Published online Jun 25, 2025. doi: 10.5501/wjv.v14.i2.106108
Revised: April 6, 2025
Accepted: April 24, 2025
Published online: June 25, 2025
Processing time: 126 Days and 16.7 Hours
The emergence of the Omicron variant (B.1.1.529) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) raised global concerns with its highly transmissible nature.
To investigate the genomic, clinical, and demographic characteristics of Omicron infections within the early outbreak cluster in western part of Sri Lanka.
We analyzed sequence data from January 2022 to April 2022 to understand variant dynamics, clinical presentation, and demographic associations.
Whole-genome sequencing of 85 nasopharyngeal and throat swab samples collected in western part of Sri Lanka between January and April 2022 identified 70 (82.34%) of it as Omicron variants. BA.2 was the most prevalent sub-lineage (57%), followed by BA.1.1 (14.20%) and majority of them were from > 12 years old individuals. Phylogenetic analysis revealed clustering into four distinct clades (21I, 21K, 21L, and 21M), suggesting potential differences in transmission chains or evolutionary pressures.
This study found BA.2 to be the predominant Omicron sub-lineage in the western part of Sri Lanka during the first quarter of 2022, aligning with global trends. Phylogenetic analysis revealed diverse introductions and local transmission. Continued genomic surveillance and robust public health measures remain crucial for managing the evolving SARS-CoV-2 landscape.
Core Tip: This study investigates into the critical period of the early Omicron outbreak in Western Sri Lanka, providing valuable insights into the variant's genomic profile and its impact on the local population. The emergence of the Omicron variant, specifically B.1.1.529, triggered global alarm due to its high transmissibility. In this retrospective study, we aimed to characterize the genomic, clinical, and demographic features of Omicron infections during the initial outbreak phase in Western Sri Lanka, spanning from January to April 2022. Methodology involved analyzing sequence data from 85 nasopharyngeal and throat swab samples. Whole-genome sequencing, conducted using the Oxford Nanopore Midnight protocol and analyzed via the Epi2Me platform, revealed that 70 samples, representing 82.34% of the total, were Omicron variants. We utilized bioinformatic tools such as Mega 11, Nextstrain, and PangoLineage for in-depth phylogenetic and lineage analysis. Demographic and clinical data were extracted from patient request forms. Findings highlighted the dominance of the BA.2 sub-lineage, accounting for 57% of the Omicron cases, followed by BA.1.1 at 14.20%. The study population primarily consisted of individuals over 12 years of age, and a male predominance was observed. Phylogenetic analysis revealed distinct clustering into clades 21K, 21L, and 21M, suggesting multiple introductions and local transmission events. The prevalence of BA.2 aligns with global trends during that period, emphasizing its enhanced transmissibility and immune evasion. The demographic data, showing a higher incidence in adults and males, raised questions about vaccine effectiveness and potential gender-specific factors. The phylogenetic clustering indicates a complex transmission dynamic, highlighting the need for continuous genomic surveillance. In conclusion, this study underscores the importance of genomic surveillance and robust public health measures in managing the evolving severe acute respiratory syndrome coronavirus 2 landscape. The dominance of BA.2 and the observed demographic patterns offer crucial insights for targeted public health interventions. However, the retrospective design and limited sample size suggest the need for future research with larger, more diverse datasets to further elucidate the impact of Omicron and its sub-lineages.
- Citation: Arachchige N, Dharmasiri R, Weerathunga A, Senanayake S, Janage N, Muthugala R. Genomic and demographic characterization of SARS-CoV-2 infections within early Omicron cluster, Western Sri Lanka. World J Virol 2025; 14(2): 106108
- URL: https://www.wjgnet.com/2220-3249/full/v14/i2/106108.htm
- DOI: https://dx.doi.org/10.5501/wjv.v14.i2.106108
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has demonstrated a remarkable ability to evolve, leading to the emergence of novel variants with distinct genetic profiles and clinical characteristics. The novel SARS-CoV-2 variant (B.1.1.529) was first reported to the World Health Organization on 24th of November 2021 and it was named as the Omicron variant[1]. This variant raised concerns due to its various mutations, especially in the spike protein region[2]. Moreover Omicron exhibits significant resistance to the neutralizing activity of vaccines, convalescent serum, and most antibody therapies[3,4]. Different from other variants of concern of SARS-CoV-2, the Omicron variant and its sub lineages exhibit increased transmissibility and immune escape from neutralizing antibodies generated through previous infection or vaccination, and have caused numerous re-infections and breakthrough infections[3,5]. According to the data from Global Initiative on Sharing All Influenza Data (GISAID) database up to 29th November 2024, 219 countries have shared 16843380 viral genome sequences from human cases of coronavirus disease 2019 (COVID-19) since 10th January 2020[6].
The Epidemiology Unit of the Ministry of Health, Sri Lanka has identified three distinct waves of COVID-19 infections since the initial detection of SARS-CoV-2 in January 2020. The third wave, spanning from April 2021, to May 2022, was characterized by the highest number of cases and associated deaths. Epidemiological data indicate that the Western Province, particularly the districts of Colombo, Gampaha, and Kalutara, experienced the most significant disease burden[7].
During the period from January 2022 to April 2022, the Omicron variant was the predominant strain circulating in Sri Lanka, as evidenced by GISAID sequence data[6]. Between November 2021 and November 2024, Sri Lanka submitted 1192 Omicron sequences to GISAID database.
While numerous studies have examined the COVID-19 pandemic and its associated factors in Sri Lanka, few have comprehensively investigated the Omicron variant, its evolutionary patterns, and its impact. This study aims to address this knowledge gap by analyzing sequence data from 2022 to understand the genetic evolution and spread of the Omicron variant within an early Omicron cluster in western part of Sri Lanka.
Investigating the clinical manifestations and outcomes associated with Omicron variant infections was essential for optimizing patient management and healthcare resources during the pandemic. While initial reports suggested milder symptoms associated with Omicron infections, the clinical impact could have vary across different populations and settings. By retrospectively analyzing genomic and clinical data from the early outbreak cluster in Western Sri Lanka, researchers can characterize the features of early Omicron clusters. By leveraging existing data and research methodologies, this study can contribute to our understanding of COVID-19 epidemiology, for the appropriate public health strategies, and ultimately help mitigate the impact of upcoming pandemics with the variant of interest.
In this context, our research aims to conduct a comprehensive study focusing on an early outbreak cluster of SARS-CoV-2 Omicron variant infections in Western Sri Lanka. By integrating genetic sequencing, clinical data analysis, and demographic profiling, we seek to provide understanding of the epidemiological and virological characteristics of Omicron within the local context.
Continuous genomic surveillance of SARS-CoV-2 is important to early identification of new variants, which might have high transmissible, high virulence or immune evasion properties. This will help in to adopt early control activities lead in to new explosive epidemics.
This was a retrospective, cross-sectional, observational study.
This study protocol was approved by the Ethics and Research Committees of the Medical Research Institute (MRI), Colombo (No. EC/19/2024).
The nasopharyngeal and throat swab samples were systemically selected for SARS-CoV-2 gene sequencing out of the samples received to the Molecular Biology Laboratory, MRI during the period between January 2022 and April 2022. Samples submitted for COVID-19 polymerase chain reaction (PCR) testing to the laboratory for Covid-19 diagnosis from hospitals and quarantine camps with a cycle threshold value below 30 and samples received directly for COVID-19 genomic sequencing by COVID-19 detection peripheral PCR laboratories were selected for the study. A total of 85 samples were sequenced during selected study period and genomic data was included for analysis.
The gene sequencing was carried out using the Oxford Nanopore Midnight protocol and the data were analysed using Epi2Me platform. Gene sequence data were submitted to the GISAID database. Whole genome sequencing data from SARS-CoV-2 isolates were retrieved in FASTA format. Consensus sequences were analyzed using the following bioinformatic tools: (1) Mega 11 software used for the multiple sequence alignment was performed with Muscle DNA; and (2) The phylogenetic tree building was performed using Neighbour Joining method. The Nextstrain platform aligned the sequences to the reference genome using a banded Waterman-Smith sequence alignment algorithm and phylogenetic tree was built using greedy parsimony tree builder. This was used for in-depth phylogenetic analysis to characterize outbreak dynamics and potential transmission routes. PangoLineage: This tool was employed for variant lineage assignment, with a specific focus on identifying Omicron infections.
Demographic and clinical data associated with the selected samples were analyzed. These data were extracted from request forms.
A total of 2005 samples were received to the laboratory from the western region of Sri Lanka between January 2022 and April 2022 and from those 85 samples were subjected whole genome sequencing. Of these, 70 (82.34 %) samples were identified as Omicron variants by both PangoLineage and NextClade databases. Others were belonged to delta variants.
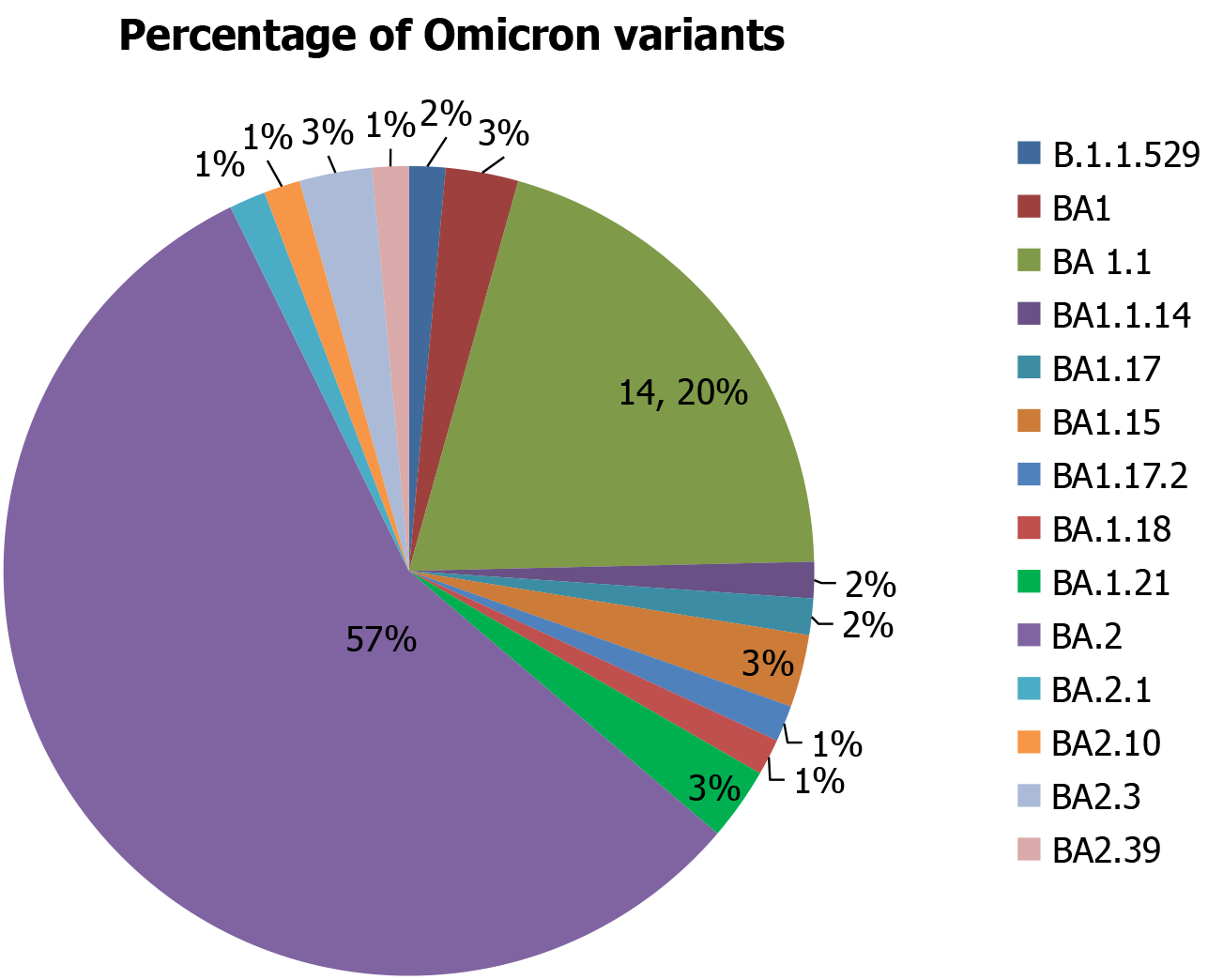
The most prevalent Omicron sub-lineage was BA.2, accounting for 57% of the identified cases. Other significant sub-lineages included BA.1.1 (14.20%), followed by B.1.1.529 (2%), BA.1 (3%), BA.1.1.14 (2%), BA.1.17 (2%), BA.1.15 (3%), BA.1.17.2 (1%), BA.1.18 (1%), BA.1.21 (3%), BA.2.1 (1%), BA.2.10 (1%), BA.2.3 (3%), and BA.2.39 (1%) (Figure 1).
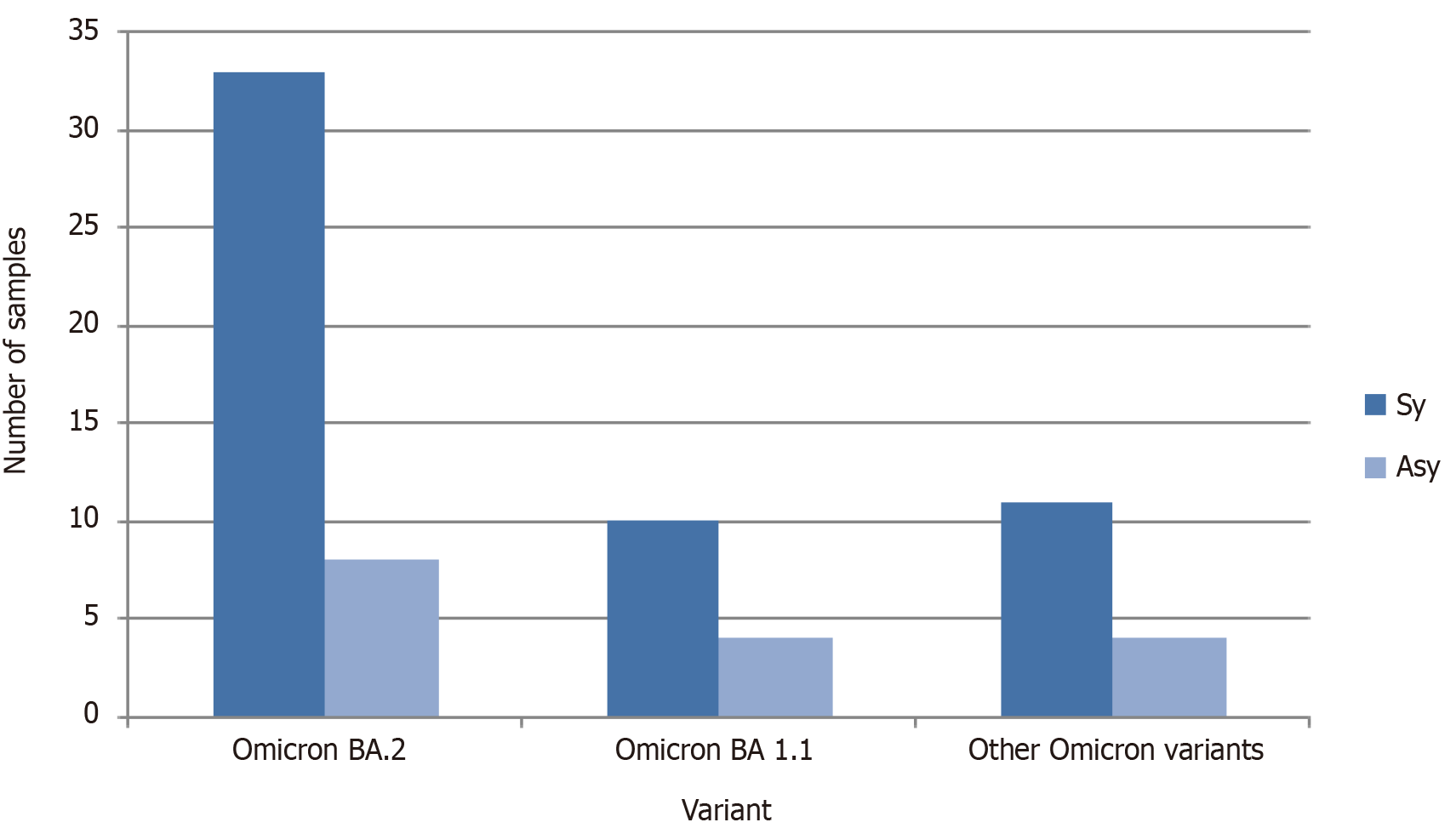
Those 70 samples were analyzed according the symptomatic and asymptomatic status (Figure 2). Majority of the patients were symptomatic and infected with omicron BA. 2.
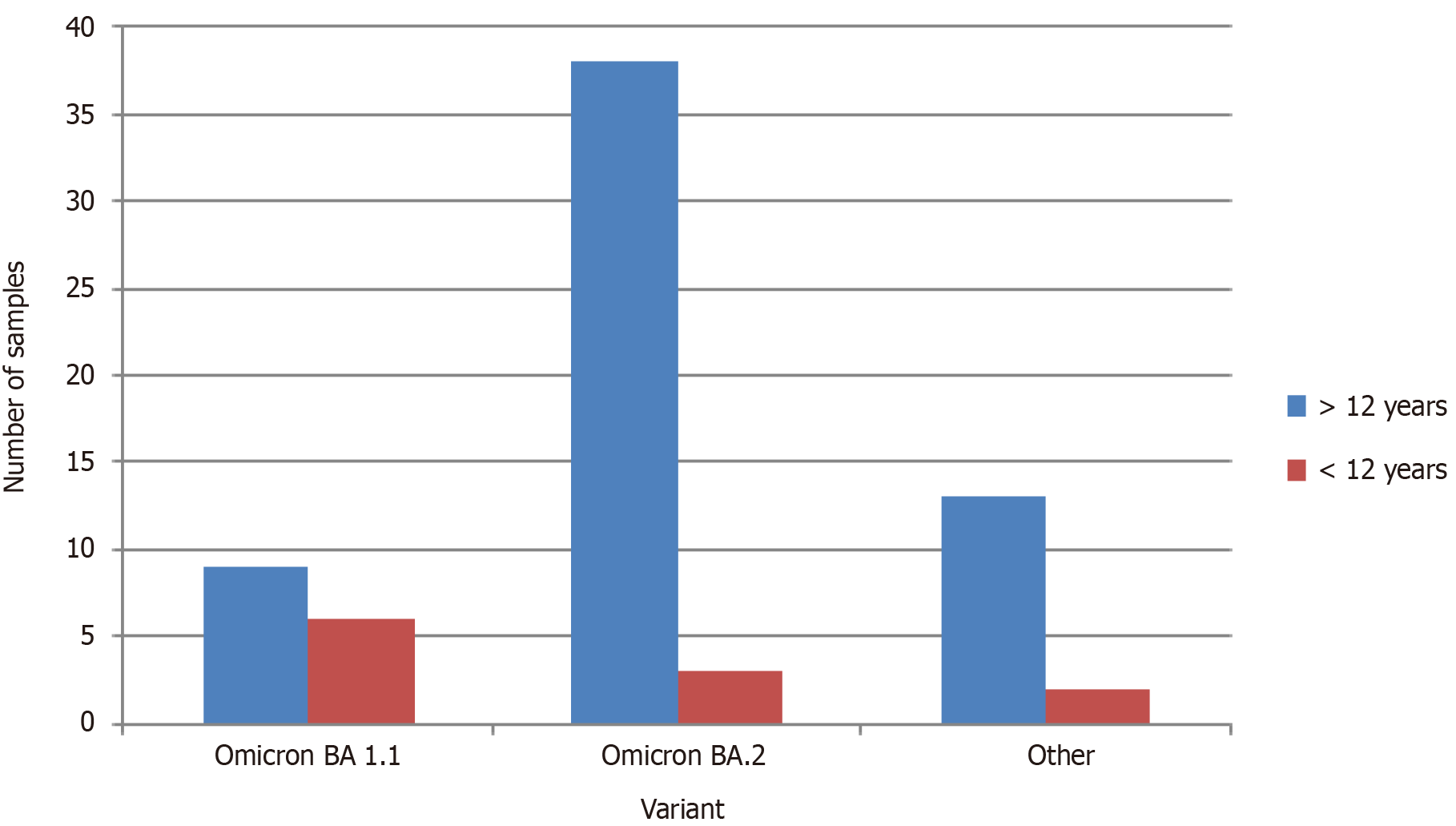
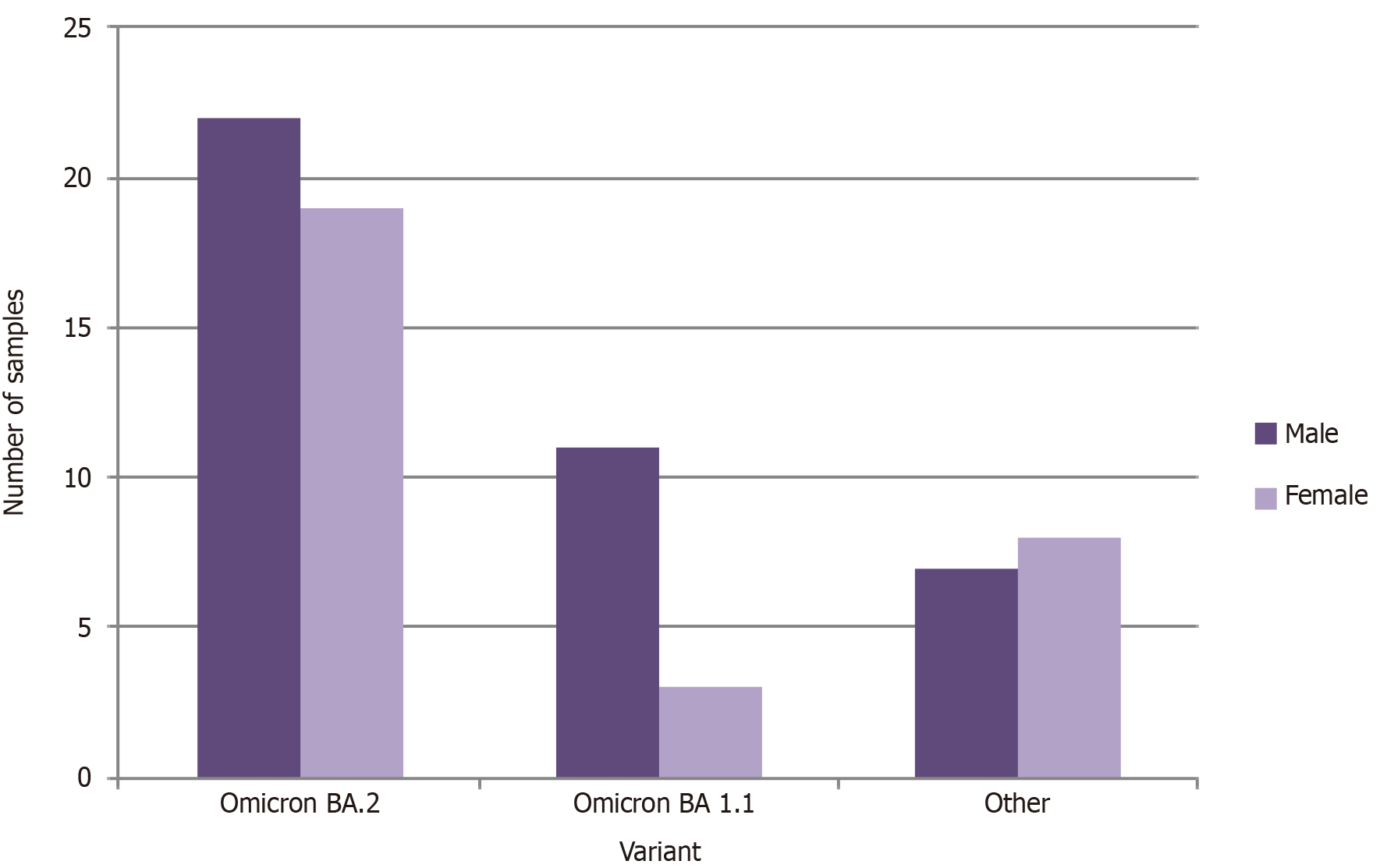
Study population was categorized in to two groups as > 12 years and < 12 years in order to observe the effect of the vaccination for the incidence of COVID-19. The vaccine was given to the individuals > 12 years of age in Sri Lanka. Omicron variant BA 1.1 and BA 2 were selected as the major two categories for the analysis since they were showing the highest prevalence (Figure 3). The majority of the individuals were > 12 years old. According to the findings of the current study, majority of the individuals from both Omicron BA 2 and Omicron BA1.1 category were male (Figure 4).
To further investigate the evolutionary dynamics of these Omicron variants, a phylogenetic tree was constructed using Mega 11 software using the neighbor joining method. The SARS-CoV-2 Wuhan-Hu-1 isolate forms a distinct lineage, clearly separated from the majority of the analyzed transcripts. This separation signifies a unique evolutionary trajectory of the virus, setting it apart from the other RNA sequences examined. While clearly distinct from the Wuhan-Hu-1 isolate, a subset of transcripts (e.g., T1136, T1131, T1126, T1195, T1246, T1181, etc.) exhibits a closer relationship to the viral sequence than others. This suggests potential homology or evolutionary relatedness. Within the transcripts, several distinct sub-clusters are observed, indicating diversification and independent evolutionary events. For instance, the cluster containing T1208, T1219, T1353, and T1139 suggests a specific group of related sequences. Further analysis is required to elucidate the functional implications of these sub-clusters. The branch lengths and topology of the tree reflect the evolutionary distances between the SARS-CoV-2 isolate and the transcripts. The relatively deep branching structure suggests that the divergence between the viral sequence and some of the transcripts is substantial, potentially indicative of distinct origins or functions (Figure 5).
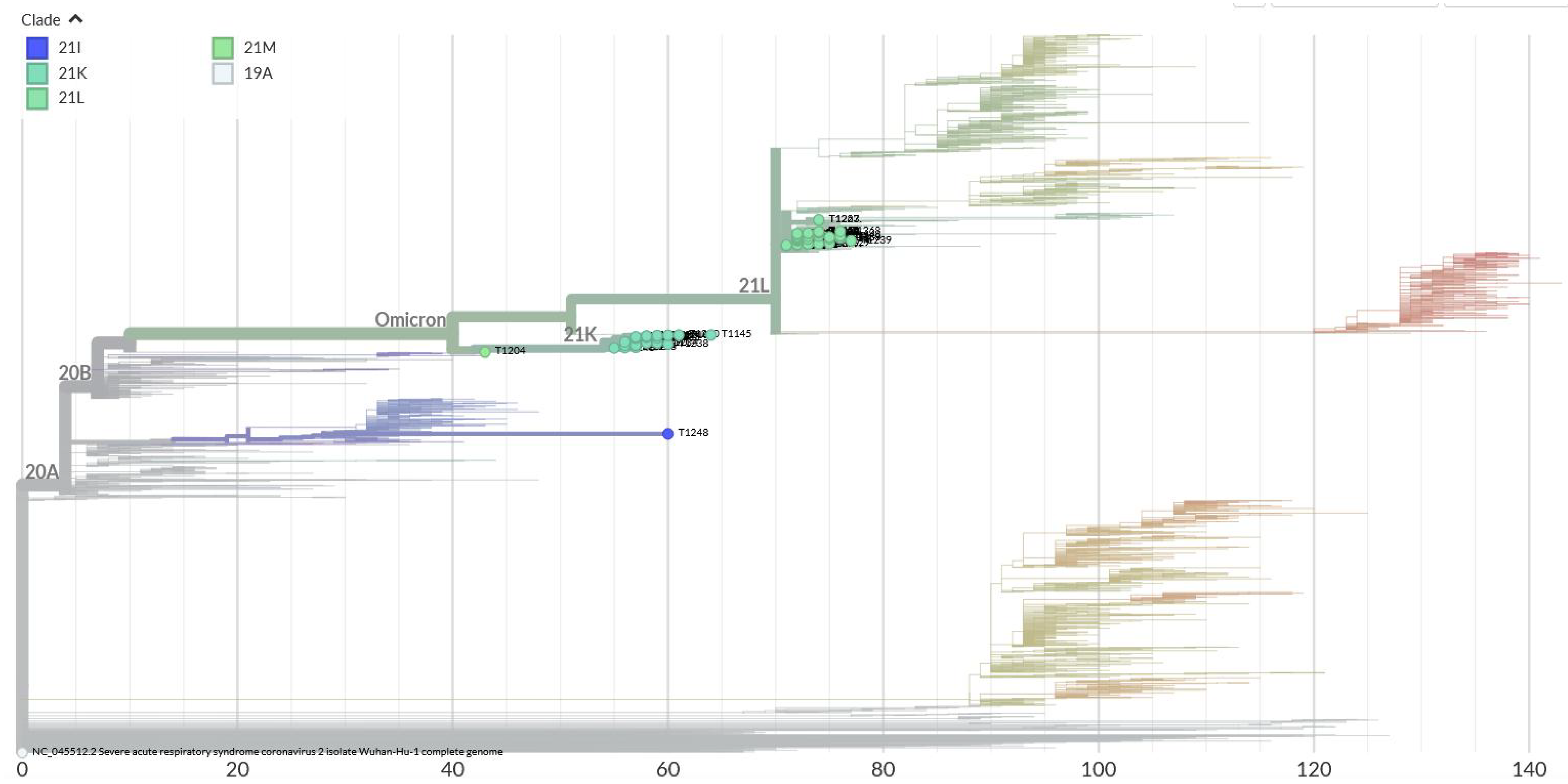
According to the phylogenetic analysis performed using the Nextclade database, 70 sequenced Omicron variants clustered into four distinct clades: (1) 21I; (2) 21K; (3) 21 L; and (4) 21M. This clustering suggests that these variants may have originated from different transmission chains or experienced unique evolutionary pressures (Figure 6).
The current study provides valuable insights into the SARS-CoV-2 Omicron variant infections in Western part of Sri Lanka during the first quarter of 2022. Our findings contribute to a growing body of evidence regarding the epidemiology, molecular characteristics, and clinical implications of Omicron sub-lineages.
The predominant Omicron variant identified was BA.2 (57%), aligning with global trends during the same period. BA.2, with its enhanced transmissibility and moderate immune evasion compared to BA.1[8,9], has been a significant driver of COVID-19 waves worldwide. The presence of other sub-lineages (BA.1.1, BA.1.17, BA.2.3, etc.) underscores the genetic diversity of Omicron, likely influenced by localized transmission dynamics and evolutionary pressures. BA.2, with eight unique mutations not found in BA.1 and lacking 13 mutations present in BA.1, shares approximately 30 mutations with BA.1[10]. Notably, these unique mutations, particularly in the spike protein, contribute to the increased transmissibility and immune evasion of BA.2[11].
Demographic analysis revealed a higher prevalence of Omicron BA.1.1 and BA.2 in individuals > 12 years old. Although the most of adult population were given vaccination by this time, we noticed high incidence of cases among adults. While some studies suggest waning vaccine efficacy over time[12-15], others support the current findings, indicating that vaccination may be less effective against BA.2 compared to other variants[16]. However, booster doses have been shown to provide substantial protection against Omicron infection and severe clinical outcomes[17]. Our study did not provide a definitive conclusion on vaccine effectiveness, but the low number of severe cases suggests that vaccination may still mitigate disease severity, particularly in older populations. A male predominance was observed across major sub-lineages, although some studies have reported a female predominance[18]. The reasons for this discrepancy remain unclear but may be related to gender-specific behaviors, occupations, or biological factors.
The majority of BA.2 cases were symptomatic, consistent with its classification as a more potentially transmissible sub-lineage than the sub-lineage BA.1. Phylogenetic analysis revealed clustering into clades 21I, 21K, 21L, and 21M, indicating multiple introductions of the virus into the region and subsequent local transmission.
These findings highlight the critical role of genomic surveillance in understanding variant dynamics and guiding public health interventions. The high prevalence of BA.2 underscores the need for continued monitoring, importance of vaccination and non-pharmaceutical interventions to control transmission. However, the retrospective design and limited sample size of this study constrain its generalizability. Future research should aim to incorporate larger, more diverse sample sets and investigate factors such as comorbidities, treatment outcomes, and vaccine effectiveness in greater detail. Expanding genomic sequencing efforts can also elucidate the emergence and spread of newer variants.
In conclusion, this study provides valuable insights into the complex interplay of epidemiological, demographic, and molecular factors shaping the Omicron variant's impact in western part of Sri Lanka. These findings can inform targeted public health strategies and guide future research into SARS-CoV-2 variant evolution.
We would like to acknowledge Prof. Andreas Nitsche and his team at the Robert Koch Institute, Germany for the support given for establishment of Oxford Nanopore Technologies (ONT) sequencing facility at the Molecular Biology Laboratory, Medical Research Institute under Identification of Emerging Agents project. We also thankful to Prof. Inoka Perera and his team at the Department of Zoology, Faculty of Science, University of Colombo, Sri Lanka to provide training and coordinating on ONT sequencing in initial part of severe acute respiratory syndrome coronavirus 2 genomic surveillance activity. Ministry of Heath Sri Lanka and World Health Organization is acknowledged for providing reagents and administrative support.
| 1. | World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available from: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern. |
| 2. | Xia S, Wang L, Zhu Y, Lu L, Jiang S. Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages. Signal Transduct Target Ther. 2022;7:241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 112] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 3. | Fan Y, Li X, Zhang L, Wan S, Zhang L, Zhou F. SARS-CoV-2 Omicron variant: recent progress and future perspectives. Signal Transduct Target Ther. 2022;7:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 466] [Cited by in RCA: 398] [Article Influence: 99.5] [Reference Citation Analysis (0)] |
| 4. | Takashita E, Kinoshita N, Yamayoshi S, Sakai-Tagawa Y, Fujisaki S, Ito M, Iwatsuki-Horimoto K, Chiba S, Halfmann P, Nagai H, Saito M, Adachi E, Sullivan D, Pekosz A, Watanabe S, Maeda K, Imai M, Yotsuyanagi H, Mitsuya H, Ohmagari N, Takeda M, Hasegawa H, Kawaoka Y. Efficacy of Antibodies and Antiviral Drugs against Covid-19 Omicron Variant. N Engl J Med. 2022;386:995-998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 308] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 5. | Dhawan M, Saied AA, Mitra S, Alhumaydhi FA, Emran TB, Wilairatana P. Omicron variant (B.1.1.529) and its sublineages: What do we know so far amid the emergence of recombinant variants of SARS-CoV-2? Biomed Pharmacother. 2022;154:113522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 6. | Global Initiative on Sharing All Influenza Data data base. Available from: https://www.epicov.org/epi3/frontend#2a1322 ; accessed on 29.11.2024. |
| 7. | Epidemiology Unit. COVID-19 disease surveillance and vaccination. https://www.epid.gov.lk/epid/public/index.php/covid-19-data; Accessed on 10.11.2024. |
| 8. | Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, Ito J, Uriu K, Tsuda M, Zahradnik J, Shirakawa K, Suzuki R, Kishimoto M, Kosugi Y, Kobiyama K, Hara T, Toyoda M, Tanaka YL, Butlertanaka EP, Shimizu R, Ito H, Wang L, Oda Y, Orba Y, Sasaki M, Nagata K, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Kuramochi J, Seki M, Fujiki R, Kaneda A, Shimada T, Nakada TA, Sakao S, Suzuki T, Ueno T, Takaori-Kondo A, Ishii KJ, Schreiber G; Genotype to Phenotype Japan (G2P-Japan) Consortium, Sawa H, Saito A, Irie T, Tanaka S, Matsuno K, Fukuhara T, Ikeda T, Sato K. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022;185:2103-2115.e19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 234] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 9. | Lyngse FP, Kirkeby CT, Denwood M, Christiansen LE, Mølbak K, Møller CH, Skov RL, Krause TG, Rasmussen M, Sieber RN, Johannesen TB, Lillebaek T, Fonager J, Fomsgaard A, Møller FT, Stegger M, Overvad M, Spiess K, Mortensen LH. Household transmission of SARS-CoV-2 Omicron variant of concern subvariants BA.1 and BA.2 in Denmark. Nat Commun. 2022;13:5760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 86] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 10. | Chen J, Wei GW. Omicron BA.2 (B.1.1.529.2): high potential to becoming the next dominating variant. Res Sq. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 11. | Kumar S, Karuppanan K, Subramaniam G. Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment. J Med Virol. 2022;94:4780-4791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 135] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 12. | Andrews N, Tessier E, Stowe J, Gower C, Kirsebom F, Simmons R, Gallagher E, Thelwall S, Groves N, Dabrera G, Myers R, Campbell CNJ, Amirthalingam G, Edmunds M, Zambon M, Brown K, Hopkins S, Chand M, Ladhani SN, Ramsay M, Lopez Bernal J. Duration of Protection against Mild and Severe Disease by Covid-19 Vaccines. N Engl J Med. 2022;386:340-350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 485] [Cited by in RCA: 504] [Article Influence: 126.0] [Reference Citation Analysis (0)] |
| 13. | Chemaitelly H, Tang P, Hasan MR, AlMukdad S, Yassine HM, Benslimane FM, Al Khatib HA, Coyle P, Ayoub HH, Al Kanaani Z, Al Kuwari E, Jeremijenko A, Kaleeckal AH, Latif AN, Shaik RM, Abdul Rahim HF, Nasrallah GK, Al Kuwari MG, Al Romaihi HE, Butt AA, Al-Thani MH, Al Khal A, Bertollini R, Abu-Raddad LJ. Waning of BNT162b2 Vaccine Protection against SARS-CoV-2 Infection in Qatar. N Engl J Med. 2021;385:e83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 668] [Cited by in RCA: 620] [Article Influence: 124.0] [Reference Citation Analysis (0)] |
| 14. | Andrews N, Stowe J, Kirsebom F, Toffa S, Rickeard T, Gallagher E, Gower C, Kall M, Groves N, O'Connell AM, Simons D, Blomquist PB, Zaidi A, Nash S, Iwani Binti Abdul Aziz N, Thelwall S, Dabrera G, Myers R, Amirthalingam G, Gharbia S, Barrett JC, Elson R, Ladhani SN, Ferguson N, Zambon M, Campbell CNJ, Brown K, Hopkins S, Chand M, Ramsay M, Lopez Bernal J. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N Engl J Med. 2022;386:1532-1546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1597] [Cited by in RCA: 1683] [Article Influence: 420.8] [Reference Citation Analysis (4)] |
| 15. | Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, Subbarao K, Kent SJ, Triccas JA, Davenport MP. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27:1205-1211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2207] [Cited by in RCA: 2891] [Article Influence: 578.2] [Reference Citation Analysis (0)] |
| 16. | Rigby J, Steenhuysen J. Omicron BA.2 variant: Here’s what we know so far. World Economic Forum. . 2022. |
| 17. | Song S, Madewell ZJ, Liu M, Longini IM, Yang Y. Effectiveness of SARS-CoV-2 vaccines against Omicron infection and severe events: a systematic review and meta-analysis of test-negative design studies. Front Public Health. 2023;11:1195908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 18. | Wang L, Berger NA, Kaelber DC, Davis PB, Volkow ND, Xu R. COVID infection rates, clinical outcomes, and racial/ethnic and gender disparities before and after Omicron emerged in the US. medRxiv. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/