Published online Sep 24, 2025. doi: 10.5306/wjco.v16.i9.111742
Revised: July 26, 2025
Accepted: August 22, 2025
Published online: September 24, 2025
Processing time: 77 Days and 5.7 Hours
Aberrant microRNAs expression and associated pathways have been proved participate in regulation vast various physiologic and pathologic processed of different human cancers including liver cancer. While, the function of miR-451a in liver cancer still indistinct.
To study the effect of miR-451a in liver cancer development.
GeneChip microarray analysis performed to detect miR-451a expression in liver cancer tissues and normal liver tissues. Reverse transcription-polymerase chain reaction was used to validate the expression of miR-451a in liver cancer cell and other tumor cell lines. Construction of liver cancer cell lines that stably overexpressed miR-451a by transfecting Lentivirus produced by Genechem company. Methylthiazolyldiphenyl-tetrazolium (MTT) bromide assay and colony formation assay to determine the effect of miR-451a in liver cancer cell proliferation. Flow cytometry used to investigate whether miR-451a involved in liver cancer cell apoptosis. Cell migration ability was measured via wound scratch assay. Target gene was explored by bioinformatic analysis, and downstream molecule of miR-451a in liver cancer identified by rescue experiments.
MiR-451a expression significantly downregulation in liver cancer tissues compared with that in normal liver tissue. MiR-451a also obviously low-expressed in liver cancer cell, colorectal carcinoma cell and esophageal carcinoma cell lines. Human hepatoblastoma G2 (HepG2) and BEL-7404 cell lines that stably overexpressed miR-451a by transfecting lentivirus constructed successfully. MTT bromide assay and colony formation assay showed that the overexpressed miR-451a inhibit HepG2 cell proliferation viability, but not BEL-7404 cell. Flow cytometry determined that miR-451a regulating proliferation not through inducing apoptosis. Wound scratch assay revealed that miR-451a overexpression suppressed HepG2 cell migration. Furthermore, mex-3 RNA binding family member C was predicted as the target gene by bioinformatic analysis, and rescue experiments confirmed the hypothesis.
Therefore, miR-451a may be candidate miRNA for understanding molecular mechanisms of liver cancer development and novel target in liver cancer cell.
Core Tip: Aberrant microRNAs expression and associated pathways have been proved participate in regulation vast various physiologic and pathologic processed of different human cancers including liver cancer. In this study, we first provided evidence that miR-451a obviously down-regulated in hepatocellular carcinoma (HCC) tissues and tumor cell lines. Overexpression of miR-451a significantly inhibit human hepatoblastoma G2 (HepG2) cell proliferation, colony formation and migration. However, miR-451a were not participated in HCC cell apoptosis. Moreover, bioinformatic analysis showed that mex-3 RNA binding family member C (MEX3C) was target gene of miR-451a, which validated by reverse transcription-polymerase chain reaction and western blotting. Furthermore, rescue experiments confirmed that overexpression of MEX3C can inhibit the effect of miR-451a in HepG2.
- Citation: Liu DZ, Wang L, Yang SK, Zhao ZJ, Cui YY, Zhao XK, Zhang F, Guan QH, Zhou L. MiR-451a targets mex-3 RNA binding family member C to regulate human hepatoblastoma G2 cell growth and metastasis. World J Clin Oncol 2025; 16(9): 111742
- URL: https://www.wjgnet.com/2218-4333/full/v16/i9/111742.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i9.111742
The common hallmark of the vast majority of liver cancer is a chronic inflammation caused by chronic hepatitis B virus (HBV) infection in China[1]. HBV infection can result in a cascade of complex reaction between oncogenes and tumor suppressor genes, owing to carcinogenesis and development of liver cancer[2].
MicroRNAs (miRNAs) are a class of small RNAs, which play as post-transcriptional gene silencers by decreasing or suppressing their target mRNA expression[3]. It has been reported that lots of miRNAs deregulation were related to many cancers’ progression including colorectal cancer[4], glioma[5], osteosarcoma[6], liver cancer[7], breast cancer[8], etc. Therefore, it is significant important to investigate the mechanism of miRNA behind the liver cancer development.
Our previous study found sheer number of dysregulated miRNAs in liver cancer tissues compared with that in normal liver tissues by GeneChip microarray analysis. The expression of miR-451a obviously decreased in liver cancer tissues, which indicated that miR-451a may play crucial role in liver cancer progression. The aim of our study was to further determine the effect of miR-451a in hepatocarcinogenesis.
A total of 100 Liver cancer samples and corresponding normal liver tissues collect from patients with liver cancer. The patients were pathological diagnosed clearly, having no preoperative adjuvant chemotherapy or radiotherapy, who underwent complete surgical resection at The Affiliated Hospital of Binzhou Medical University. Informed consent was provided by all patients, and the experiment approved by our Ethics Committee of The Affiliated Hospital of Binzhou Medical University.
Human liver cancer cell lines [including SMMC-7721, Huh7, human hepatoblastoma G2 (HepG2), Hep3B, BEL-7402, BEL-7404], human colorectal carcinoma cell lines (HCT-116, RKO) and human esophageal carcinoma cell lines (ECA109, TE-1) were purchased from the American Type Culture Collection. All the cell lines were cultured in Dulbeccos’ Modified Eagle’s Medium (Hyclone, NJ, United States) containing 10% fetal bovine serum, 100 units/mL of penicillin and 100 μg/mL streptomycin. For transfection, lentivirus system for miR-451a stable overexpression and pcDNA-mex-3 RNA binding family member C (MEX3C) plasmid overexpressing MEX3C were created by the Genechem Company (Shanghai, China). All the cell lines in this study authenticated by STR profiling.
Total RNA was purified from tissue and cell lines using Trizol reagent (Invitrogen, United States). The cDNA was synthesized with promega M-MLV Reverse Transcriptase kit. The quantitative polymerase chain reaction (qPCR) was performed using qPCR SYBR Green master mix (Takara, Dalian, Liaoning Province, China) on a Real-time PCR System (Applied Biosystems). MiR-451a expression was normalized to U6 mRNA level, and gene expression levels normalized to GAPDH. Each reaction was performed in duplicate. MiRNA-specific primers designed by RiboBio (Guangzhou, Guangdong Province, China). Primers used for amplification were as follows: (1) GAPDH: Forward primer: 5’-TG ACTTCAACAGCGACACCCA-3’, reverse primer: 5’-CACCCTGTTGCTGTAGCCAAA-3’; and (2) MEX3C: Forward primer: 5’-ATGAGGTTATTGCTGCCCTAG-3’, reverse primer: 5’-GTGAATTTGGATTGCCTGAGTA-3’. Detailed primers for target genes shown in Table 1.
| Gene ID | Forward primer sequence | Reverse primer sequence | Amplification fragment (bp) |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA | 121 |
| TSC1 | CAACAAGCAAATGTCGGGGAG | CATAGGGCCACGGTCAGAA | 114 |
| OSR1 | CACAGTGCCTTCGGACCATCT | CTCGGGCTTGGGTTGAATGAC | 211 |
| EMSY | ATGACTTCCAAGCCCAACGTG | TTCCCTTGCTGCCCATAAACA | 275 |
| SAMD4B | TGGGCATTCGGCTCCAACTCG | CATTGGGAACTGCCGCTGGGT | 84 |
| MEX3C | ATGAGGTTATTGCTGCCCTAG | GTGAATTTGGATTGCCTGAGTA | 134 |
| PSMB8 | CTGGGTCCTACATTAGTGCCT | CCATTTCGCAGATAGTACAGC | 148 |
| PMM2 | ATCAAAATCGGAGTGGTAGGC | CAGCTTCTTCCAATAGGGGAC | 293 |
| CAB39 | GACAAAAGTCGCAACATCCAG | TAAACTGCTCATCCTCCGTCC | 166 |
| TTN | GGGAGGAAAGGCTTACAGAG | ATGGATACCATCATAGCGAACT | 261 |
HepG2 and BEL-7404 cells transfected with lentivirus-negative control (LV-NC) or lentivirus miR-451a (LV-miR-451a) were seeded at 2 × 103 per well into 96-well culture plates and incubated in humidified incubators at 37 ºC with 5% CO2. The Methylthiazolyldiphenyl-tetrazolium (MTT) bromide solution (Genechem, Shanghai, China) were added to cells, and then incubated for 4 hours. Following incubation, 150 μL DMSO (Sigma-Aldrich, St. Louis, MO, United States) was added into each well and shaken for 15 minutes. Optical density value was measured by a microplate reader at the wavelength of 490 nm. All experiments were independently repeated at least 3 times.
Cells transfected with LV-NC or LV-miR-451a treatment for 3 days, and then seeded into 60 mm culture plates at a concentration of 800 cells per well. The medium changed every 3 days and colonies were fixed in methanol and stained with crystal violet solution (Beyotime) 10 days later and quantified. The experiment was repeated three times.
After 72 hours, LV-NC or LV-miR-451a transfection, HepG2 or BEL-7404 cells were harvested, washed and resuspended in the binding buffer. The apoptosis assay kit (eBioscience, Inc.) was used to examine the cells according to the instruction, and stained cells detected by flow cytometry (FACS Guava easyCyte HT, Millipore, MA, United States). Each experiment was performed three times.
HepG2 and BEL-7404 cells were seeded into 6-well plates and transfected with LV-NC or LV-miR-451a for 24 hours. A sterile pipette tip used to make the wounds in the cell monolayer as beginning gap length at 0 hour, while the residual gap length at 48 hours were detected using the Image Analysis and Detection System (Leica Microsystems GmbH, Wetzlar, Germany). The experiments repeated at least three times.
Transfected HepG2 or BEL-7404 cells were ground and lysed in RIPA buffer on ice and then subjected to Western blot analysis. Equal amounts of proteins were loaded onto sodium dodecyl sulfate-polyacrylamide gels, and the resulting bands were transferred to a polyvinylidene difluoride membranes (Millipore, MA, United States). The membrane was blocked and then incubated with primary antibodies of MEX3C and β-actin overnight. Next day, the membranes were washed and incubated with corresponding secondary antibodies. Signals were detected on X-ray film using an electrochemiluminescence detection system (Pierce, Rockford, IL, United States).
Data are presented as mean ± SD. The student’s t-test was used to compare the difference between two groups. P < 0.05 was considered to be significant. All analyses were performed using Statistical Package for the Social Sciences version 24.0 (SPSS, Inc., Chicago, IL, United States).
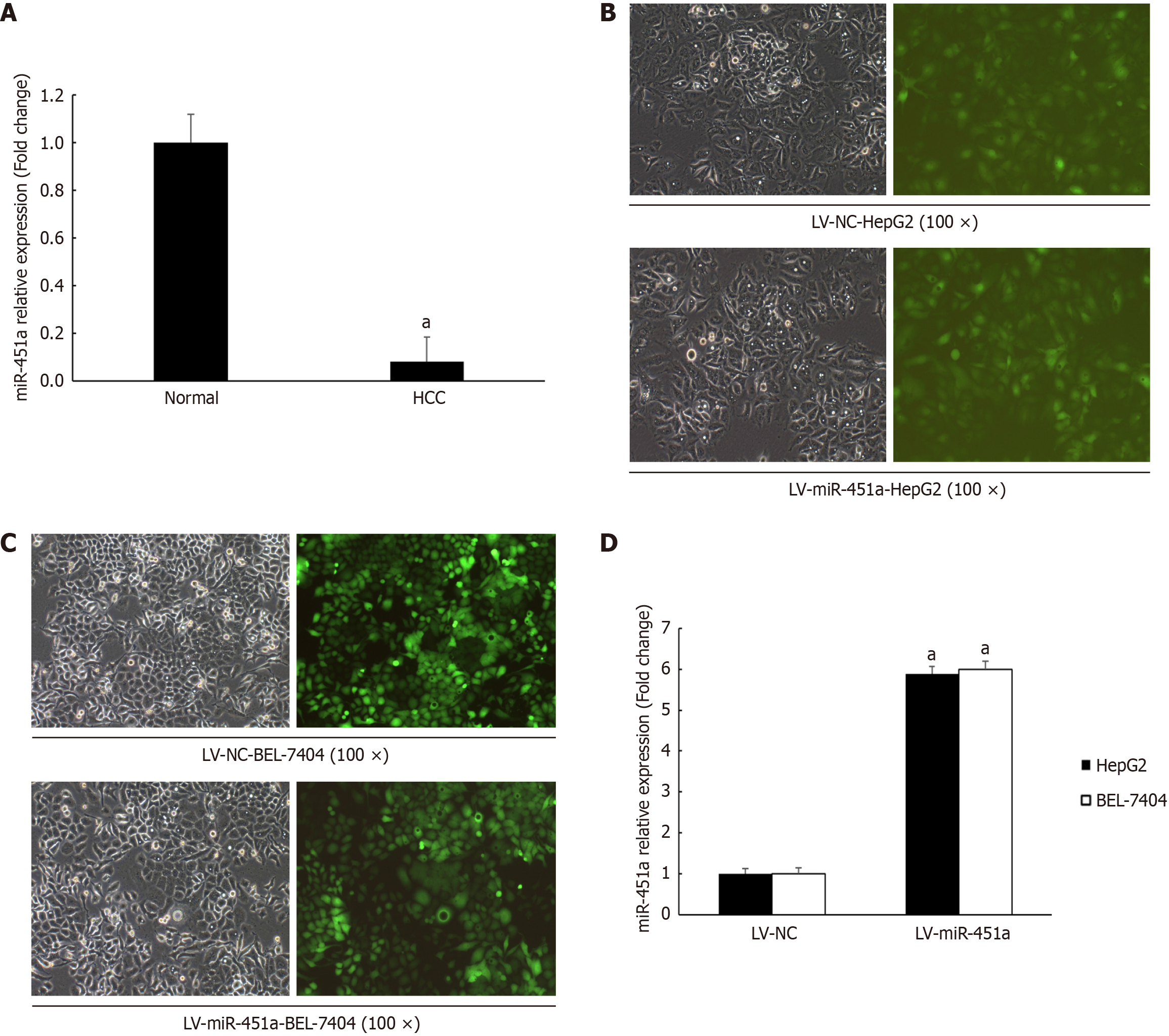
GeneChip microarray analysis performed between 10 Liver cancer tissues and paired normal liver tissues. It was found that miR-451a down-regulated more than 12-fold compared with that in normal liver tissues (Figure 1A). And the result confirmed by real-time quantitative reverse transcription-PCR (RT-PCR) analysis in liver cancer cell lines (including SMMC-7721, Huh7, HepG2, Hep3B, BEL-7402, BEL-7404) (Table 2). The data indicated that miR-451a expression decreased significantly in liver cancer cell lines and tissues. In order to confirm this result, some other tumor cell lines (HCT-116, RKO, ECA109, TE-1) also examined (Table 3). Consistent with the expression of miR-451a in liver cancer tissues and cell lines, miR-451a expression also hard to detect in colorectal cancer cell and esophageal squamous cancer cells. It was indicated that down-regulation of miR-451a in liver cancer may contribute to liver cancer carcinogenesis and progression.
| Cell type | U6 | hsa-miR-451a, Ct value | ∆Ct |
| Human hepatoblastoma G2 | 10.84 | 35 | 24.16 |
| 10.72 | 35 | 24.28 | |
| 10.77 | 35 | 24.23 | |
| BEL-7402 | 13.19 | 35 | 21.81 |
| 13.09 | 35 | 21.91 | |
| 13.2 | 35 | 21.8 | |
| BEL-7404 | 11.67 | 35 | 23.33 |
| 11.61 | 35 | 23.39 | |
| 11.53 | 35 | 23.47 | |
| Huh-7 | 10.82 | 35 | 24.18 |
| 10.84 | 35 | 24.16 | |
| 10.74 | 35 | 24.26 | |
| SMMC-7721 | 12.56 | 35 | 22.44 |
| 12.58 | 35 | 22.42 | |
| 12.66 | 35 | 22.34 | |
| Hep3B | 13.55 | 35 | 21.45 |
| 13.53 | 35 | 21.47 | |
| 13.51 | 35 | 21.49 |
| Cell type | U6 | hsa-miR-451a, Ct value | ∆Ct |
| HCT116 | 11.74 | 35 | 23.26 |
| 11.56 | 35 | 23.44 | |
| 11.68 | 35 | 23.32 | |
| RKO | 11.77 | 35 | 23.23 |
| 11.7 | 35 | 23.3 | |
| 11.79 | 35 | 23.21 | |
| ECA109 | 11.04 | 35 | 23.96 |
| 11.07 | 35 | 23.93 | |
| 11.06 | 35 | 23.94 | |
| TE-1 | 11.79 | 35 | 23.21 |
| 11.81 | 35 | 23.19 | |
| 11.79 | 35 | 23.21 |
To determine potential function of miR-451a, lentivirus system (LV-NC or LV-miR-451a) produced by Genechem Company transfecting HepG2 and BEL-7404 cell line (Figure 1B and C). The transfection efficiency was evaluated using RT-qPCR, as shown in Figure 1D, the miR-451a expression in HepG2 and BEL-7404 up-regulated 5.886-fold and 5.994- fold, respectively. Subsequently, HepG2 and BEL-7404 with stable up-expression of miR-451a were used to explore role of miR-451a in liver cancer progression.
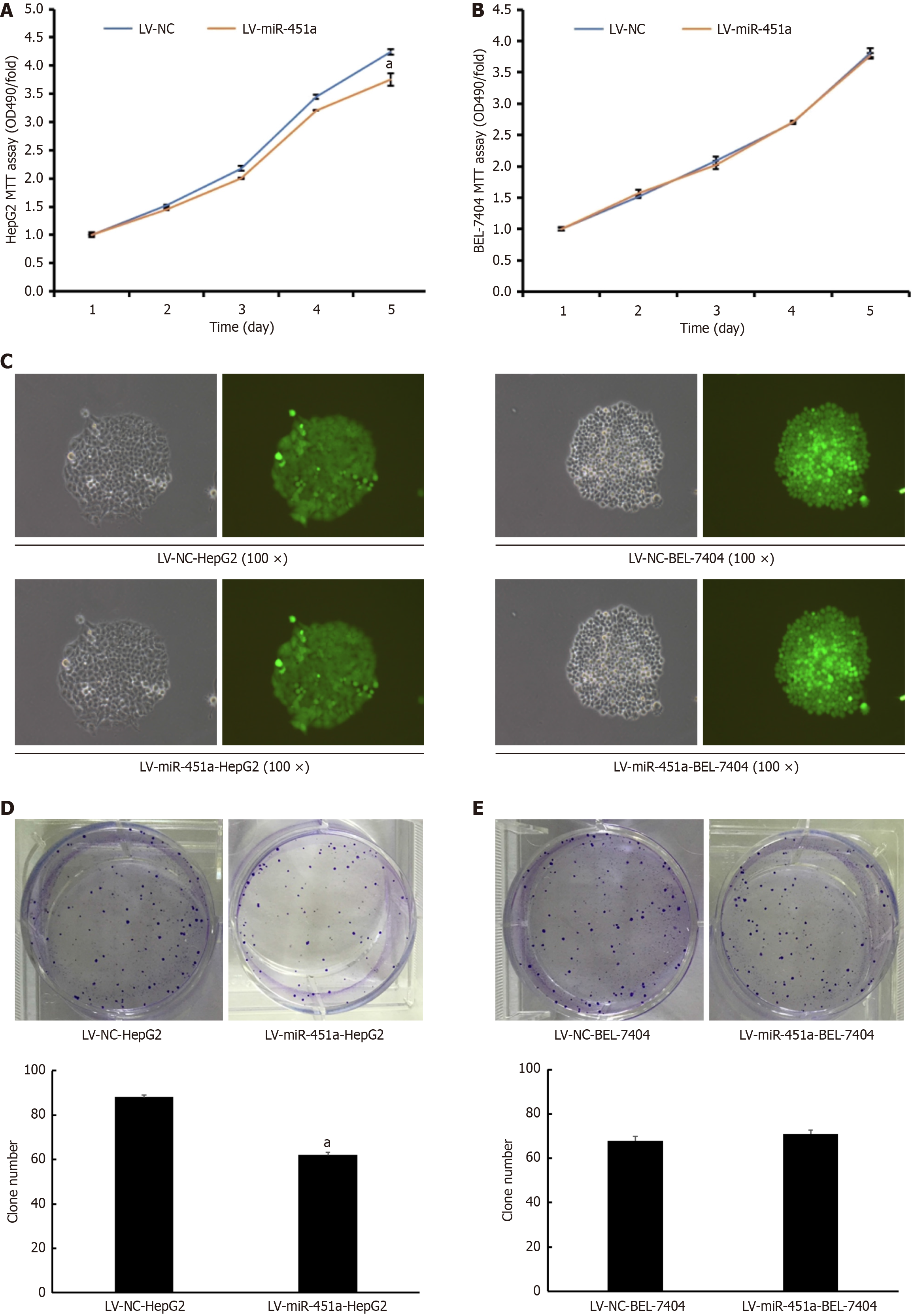
Having found decreased expression of miR-451a in liver cancer, we then explored whether miR-451a functions as a tumor suppressor. The effect of miR-451a on liver cancer cell growth was observed. The absorptivity of liver cancer cells was determined by MTT bromide assay, after transfection for 0 day, 1 day, 2 days, 3 days, 4 days and 5 days. The absorptivity of the HepG2 cell decreased at the 490 nm from 48 hours. It was indicated that the growth of HepG2 cell transfected with LV-miR-451a significantly decreased as compared with that of LV-NC group (P < 0.05) (Figure 2A). By contrast, there was no difference in BEL-7404 cell line between LV-NC group and LV-miR-451a group (Figure 2B). Figure 2C showed that lentivirus transfection efficiency of colony formation assay for HepG2 cell and BEL-7404 cell line. Meanwhile, the number of colonies in LV-miR-451a group for HepG2 cell line was obviously less than those in LV-NC group (Figure 2D). Consistent with the MTT bromide assay result, in BEL-7404 cell line, the number of colonies have no significant difference between LV-miR-451a group and LV-NC group (Figure 2E).
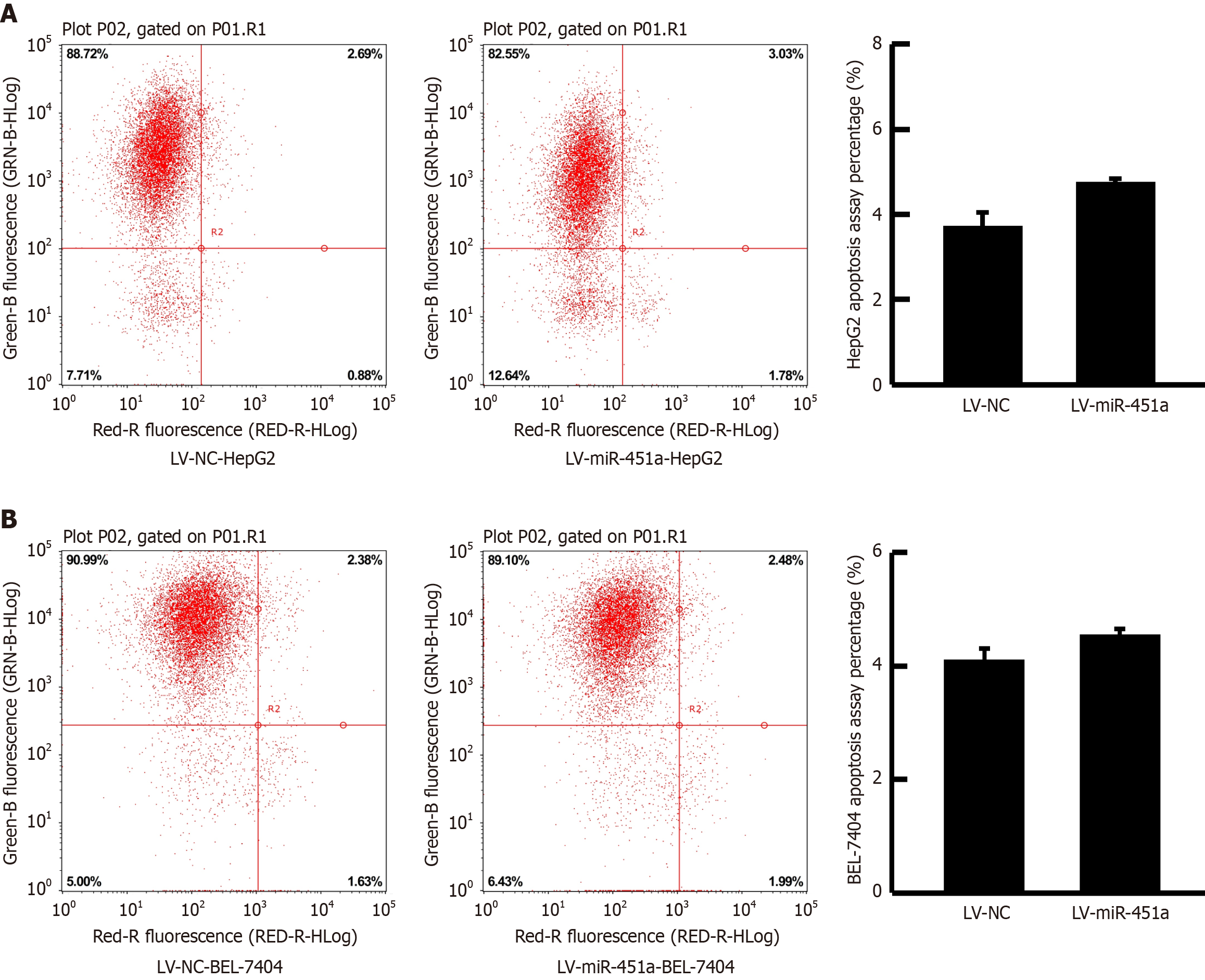
Because overexpression of miR-451a can prohibit HepG2 cell proliferation, it was then investigated whether the role of miR-451a is related to the cell apoptosis by flow cytometry. However, for both HepG2 cell and BEL-7404 cell line, up-regulation of miR-451a had no influence on liver cancer cell apoptosis (Figure 3). This result indicated that miR-451a was not via induction of apoptosis to suppress liver cancer cell proliferation.
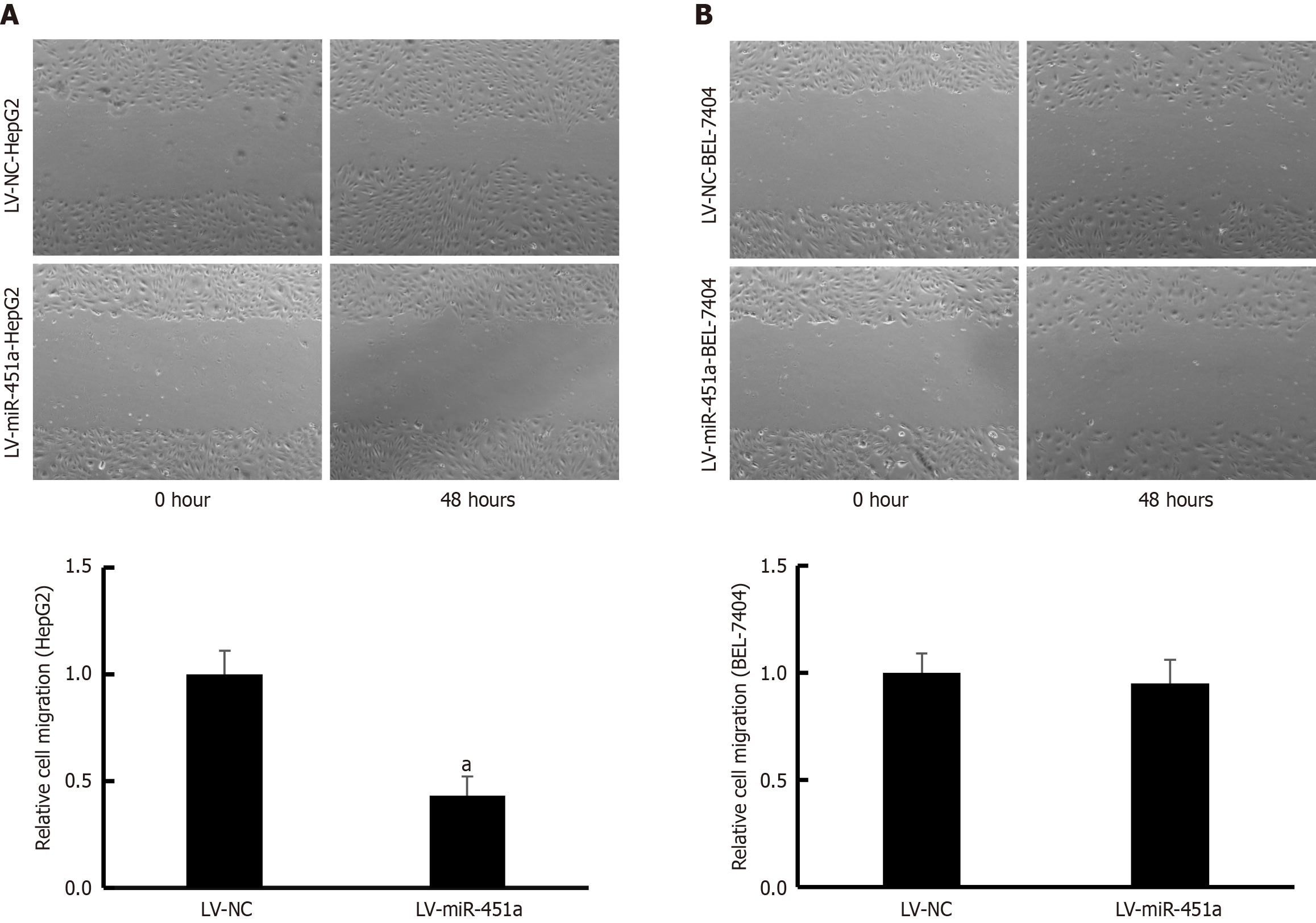
Wound scratch assay was used to evaluate migration ability of HepG2 and BEL-7404 cell lines transfected with LV-miR-451a or LV-NC virus. The distance of cell migration in HepG2 cells transfected with LV-miR-451a was obviously shorter compared with cells treated with LV-NC virus (P < 0.05; Figure 4A). However, the distance of cell migration of BEL-7404 cell showed no significant difference treated with LV-miR-451a/LV-NC groups (Figure 4B).
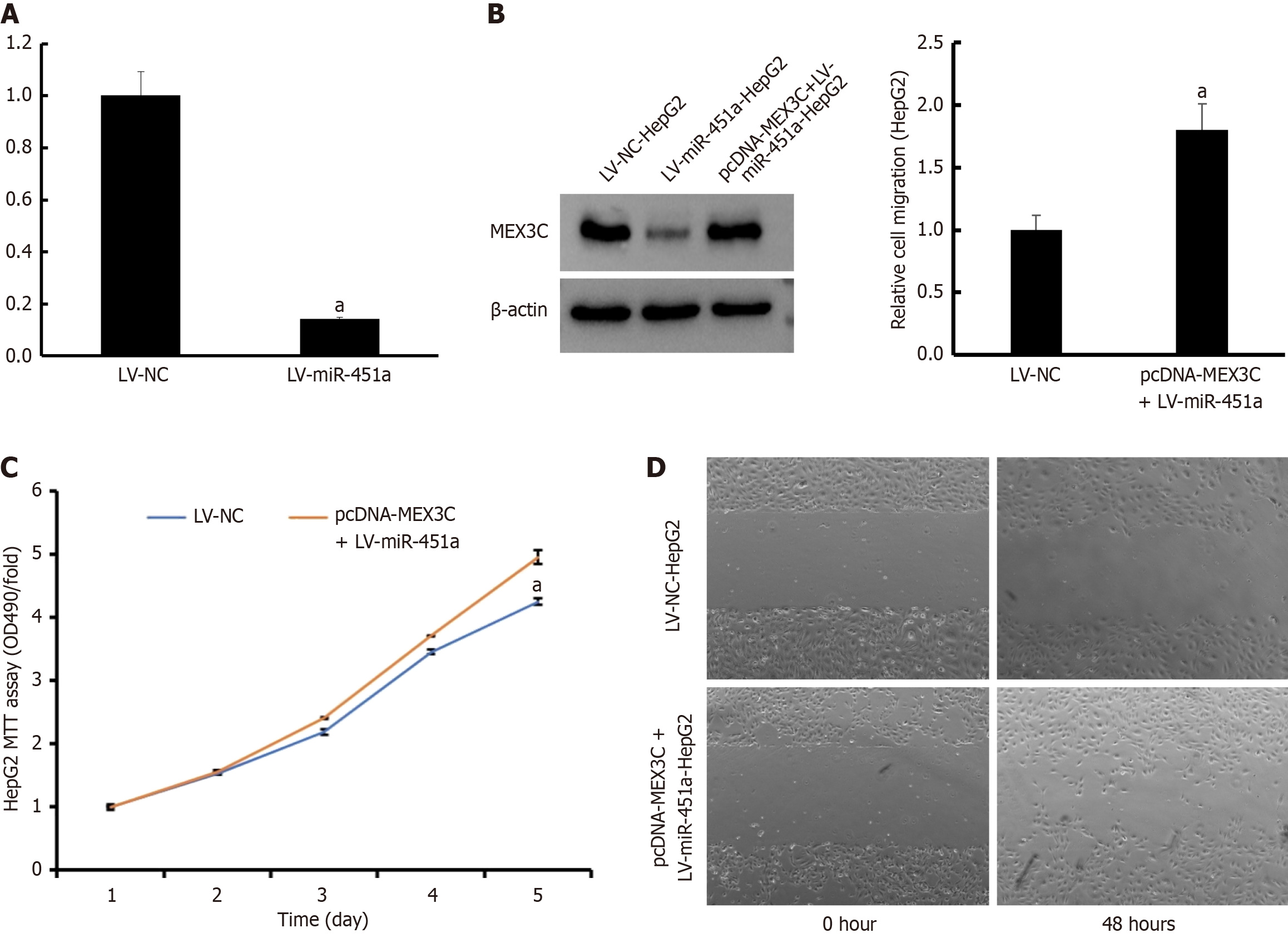
To investigate the underlying molecular mechanisms of miR-451a in proliferation and migration, potential target genes for miR-451a explored using the Targetscan, microRNA. ORG and miRDB database. As a result, MEX3C was predicted as potential target for miR-451a. Subsequently, RT-PCR and western blotting performed to confirm MEX3C expression level (Figure 5A and B). After co-transfection with LV-miR-451a and pcDNA-MEX3C, the protein level of MEX3C increased. Moreover, rescue experiments were designed by over-expression MEX3C in HepG2 cells via using pcDNA-MEX3C. The cell proliferation and invasion ability of HepG2 cell compared between LV-NC group and LV-miR-451a+pcDNA-MEX3C group. MTT bromide assay found that HepG2 cell growth inhibited by LV-miR-451a was rescued by MEX3C recovery (P < 0.05; Figure 5C). Meanwhile, cell migration capability was also restored by pcDNA-MEX3C (P < 0.05; Figure 5D). These results demonstrated that miR-451a inhibited HepG2 cell proliferation and migration via suppressing MEX3C expression.
Our present study mainly to determine the miR-451a expression in liver cancer cell lines, by contrast, miR-451a expression in colorectal cancer cell and esophageal squamous cancer cell were also detected. In order to investigate the function of miR-451a, HepG2 and BEL-7404 cell transfected with LV-miR-451a to construct stably overexpressed miR-451a liver cancer cell line. MTT bromide assay, colony formation assay and wound scratch assay showed that miR-451a overexpression can inhibit HepG2 cell proliferation and migration, but not for BEL-7404 cell line. This discrepancy is likely attributable to inherent differences between the two cell types in their molecular profiling, target gene expression, and compensatory capacity within signaling networks. Resolving this issue necessitates further elucidation of the specific signal-dependent mechanisms unique to BEL-7404 cells.
Concurrently, our findings demonstrate that miR-451a does not influence apoptosis in this system. This is likely because, in HepG2 cells, miR-451a suppresses proliferation by inhibiting its target(s), thereby downregulating associated signaling pathways and ultimately inducing cell cycle arrest at the G1 phase. In contrast, in BEL-7404 cells, this G1 arrest mechanism cannot be effectively activated, potentially due to the absence of key target pathways, p53 mutation, and/or the presence of compensatory signaling pathways. Elucidating the precise growth inhibitory mechanisms involved will also constitute one of the key foci of our future investigations.
Meanwhile, 9 target gene for miR-451a explored by bioinformatics analysis and validated through RT-PCR and western blotting. Our preliminary literature review identified MEX3C as a frequently dysregulated target gene across multiple cancer types. Critically, its direct regulatory role in cell cycle control exhibits high mechanistic plausibility and logical coherence with the miR-451a-induced proliferation suppression phenotype. Hence, MEX3C was prioritized as a candidate target for further investigation. Moreover, rescue experiments were designed to confirm the predicted target gene MEX3C. Therefore, the results of our study suggested the importance of miR-451a in HepG2 cells by prohibiting cell proliferation and migration.
It has been reported that miR-451a act as various aspect role in several kinds of cancers. For example, it is proved that miR-451a upregulated in whole blood samples collected from luminal A breast cancer patients. While, up-regulation of miR-451a can suppress the resistance to tamoxifen through regulating autophagy, 14-3-3ζ expression and Erα[9]. It was also found that miR-451a was preferentially expressed in the extracellular vesicles (EVs) of T lymphocytes, which indicated that miR-451a interact with the tumor immune environment[10]. Moreover, EVs-encapsulated miR-451a can be performed as circulating biomarker of breast cancer[11]. In non-small cell lung cancer (NSCLC), miR-451 was low-expression in NSCLC tissues and A549 and NCI-H460 cell lines. Moreover, miR-451a/activating transcription factor 2 (ATF2) axis through ATF2 regulation to inhibited NSCLC cells migration and invasive ability[12]. Interestingly, the expression of miR-451a significantly different in NSCLC cell lysates from NSCLC EVs[13], furthermore, plasma exosomal miR-451a can act as biomarker for early prediction of recurrence and prognosis of NSCLC[14]. GeneChip microarray analysis was used for colorectal cancer showed that miR-451a had decreased expression with more advanced tumors[15]. And miR-451a can inhibit proliferation and increase apoptosis of colorectal cancer cell through inducing endoplasmic reticulum stress by binding to the 5’-UTR of BAP31[16]. In addition, radiation may induce miR-451a up-regulation and may influence colorectal carcinoma proliferation via CAB39 and EMSY pathways[17]. MiR-451a expression both in papillary thyroid carcinoma (PTC) tissues and plasma significantly higher than in those with control group, which suggested the valuable of miR-451a for the diagnosis of PTC[18]. The mechanism study showed that miR-451a can suppress PTC cell growth, epithelial-mesenchymal transition and induces apoptosis via targeting PSMB8[19]. In osteosarcoma, miR-451a was found to be downregulated and inhibited the growth and invasion of osteosarcoma cell by targeting TRIM66[20]. MiR-451a expression also significantly low-expression in renal cell carcinoma (RCC), which closely related to poor prognosis of RCC patients[21]. In our study, 9 possible target genes were found by bioinformatic analysis. Except for PSMB8, the other 8 target genes downregulated significantly, which indicated possible mechanism underlying the function of miR-451a.
However, this study has several limitations. These include the lack of patient demographic characteristics, clinical staging, treatment modalities, and prognostic outcome details for the source specimens. Additionally, the downstream pathways through which MEX3C influences proliferation and migration remain unexplored. Consequently, conducting further follow-up studies and delineating the downstream pathways involved will constitute the primary focus of our subsequent work. We plan to perform a comprehensive analysis and mechanistic dissection of these aspects in future investigations.
In summary, this study found that miR-451a obviously down-regulated in liver cancer tissues and tumor cell lines. Overexpression of miR-451a significantly inhibit HepG2 cell proliferation, colony formation and migration. However, miR-451a were not participated in liver cancer cell apoptosis. Moreover, bioinformatic analysis showed that MEX3C was target gene of miR-451a, which validated by RT-PCR and western blotting. Furthermore, rescue experiments confirmed that overexpression of MEX3C can inhibit the effect of miR-451a in HepG2 cell. Therefore, our research provides new insight for understanding molecular mechanisms of liver cancer development and progression to support diagnostic and therapeutic strategies.
| 1. | Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 3113] [Article Influence: 444.7] [Reference Citation Analysis (17)] |
| 2. | Keenan BP, Fong L, Kelley RK. Immunotherapy in hepatocellular carcinoma: the complex interface between inflammation, fibrosis, and the immune response. J Immunother Cancer. 2019;7:267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 168] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 3. | Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015;25:137-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 405] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 4. | Zhang WW, Ming XL, Rong Y, Huang CQ, Weng H, Chen H, Bian JM, Wang FB. Diagnostic Value Investigation and Bioinformatics Analysis of miR-31 in Patients with Lymph Node Metastasis of Colorectal Cancer. Anal Cell Pathol (Amst). 2019;2019:9740475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Geng F, Lu GF, Ji MH, Kong DY, Wang SY, Tian H, Xie ZM, Pan M, Gong NL. MicroRNA-26b-3p/ANTXR1 signaling modulates proliferation, migration, and apoptosis of glioma. Am J Transl Res. 2019;11:7568-7578. [PubMed] |
| 6. | Huang WT, Liu AG, Cai KT, He RQ, Li Z, Wei QJ, Chen MY, Huang JY, Yan WY, Zhou H, Chen G, Ma J. Exploration and validation of downregulated microRNA-199a-3p, downstream messenger RNA targets and transcriptional regulation in osteosarcoma. Am J Transl Res. 2019;11:7538-7554. [PubMed] |
| 7. | Zhou L, Du Y, Kong L, Zhang X, Chen Q. Identification of molecular target genes and key pathways in hepatocellular carcinoma by bioinformatics analysis. Onco Targets Ther. 2018;11:1861-1869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 8. | Arabkari V, Clancy E, Dwyer RM, Kerin MJ, Kalinina O, Holian E, Newell J, Smith TJ. Relative and Absolute Expression Analysis of MicroRNAs Associated with Luminal A Breast Cancer- A Comparison. Pathol Oncol Res. 2020;26:833-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 9. | Liu ZR, Song Y, Wan LH, Zhang YY, Zhou LM. Over-expression of miR-451a can enhance the sensitivity of breast cancer cells to tamoxifen by regulating 14-3-3ζ, estrogen receptor α, and autophagy. Life Sci. 2016;149:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 10. | Liu Z, Miao T, Feng T, Jiang Z, Li M, Zhou L, Li H. miR-451a Inhibited Cell Proliferation and Enhanced Tamoxifen Sensitive in Breast Cancer via Macrophage Migration Inhibitory Factor. Biomed Res Int. 2015;2015:207684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Moloney BM, Gilligan KE, Joyce DP, O'Neill CP, O'Brien KP, Khan S, Glynn CL, Waldron RM, Maguire CM, Holian E, Naughton E, Elhadi M, Grealish AB, Malone C, McDermott E, Dockery P, Ritter T, Prina-Mello A, Kerin MJ, Dwyer RM. Investigating the Potential and Pitfalls of EV-Encapsulated MicroRNAs as Circulating Biomarkers of Breast Cancer. Cells. 2020;9:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Shen YY, Cui JY, Yuan J, Wang X. MiR-451a suppressed cell migration and invasion in non-small cell lung cancer through targeting ATF2. Eur Rev Med Pharmacol Sci. 2018;22:5554-5561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 13. | Li C, Qin F, Hu F, Xu H, Sun G, Han G, Wang T, Guo M. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018;8:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 14. | Kanaoka R, Iinuma H, Dejima H, Sakai T, Uehara H, Matsutani N, Kawamura M. Usefulness of Plasma Exosomal MicroRNA-451a as a Noninvasive Biomarker for Early Prediction of Recurrence and Prognosis of Non-Small Cell Lung Cancer. Oncology. 2018;94:311-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 15. | Slattery ML, Herrick JS, Mullany LE, Valeri N, Stevens J, Caan BJ, Samowitz W, Wolff RK. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int J Cancer. 2015;137:428-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 119] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 16. | Xu K, Han B, Bai Y, Ma XY, Ji ZN, Xiong Y, Miao SK, Zhang YY, Zhou LM. MiR-451a suppressing BAP31 can inhibit proliferation and increase apoptosis through inducing ER stress in colorectal cancer. Cell Death Dis. 2019;10:152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 17. | Ruhl R, Rana S, Kelley K, Espinosa-Diez C, Hudson C, Lanciault C, Thomas CR Jr, Liana Tsikitis V, Anand S. microRNA-451a regulates colorectal cancer proliferation in response to radiation. BMC Cancer. 2018;18:517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 18. | Li M, Song Q, Li H, Lou Y, Wang L. Circulating miR-25-3p and miR-451a May Be Potential Biomarkers for the Diagnosis of Papillary Thyroid Carcinoma. PLoS One. 2015;10:e0132403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 19. | Fan X, Zhao Y. miR-451a inhibits cancer growth, epithelial-mesenchymal transition and induces apoptosis in papillary thyroid cancer by targeting PSMB8. J Cell Mol Med. 2019;23:8067-8075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 20. | Ma X, Li D, Gao Y, Liu C. miR-451a Inhibits the Growth and Invasion of Osteosarcoma via Targeting TRIM66. Technol Cancer Res Treat. 2019;18:1533033819870209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Yamada Y, Arai T, Sugawara S, Okato A, Kato M, Kojima S, Yamazaki K, Naya Y, Ichikawa T, Seki N. Impact of novel oncogenic pathways regulated by antitumor miR-451a in renal cell carcinoma. Cancer Sci. 2018;109:1239-1253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/