Published online Oct 19, 2025. doi: 10.5498/wjp.v15.i10.109425
Revised: June 27, 2025
Accepted: August 8, 2025
Published online: October 19, 2025
Processing time: 139 Days and 1.2 Hours
Major depressive disorder (MDD) is a significant psychiatric condition that poses a serious threat to human life, primarily due to its association with suicidal beha
To reveal the biological and pathological significance of B4GALT6 in the brain and MDD.
An inflammation-associated depression mouse model was developed by treating mice with lipopolysaccharides. B4GALT6-like immunoreactivity distribution and expression level was examined throughout the brain.
B4GALT6-like immunoreactivity increased in the hippocampus, PrL, and the vi
Although the neuropathological role of B4GALT6 in the brain remains to be fully characterized, B4GALT6 may be a potential therapeutic target for MDD.
Core Tip: Major depressive disorder (MDD) poses a serious threat to human life. The single nucleotide polymorphism rs372369000 located within the B4GALT6 gene is a risk locus for MDD. However, the pathological mechanism of B4GALT6 in the brain in relation to MDD remains unclear. We developed an inflammation-associated depression model and examined B4GALT6-like immunoreactivity distribution and levels throughout the brain. The expression of B4GALT6-like immunoreactivity increased in the hippocampus, PrL, and the visual cortex V1 region of MDD mice. This elevation varied across different brain subregions and cell types. Taken together, our data suggest that B4GALT6 may be a potential therapeutic target for MDD.
- Citation: Zhang SN, Luan D, Sheng BY, Xie B, Xiao L. Brain region and cell specificity of B4GALT6 in mice with depressive-like behavior: A pilot study. World J Psychiatry 2025; 15(10): 109425
- URL: https://www.wjgnet.com/2220-3206/full/v15/i10/109425.htm
- DOI: https://dx.doi.org/10.5498/wjp.v15.i10.109425
Major depressive disorder (MDD) is characterized by significant and persistent depressive symptoms that stem from diverse etiologies. The core features of MDD are disproportionate depressive mood and anhedonia. MDD is a complex psychiatric disorder with approximately 37% heritability[1]. Existing research shows that an intronic variant, rs372369000, located within the B4GALT6 gene, constitutes a risk locus for MDD[2].
B4GALT6 encodes for a type II membrane-bound glycoprotein, which plays a crucial role in lactosylceramide synthesis by transferring galactose from UDP-galactose to glucosylceramide[3-5]. Lactosylceramide serves as a critical precursor for ganglioside biosynthesis, which is essential for neuronal maturation and axon and myelin formation[6]. Studies have shown that abnormal interactions of glycosyltransferases/glycosidases such as B4GALT6 can cause imbalances in key glycosylation modifications, thereby driving the disease process through a triple cascade reaction: (1) Galactosylation defects of neuroadhesion proteins (such as integrins, NCAMs) lead to conformational and functional disorders, blocking integrin-mediated extracellular matrix signal transduction; (2) The impaired integrin signal further weakens downstream pathway activation of f neurotrophic factors (especially BDNF), inactivating pro-survival signals such as PI3K/Akt and ERK/CREB; and (3) The collapse of the above-mentioned signal network ultimately inhibits neural stem cell proliferation and differentiation in the subgranular region of the dentate gyrus in the hippocampus, resulting in a significant reduction in neurogenesis. This phenomenon has been confirmed to be the core pathological basis for hippocampal volume atrophy and cognitive and emotional dysfunction in patients with depression and is directly associated with behavioral de
In the current investigation, bioinformatics analysis revealed that rs372369000 was located in a genomic region that governs transcriptional activity. We utilized an lipopolysaccharides (LPS)-induced depressive-like mouse model and found that B4GALT6 expression in the hippocampus, PrL, and V1 brain regions of LPS-treated mice was significantly elevated, and this upregulation was cell subtype-specific. These findings suggest a potential role for B4GALT6 in MDD pathophysiology and further highlight the importance of exploring underlying cell-specific mechanisms in the etiology of this complex disorder.
RegulomeDB[7,8], a tool for functional variation annotation, was employed to assess whether rs372369000 possesses the capability to regulate gene expression. This tool assigned a probability score for the variant, spanning from 0 to 1, with a score closer to 1 signifying a higher likelihood that the variant has a regulatory function. The Epigenome Roadmap[9] and ENCODE[10-12] databases were utilized to ascertain whether rs372369000 is located in a transcriptionally active region of DNA and unravel potential regulatory mechanisms.
Thirty male C57BL/6J mice aged 4-6 weeks were included in the experimental study. These mice were obtained from Changzhou Cavens Experimental Animal Co., Ltd. The experimental animal use was approved by the Experimental Animal Ethics Committee of Southeast University, with approval number 20220104003.
A sucrose preference test was used to assess the core symptom of MDD, known as anhedonia. The tail suspension test and forced swim test were used to evaluate depression mood. Weight measurements were used to evaluate symptoms of weight gain/Loss. The ANY-maze animal behavior analysis system was used to assess animal behavior. The depression model utilized in this study was based on the LPS-induced depressive-like behavior paradigm[13].
After acclimating to the environment for 1 week, baseline behavioral data of the mice were collected. Five mice were excluded as outliers because their baseline data were not within the range of two standard deviations plus or minus the average value. The remaining 25 mice were randomly assigned to the LPS and phosphate-buffered saline (PBS) groups. After no statistically significant difference was observed in two independent samples t-tests, the subsequent experimental process was initiated. LPS (Sigma-Aldrich, No. L2630) was dissolved in PBS and administered intraperitoneally at a dose of 1.0 mg/kg for 7 consecutive days. The PBS group received an equivalent volume of PBS intraperitoneally.
Western blots were performed as described previously[2,14]. In brief, RIPA lysis buffer (Servicebio, No. G2002) containing PMSF (Servicebio, No. G2008), NaF (Servicebio, No. G2006), and Na3VO4 (Servicebio, No. G2007) was added to mouse brain tissue samples. SDS-PAGE kits (Servicebio, No. G2003) were applied to prepare 10% separating gel and 5% concentrating gel. The FDbio-Dura ECL Kit (Fdbio science, No. FD8030) was used for visualization. The primary antibody was rabbit anti-B4GALT6 (Proteintech, No. 20148-1-AP). The secondary antibody was HRP-conjugated anti-rabbit (Proteintech, No. SA00001-2).
Immunofluorescence was performed as described previously[14]. Briefly, mouse brain tissue was dehydrated and embedded in paraffin. Paraffin sections were dewaxed and subjected to antigen retrieval. After sequentially incubating the primary and secondary antibodies, DAPI staining was performed. The primary antibodies used are as follows: B4GALT6 (Proteintech, No. 20148-1-AP), GFAP (Servicebio, No. GB12096-100), IBA1 (Servicebio, No. GB12105-100), and NeuN (Servicebio, No. GB15138-100). The secondary antibodies used are as follows: Cy3 (Servicebio, No. GB21301) and Alexa Fluor® 488 (Servicebio, No. GB25303).
Experimental results were expressed as mean ± SD and were analyzed using two-sample t-tests.
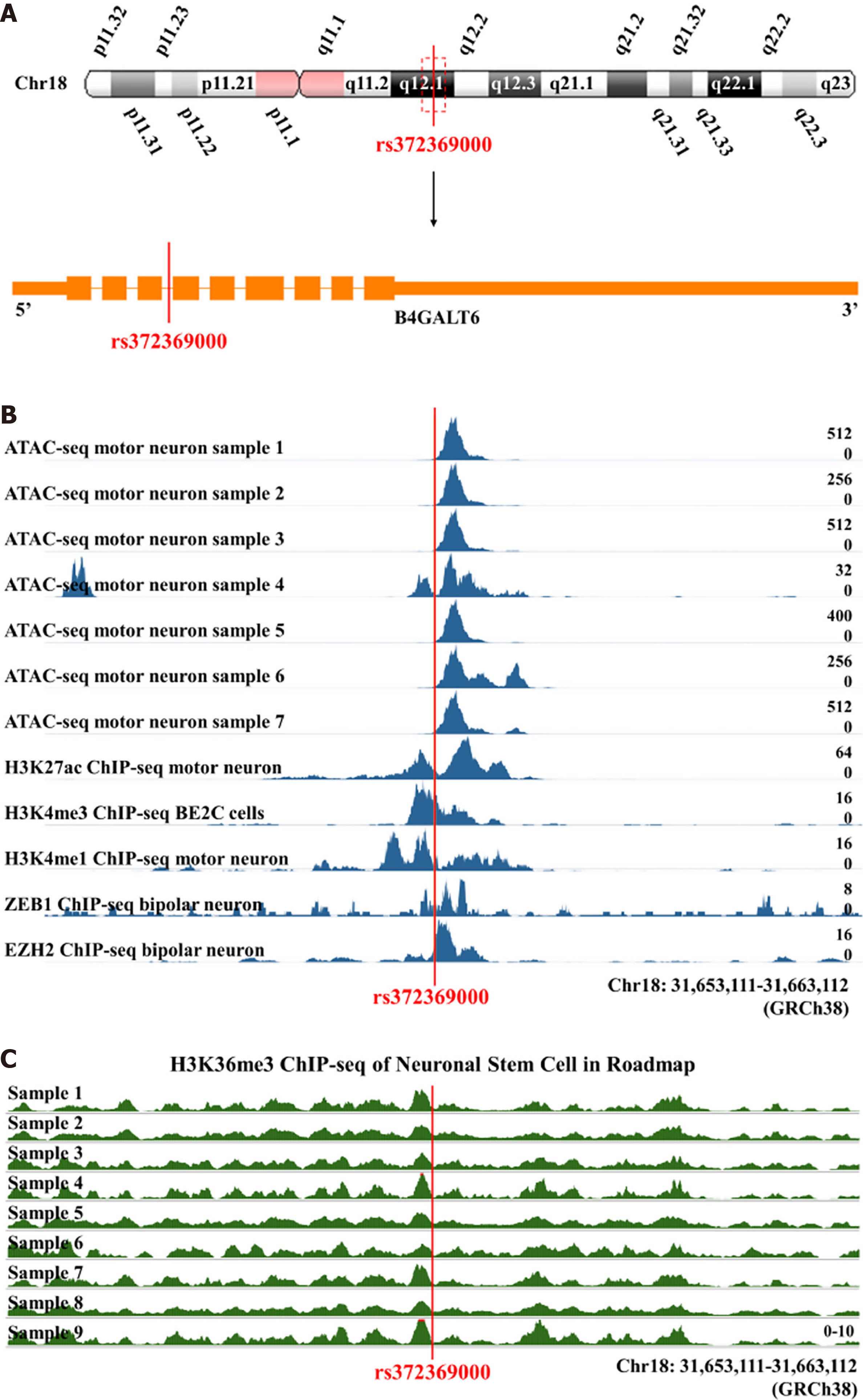
The rs372369000 SNP was located on q12.1 of chromosome 18 in the intronic region of the B4GALT6 gene (Figure 1A). The RegulomeDB probability score for rs372369000 was 0.82805, suggesting a high probability that this SNP regulates gene expression. ENCODE chromatin accessibility data indicated that rs372369000 was located in the open chromatin region of motor neurons (Figure 1B, Supplementary Figure 1). The results of ENCODE chromatin states analysis showed that the main function of rs372369000 was to weaken transcription (Supplementary Figure 2). ENCODE transcription factor chromatin immunoprecipitation sequencing (ChIP-seq) results showed that rs372369000 primarily bound to the transcription factor POLR2A and bound to transcription factors ZEB1 and EZH2 in bipolar neurons (Figure 1B, Supplementary Figure 3). ChIP-seq results of epigenetic modifications in ENCODE showed that the rs372369000 region had H3k27ac and H3k4me3 signal peaks in motor neurons and H3k4me1 signal peaks in BE2C cells (Figure 1B). ChIP-seq results of epigenetic modifications in Roadmap showed that the rs372369000 region had H3k36me3 signal peaks in neural stem cells (Figure 1C). These results suggest that rs372369000 is indeed a functional variant with transcriptional regulation activity.
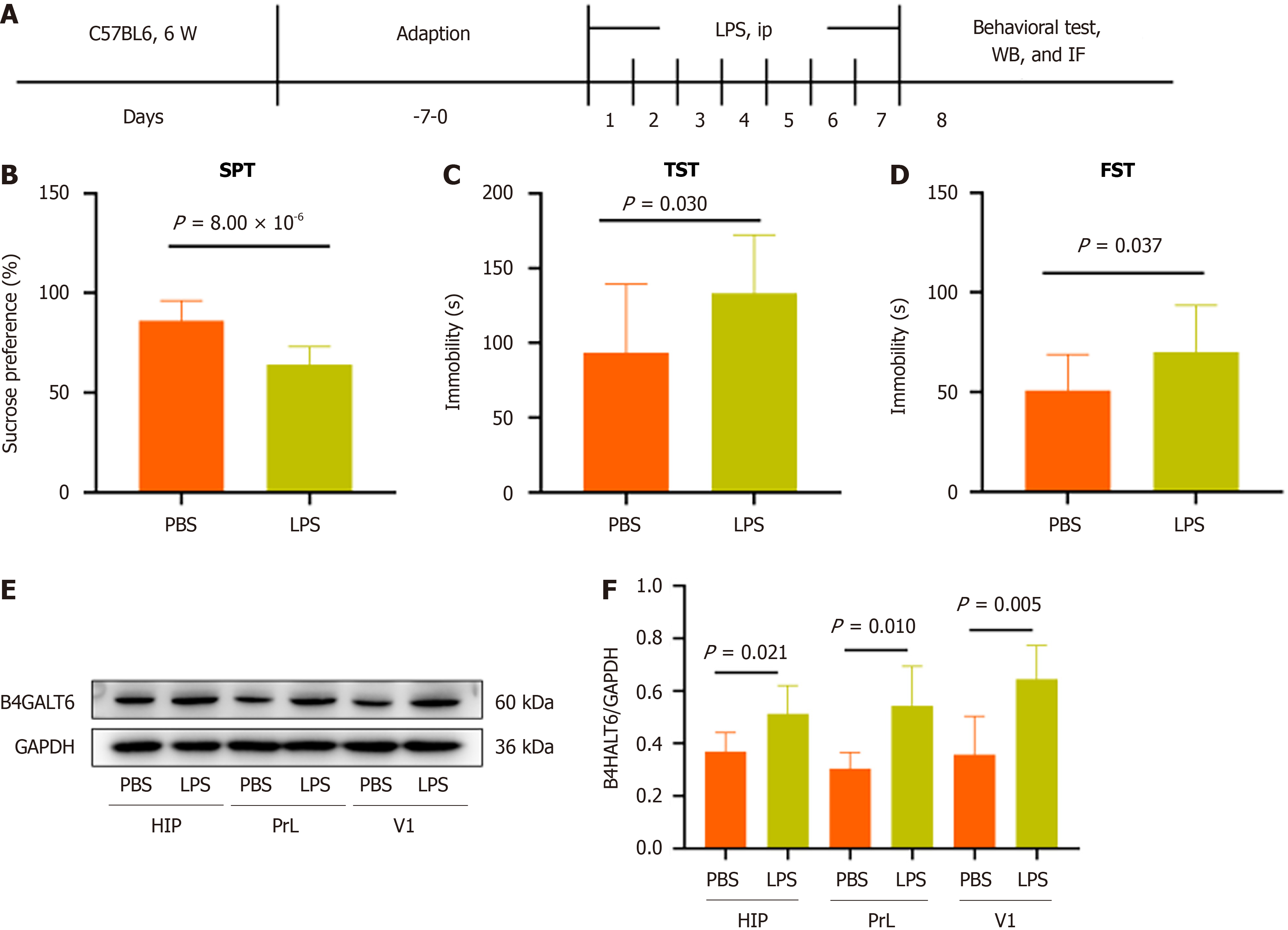
To assess whether B4GALT6 expression is altered in depressive-like behavior, we generated an LPS-induced depressive-like behavior mouse model (Figure 2A-D). We then examined B4GALT6 expression in the hippocampus, PrL, and V1 regions of control and LPS-treated mice by Western blot. B4GALT6 expression significantly increased in the hippocampus, PrL, and V1 regions of the LPS group (Figure 2E and F).
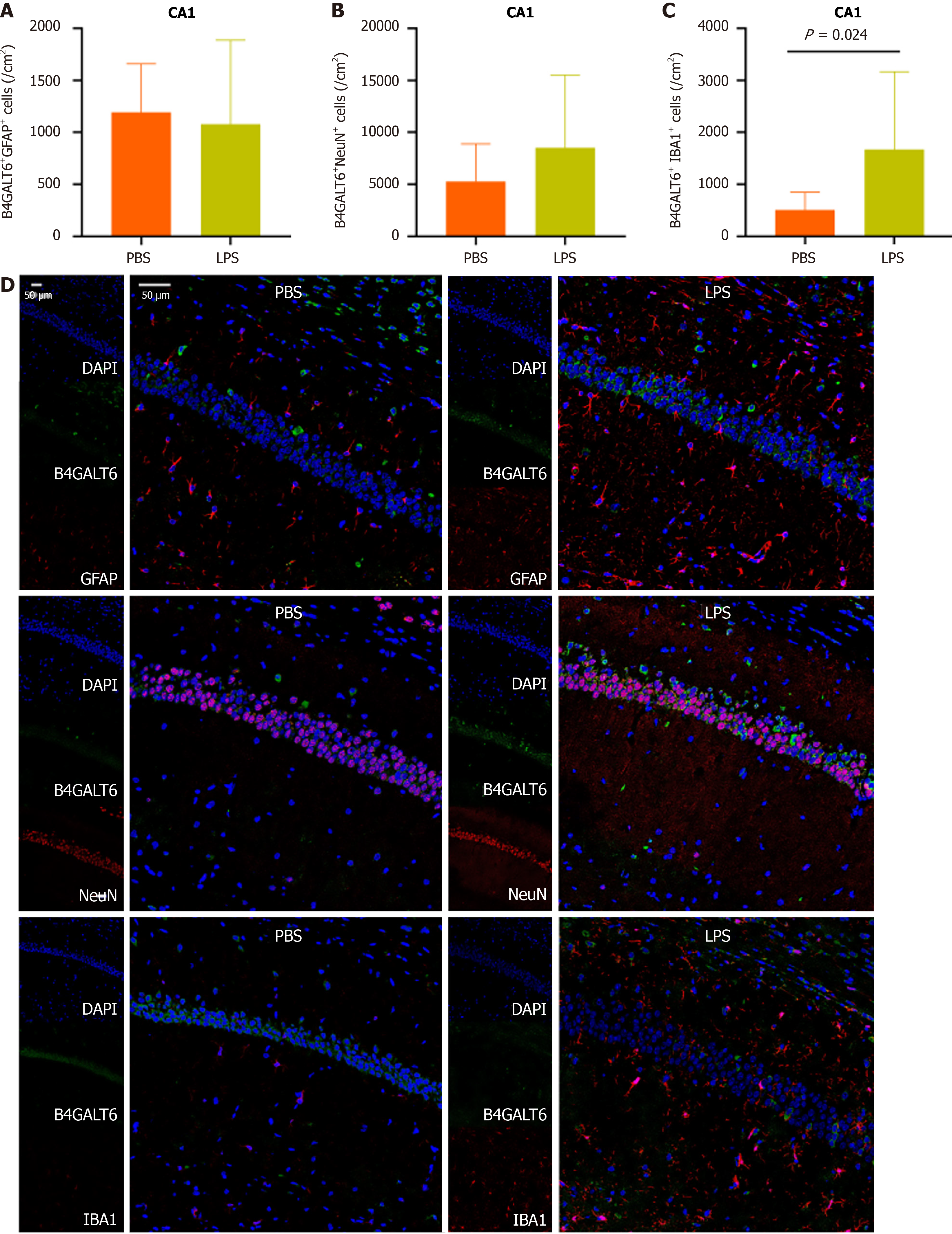
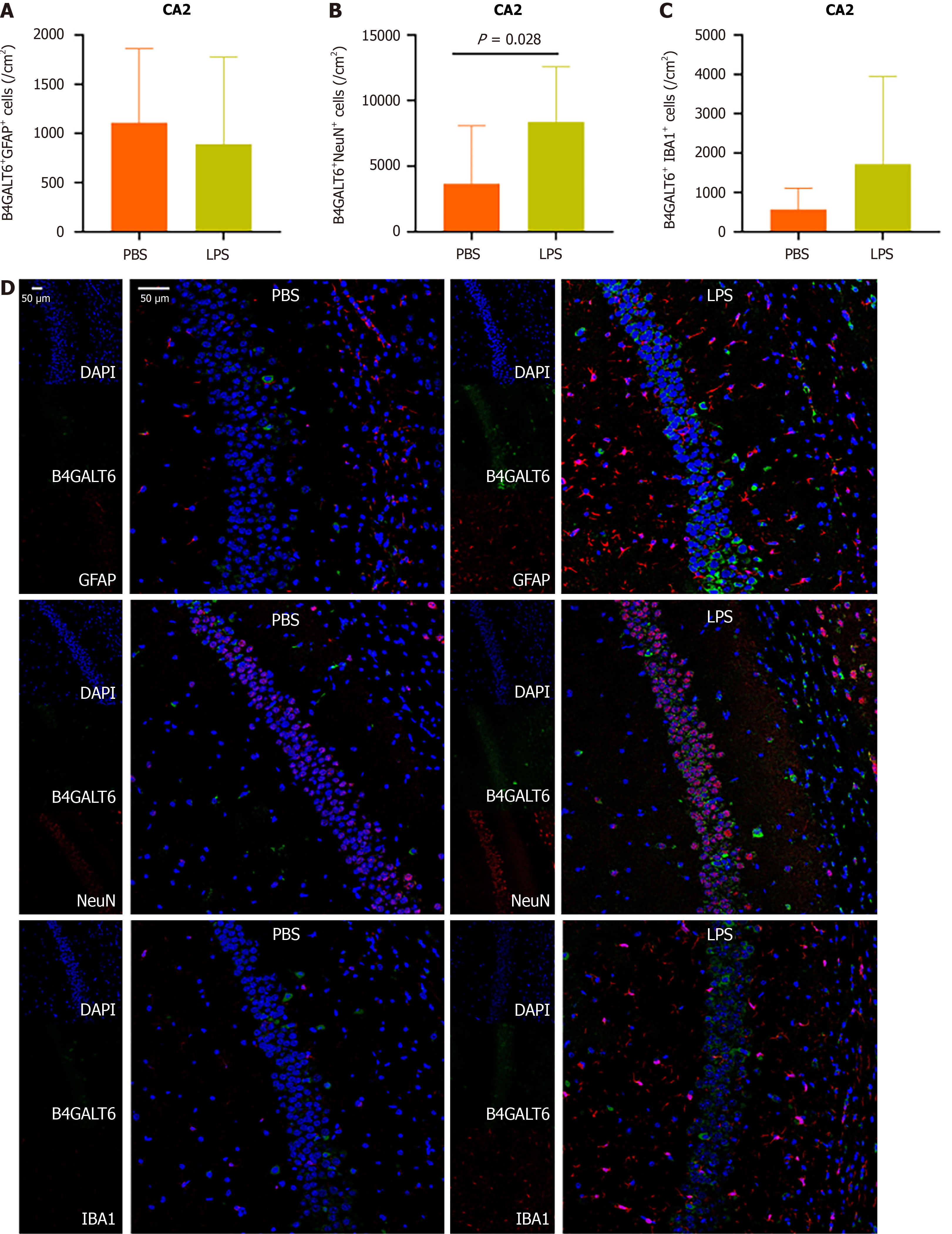
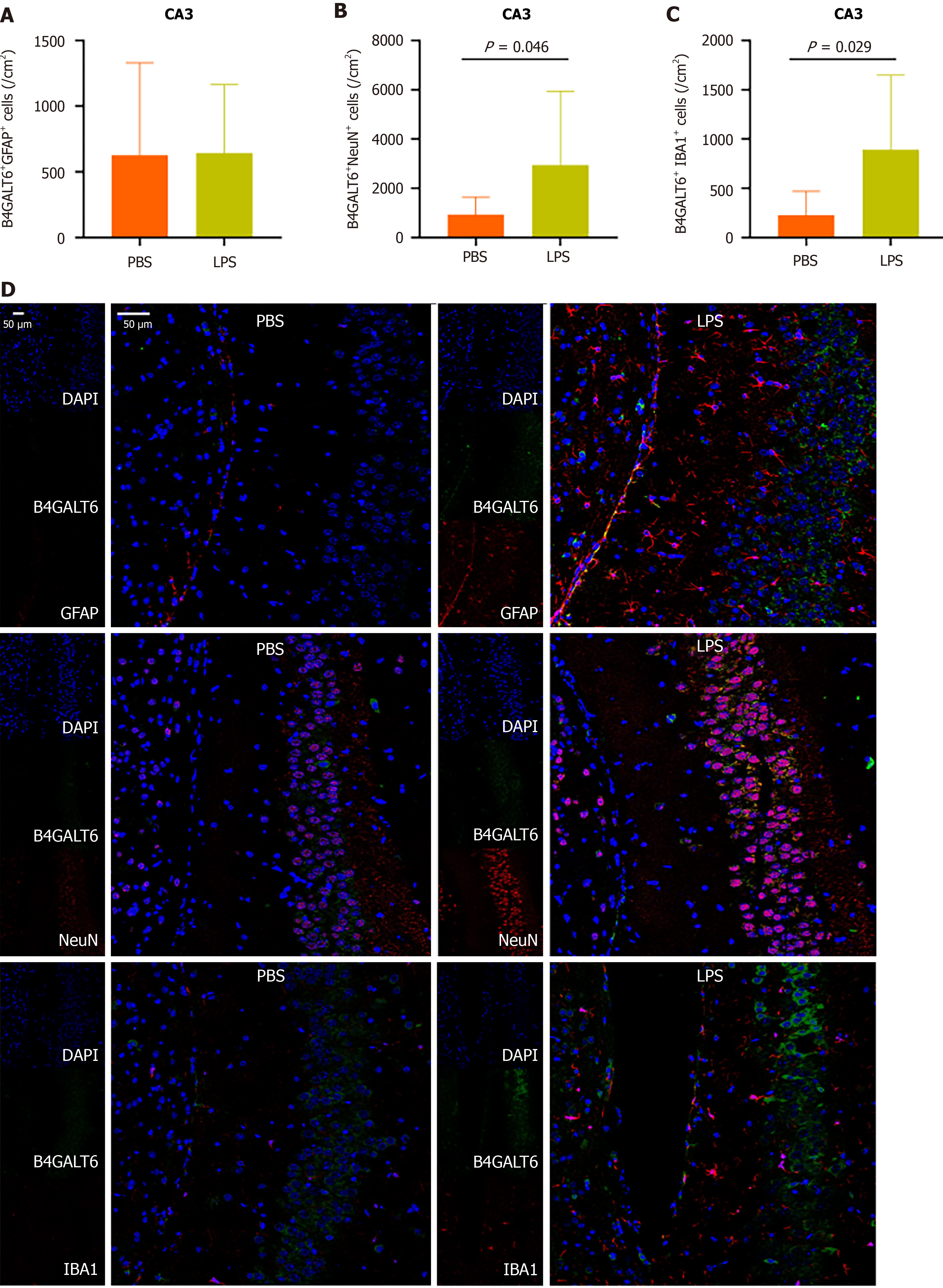
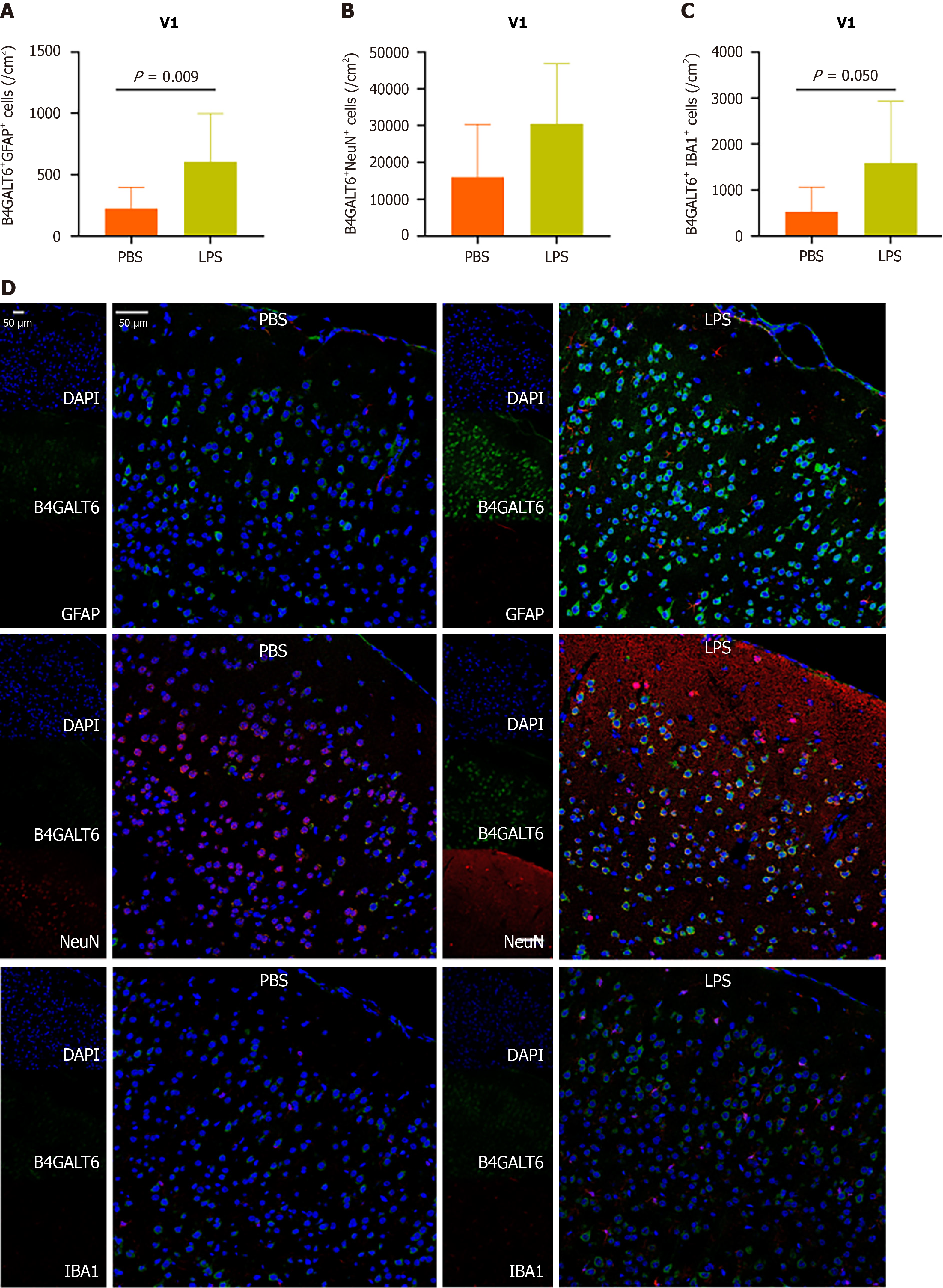
To assess the cell subtype specificity of B4GALT6 expression in depressive-like behavioral pathology, we costained brain sections with B4GALT6 and IBA1, GFAP, and NeuN using immunofluorescence to identify any differences in cell subtype density. The LPS group had increased cell density of B4GALT6+/IBA1+ cells within the hippocampal CA1 region (Figure 3) and B4GALT6+/NeuN+ cells in the hippocampal CA2 region (Figure 4). The densities of B4GALT6+/NeuN+ neurons and B4GALT6+/IBA1+ glia were both increased in the hippocampal CA3 region (Figure 5). There was no statistically significant difference in cell subtypes in the dentate gyrus of the hippocampus (Supplementary Figure 4) or the PrL brain region between the PBS and LPS groups (Supplementary Figure 5). Compared to the PBS group, the density of B4GALT6+GFAP+ and B4GALT6+IBA1+ cells significantly increased in the LPS group (Figure 6). Bioinformatics analysis revealed that B4GALT6 is evolutionarily conserved across jawed vertebrates, as shown in Supplementary Figure 6. Human brain tissue expression analysis showed widespread B4GALT6 expression, as detailed in Supplementary Figure
In this study, we observed increased expression of B4GALT6 in the hippocampus, PrL, and V1 brain regions of mice with LPS-induced depressive-like behavior. This elevation was distinct within brain subregions and cell subtypes. Specifically, increased B4GALT6 expression in CA1 microglial cells, CA2 neuronal cells, CA3 microglial and neuronal cells, and V1 microglial and astrocyte cells may be implicated in depressive pathology.
MDD patients typically exhibit smaller hippocampal volumes and compromised hippocampal function[15]. Depressed patients display phenomena including diminished neuron numbers, reduced neuronal branching, and inhibited neuronal generation within the hippocampus[16,17]. Antidepressant medication can facilitate neuronal growth and augment hippocampal volume[18]. The PrL brain region in mice corresponds to the dorsolateral prefrontal cortex (DLPFC) in humans, and the DLPFC in patients with MDD serves as a target for non-invasive brain stimulation therapy[19]. Visual cortex dysfunction plays a role in the pathophysiology and treatment of depression. MDD patients exhibit abnormal visual cortex function, and the effects of many antidepressants were aligned with the enhancement of visual cortex structure and synaptic function[20]. Our research revealed the cell subtype-specific expression of B4GALT6 in the hippocampus and V1 brain region of mice exhibiting depressive-like behaviors. While Western blot analysis indicated heightened B4GALT6 expression in the PrL brain region of these mice, no cell subtype specificity was noted, suggesting that B4GALT6 originating from microglia, astrocytes, and neurons in the PrL brain region collectively contributes to depression.
B4GALT6 is conserved across jawed vertebrates (Supplementary Figure 6) and exhibits widespread expression in human brain tissue (Supplementary Figure 7). B4GALT6 is essential for the synthesis of lactosylceramide, pivotal in neuronal generation and myelin formation[6,21]. B4GALT6 precisely regulates the ratio of gangliosides (such as GM1/GM3) and glycoprotein glycan chain structure by dynamic interaction with glycosphingolipid synthase (UGCG, ST3GAL5) and glycosidase (NEU1). This network imbalance leads to three pathological effects: (1) Synaptic dysfunction: GM1 deficiency destroys the lipid raft microdomain, resulting in abnormal localization of the TrkB receptor, weakening BDNF signal transmission and damaging synaptic plasticity of the prefrontal-hippocampus neural circuit; (2) Neuroinflammatory activation: NEU1-mediated sialidase hydrolysis increased, exposing galactose residues and activating the Galectin-3-TLR4 pathway, triggering the release of glial inflammatory factors (IL-6, TNF-α); and (3) HPA axis dysregulation: Glucolipid metabolism disorder in paraventricular nucleus of the hypothalamus leads to abnormal glycosylation modification of glucocorticoid receptor, which hinders the negative feedback regulation and ultimately leads to sustained stress response hyperactivity. Together, these mechanisms may contribute to the pathological progression of MDD.
Additionally, B4GALT6 is implicated in numerous neurological and psychiatric diseases. Postmortem brain studies reveal reduced B4GALT6 expression in the prefrontal cortex of patients with schizophrenia compared to controls[22]. Loss of B4GALT6 alters developmentally timed Caenorhabditis elegans sleep[23]. In multiple sclerosis, brain tissue analysis revealed elevated levels of B4GALT6 and its product lactosylceramide compared to controls[24,25]. The lactosylceramide produced by B4GALT6 enhances CCL2 expression via the NF-κb and IRF-1 signaling pathways, which in turn activates astrocytes through an autocrine manner, exacerbating conditions in experimental autoimmune encephalomyelitis[24,25]. Additionally, B4GALT6 regulates microglia activation and plays a role in central nervous system inflammation by enha
We utilized the gene and protein function prediction tools GeneMANIA[28] and STRING[29] to predict the function of B4GALT6 (Supplementary Figure 8). At the genetic and protein levels, B4GALT6 functions through other glycosyltransferases and glycosidases. Further research is needed to determine whether B4GALT6 participates in the patho
Our study has several limitations. First, the absence of systematic functional genomics experiments has hindered a full elucidation of how rs372369000 affects B4GALT6 expression. Second, the absence of knockout and conditional knockout mice hinders a more detailed elucidation of the brain region and cell specificity of B4GALT6 in the pathology of MDD. Third, this study did not explore the specific mechanism of B4GALT6 leading to MDD and its effect on cognitive function. Finally, this research serves as a pilot study, and future research should delve into the specific mechanisms of B4GALT6 in models like chronic unpredictable mild stress (CUMS), chronic social defeat stress (CSDS), and chronic restraint stress (CRS). In addition, future studies should comprehensively assess behavioral phenotypes through cognitive function tests such as the Y-maze and the Morris water maze and explore the impact of B4GALT6 Levels on the cognitive functions of brain regions.
In summary, we have reported the brain region and cell-specific characteristics of B4GALT6 expression in mice exhibiting depressive-like behaviors. These findings may broaden our understanding and help develop new potential targets for diagnosis and treatment of MDD.
| 1. | Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 918] [Cited by in RCA: 875] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 2. | Luan D, You D, Wu Y, Wu F, Xu Z, Li L, Jiao J, Zhang A, Feng H, Kong Y, Zhao Y, Zhang Z. Effects of interaction between single nucleotide polymorphisms and psychosocial factors on the response to antidepressant treatment in patients with major depressive disorder. J Genet Genomics. 2022;49:587-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 3. | Chatterjee S, Castiglione E. UDPgalactose:glucosylceramide beta 1----4-galactosyltransferase activity in human proximal tubular cells from normal and familial hypercholesterolemic homozygotes. Biochim Biophys Acta. 1987;923:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 4. | Chatterjee S, Ghosh N, Khurana S. Purification of uridine diphosphate-galactose:glucosyl ceramide, beta 1-4 galactosyltransferase from human kidney. J Biol Chem. 1992;267:7148-7153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Yamaji T, Hanada K. Establishment of HeLa cell mutants deficient in sphingolipid-related genes using TALENs. PLoS One. 2014;9:e88124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 6. | Yoshihara T, Satake H, Nishie T, Okino N, Hatta T, Otani H, Naruse C, Suzuki H, Sugihara K, Kamimura E, Tokuda N, Furukawa K, Fururkawa K, Ito M, Asano M. Lactosylceramide synthases encoded by B4galt5 and 6 genes are pivotal for neuronal generation and myelin formation in mice. PLoS Genet. 2018;14:e1007545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 7. | Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790-1797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1866] [Cited by in RCA: 2222] [Article Influence: 170.9] [Reference Citation Analysis (0)] |
| 8. | Dong S, Zhao N, Spragins E, Kagda MS, Li M, Assis P, Jolanki O, Luo Y, Cherry JM, Boyle AP, Hitz BC. Annotating and prioritizing human non-coding variants with RegulomeDB v.2. Nat Genet. 2023;55:724-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 124] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 9. | Satterlee JS, Chadwick LH, Tyson FL, McAllister K, Beaver J, Birnbaum L, Volkow ND, Wilder EL, Anderson JM, Roy AL. The NIH Common Fund/Roadmap Epigenomics Program: Successes of a comprehensive consortium. Sci Adv. 2019;5:eaaw6507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 10. | ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14554] [Cited by in RCA: 13477] [Article Influence: 962.6] [Reference Citation Analysis (0)] |
| 11. | Hitz BC, Lee JW, Jolanki O, Kagda MS, Graham K, Sud P, Gabdank I, Strattan JS, Sloan CA, Dreszer T, Rowe LD, Podduturi NR, Malladi VS, Chan ET, Davidson JM, Ho M, Miyasato S, Simison M, Tanaka F, Luo Y, Whaling I, Hong EL, Lee BT, Sandstrom R, Rynes E, Nelson J, Nishida A, Ingersoll A, Buckley M, Frerker M, Kim DS, Boley N, Trout D, Dobin A, Rahmanian S, Wyman D, Balderrama-Gutierrez G, Reese F, Durand NC, Dudchenko O, Weisz D, Rao SSP, Blackburn A, Gkountaroulis D, Sadr M, Olshansky M, Eliaz Y, Nguyen D, Bochkov I, Shamim MS, Mahajan R, Aiden E, Gingeras T, Heath S, Hirst M, Kent WJ, Kundaje A, Mortazavi A, Wold B, Cherry JM. The ENCODE Uniform Analysis Pipelines. Res Sq. 2023;rs.3.rs-3111932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, Baymuradov UK, Graham K, Litton C, Miyasato SR, Strattan JS, Jolanki O, Lee JW, Tanaka FY, Adenekan P, O'Neill E, Cherry JM. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020;48:D882-D889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 702] [Article Influence: 117.0] [Reference Citation Analysis (0)] |
| 13. | Li W, Ali T, Zheng C, He K, Liu Z, Shah FA, Li N, Yu ZJ, Li S. Anti-depressive-like behaviors of APN KO mice involve Trkb/BDNF signaling related neuroinflammatory changes. Mol Psychiatry. 2022;27:1047-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 14. | Luan D, Zhang Y, Yuan L, Chu Z, Ma L, Xu Y, Zhao S. MST4 modulates the neuro-inflammatory response by regulating IκBα signaling pathway and affects the early outcome of experimental ischemic stroke in mice. Brain Res Bull. 2020;154:43-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Belleau EL, Treadway MT, Pizzagalli DA. The Impact of Stress and Major Depressive Disorder on Hippocampal and Medial Prefrontal Cortex Morphology. Biol Psychiatry. 2019;85:443-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 393] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 16. | Sheline YI, Mittler BL, Mintun MA. The hippocampus and depression. Eur Psychiatry. 2002;17 Suppl 3:300-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 122] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 17. | Larosa A, Wong TP. The hippocampus in stress susceptibility and resilience: Reviewing molecular and functional markers. Prog Neuropsychopharmacol Biol Psychiatry. 2022;119:110601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 31] [Reference Citation Analysis (0)] |
| 18. | Fang S, Wu Z, Guo Y, Zhu W, Wan C, Yuan N, Chen J, Hao W, Mo X, Guo X, Fan L, Li X, Chen J. Roles of microglia in adult hippocampal neurogenesis in depression and their therapeutics. Front Immunol. 2023;14:1193053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 62] [Reference Citation Analysis (0)] |
| 19. | Hsu TW, Yeh TC, Kao YC, Thompson T, Brunoni AR, Carvalho AF, Hsu CW, Tu YK, Liang CS. The dose-effect relationship of six stimulation parameters with rTMS over left DLPFC on treatment-resistant depression: A systematic review and meta-analysis. Neurosci Biobehav Rev. 2024;162:105704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 20. | Wu F, Lu Q, Kong Y, Zhang Z. A Comprehensive Overview of the Role of Visual Cortex Malfunction in Depressive Disorders: Opportunities and Challenges. Neurosci Bull. 2023;39:1426-1438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 21. | Tokuda N, Numata S, Li X, Nomura T, Takizawa M, Kondo Y, Yamashita Y, Hashimoto N, Kiyono T, Urano T, Furukawa K, Furukawa K. β4GalT6 is involved in the synthesis of lactosylceramide with less intensity than β4GalT5. Glycobiology. 2013;23:1175-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 22. | Narayan S, Head SR, Gilmartin TJ, Dean B, Thomas EA. Evidence for disruption of sphingolipid metabolism in schizophrenia. J Neurosci Res. 2009;87:278-288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 23. | Huang H, Zhu Y, Eliot MN, Knopik VS, McGeary JE, Carskadon MA, Hart AC. Combining Human Epigenetics and Sleep Studies in Caenorhabditis elegans: A Cross-Species Approach for Finding Conserved Genes Regulating Sleep. Sleep. 2017;40:zsx063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Mayo L, Trauger SA, Blain M, Nadeau M, Patel B, Alvarez JI, Mascanfroni ID, Yeste A, Kivisäkk P, Kallas K, Ellezam B, Bakshi R, Prat A, Antel JP, Weiner HL, Quintana FJ. Regulation of astrocyte activation by glycolipids drives chronic CNS inflammation. Nat Med. 2014;20:1147-1156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 394] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 25. | Rostami A, Ciric B. Astrocyte-derived lactosylceramide implicated in multiple sclerosis. Nat Med. 2014;20:1092-1093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Ruiz M, Jové M, Schlüter A, Casasnovas C, Villarroya F, Guilera C, Ortega FJ, Naudí A, Pamplona R, Gimeno R, Fourcade S, Portero-Otín M, Pujol A. Altered glycolipid and glycerophospholipid signaling drive inflammatory cascades in adrenomyeloneuropathy. Hum Mol Genet. 2015;24:6861-6876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 27. | Ghatnatti V, Vastrad B, Patil S, Vastrad C, Kotturshetti I. Identification of potential and novel target genes in pituitary prolactinoma by bioinformatics analysis. AIMS Neurosci. 2021;8:254-283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214-W220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2336] [Cited by in RCA: 3456] [Article Influence: 216.0] [Reference Citation Analysis (0)] |
| 29. | Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, Bork P, Jensen LJ, von Mering C. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638-D646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1815] [Cited by in RCA: 5190] [Article Influence: 1730.0] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/