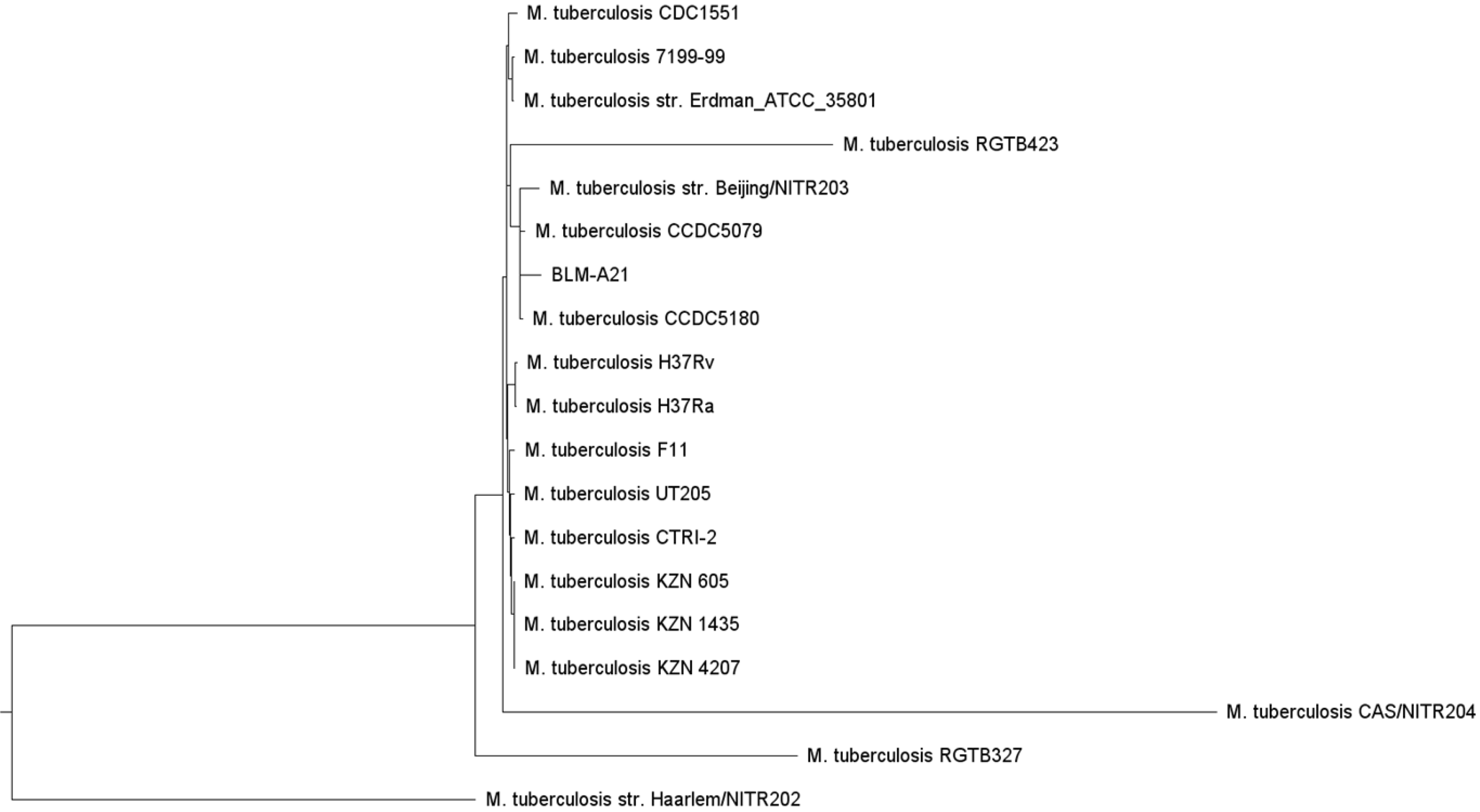
Figure 2 Whole-genome evolutionary tree of Mycobacterium tuberculosis.
Additional Mycobacterium tuberculosis genomes were selected from the Virulence Factor Database (VFDB) (http://www.mgc.ac.cn/cgi-bin/VFs/genus.cgi?Genus=Mycobacterium), developed by the bioinformatics research team of the Institute of Pathogenic Biology, Chinese Academy of Medical Science, and a phylogenetic tree was built using the PhyML (maximum likelihood) software. The whole genomes of several strains, including CCDC5079, CCDC5180, Beijing NTR203, CDC1551, and classic H37Rv and H37Ra, were analysed for subsequent genome comparison, as these strains have previously shown strong dissemination ability. The patient’s strain (BLM-A21) was closer to CCDC5079, CCDC5180, and Beijing NTR203 as shown on the evolutionary tree.
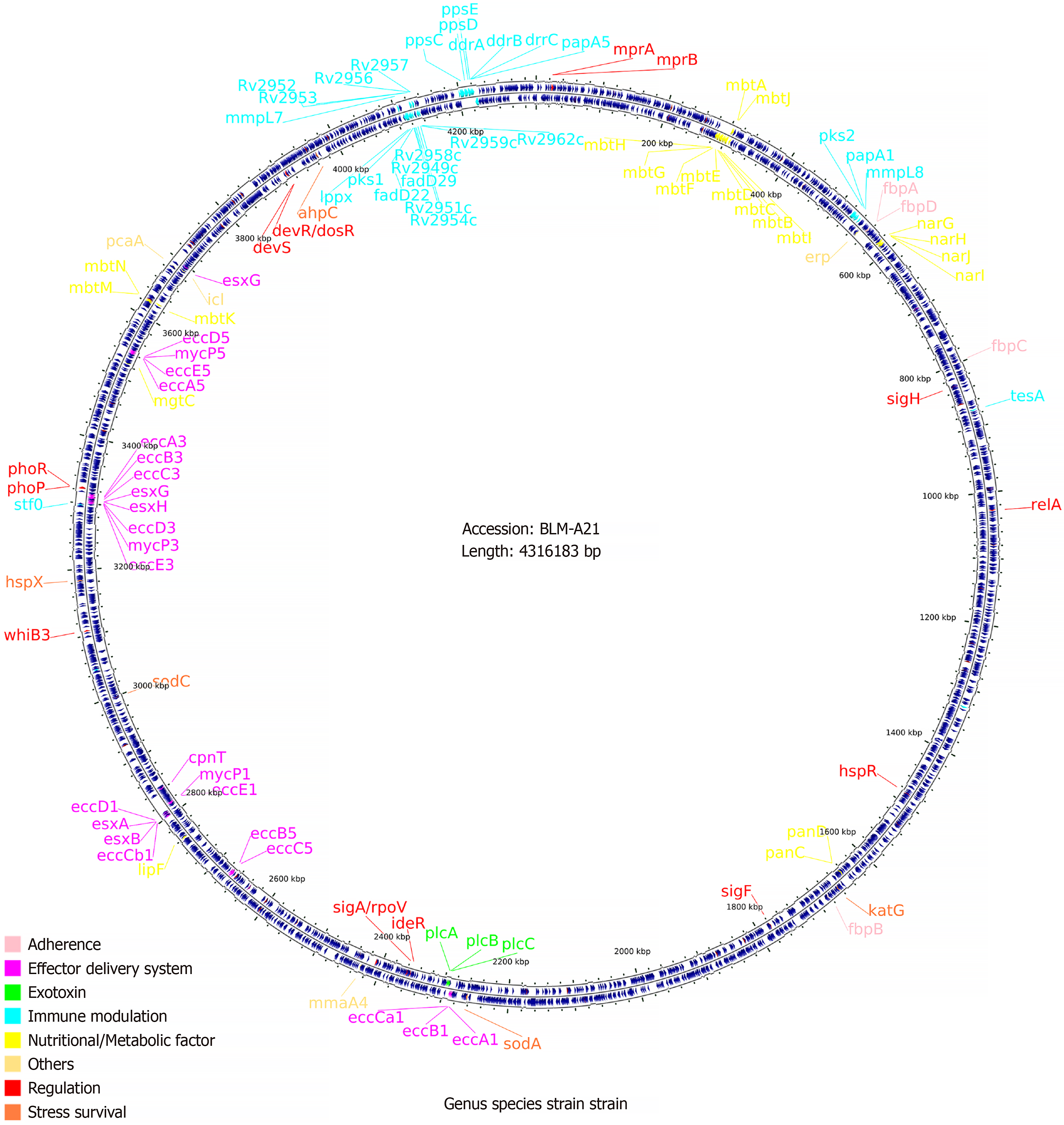
Figure 3 Chromosome genome circle map and virulence factor annotation of Mycobacterium tuberculosis strain BLM-A21.
The BLASTP performance comparison algorithm and the virulence factor protein reference sequence of the Virulence Factor Database was used to annotate genes of these species, and the virulence gene classification and genome position information was mapped using CGView.
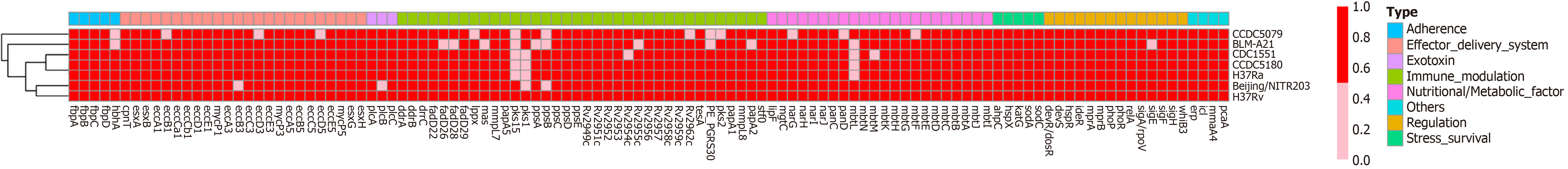
Figure 4 Virulence genes of Mycobacterium tuberculosis strains and their related genomes.
The red color indicates that the presence of the virulence gene, whereas the pink color indicates the absence of the virulence gene in the genome. As the figure shows, CDC1551 Lacks genes pks1–15. BLM-A21 carries the pks1 gene but lacks pks15; nevertheless, the number of other virulence genes is essentially consistent with that of CDC1551 and H37Rv.
- Citation: Wu F, Yang B, Xiao Y, Ren LL, Chen HY, Hu XL, Pan YY, Chen YS, Li HR. psk1 virulence gene-induced pulmonary and systemic tuberculosis in a young woman with normal immune function: A case report. World J Clin Cases 2024; 12(35): 6826-6833
- URL: https://www.wjgnet.com/2307-8960/full/v12/i35/6826.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i35.6826