Published online Feb 24, 2025. doi: 10.5306/wjco.v16.i2.99839
Revised: October 10, 2024
Accepted: November 13, 2024
Published online: February 24, 2025
Processing time: 132 Days and 19.1 Hours
In recent years, many studies have shown that proteasome 26S subunit non-ATPase 6 (PSMD6) plays an important role in the occurrence and development of malignant tumours. Unfortunately, there are no reports on the evaluation of the potential role of PSMD6 in hepatocellular carcinoma (HCC).
To comprehensively evaluate the overexpression pattern and clinical significance of PSMD6 in HCC tissues.
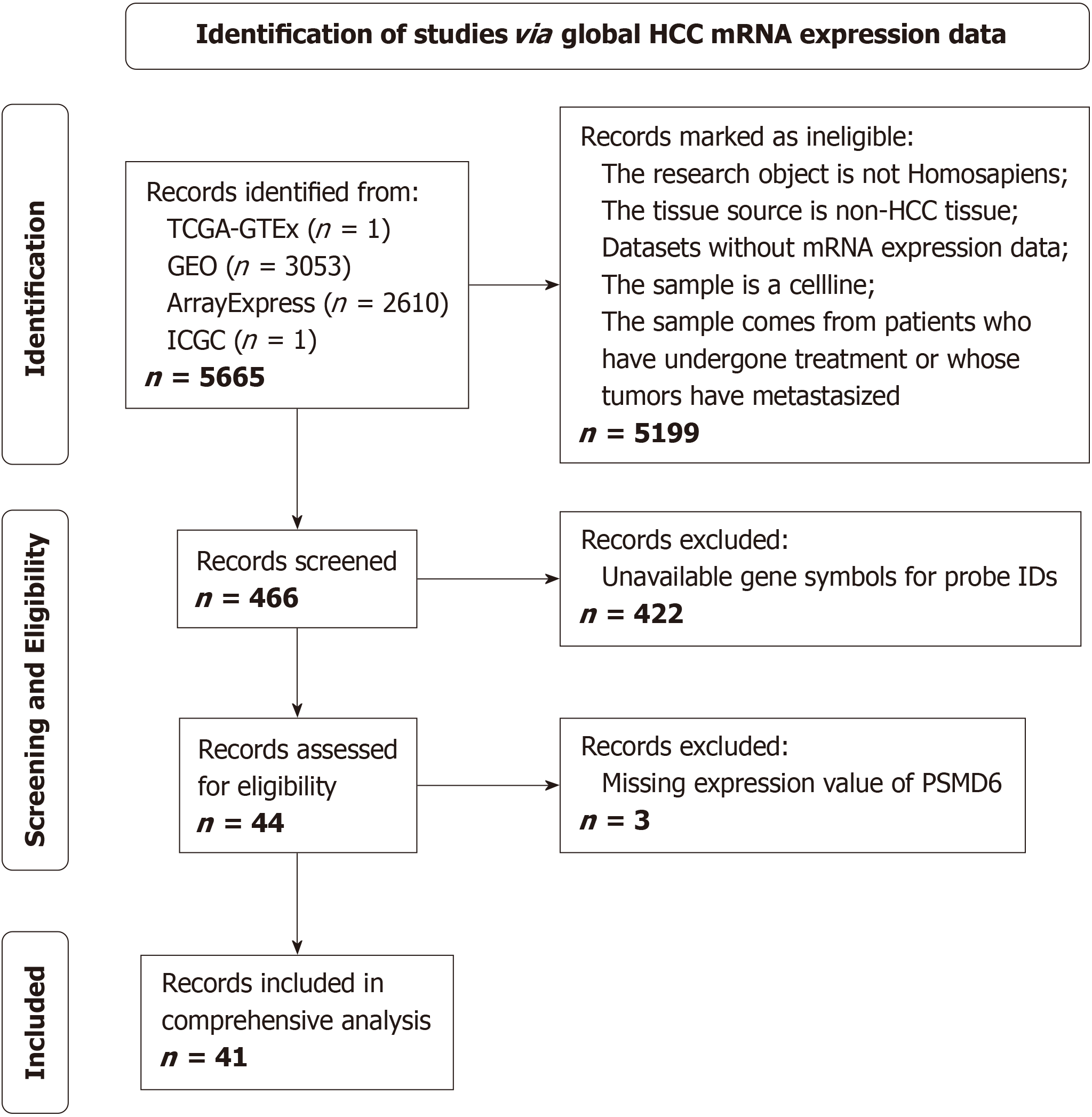
This study integrated PSMD6 mRNA expression profiles from 4672 HCC and 3667 non-HCC tissues, along with immunohistochemical scores from 383 HCC and adjacent tissues, to assess PSMD6 overexpression in HCC. Clustered regularly interspaced short palindromic repeats knockout technology evaluated PSMD6’s essential role in HCC cell growth. Functional enrichment analysis explored the molecular mechanism of PSMD6 abnormalities in HCC. Drug sensitivity analysis and molecular docking analysed the effect of abnormal expression of PSMD6 on the drug sensitivity of HCC cells.
The results of 41 external and two internal datasets showed that PSMD6 mRNA (SMD = 0.26, 95%CI: 0.09-0.42, P < 0.05) and protein (SMD = 2.85, 95%CI: 1.19-4.50, P < 0.05) were significantly overexpressed in HCC tissues. The integrated analysis results showed that PSMD6 had a significant overexpression pattern in HCC tissues (SMD = 0.40, 95%CI: 0.15-0.66, P < 0.05). PSMD6 knockout inhibited HCC cell growth (chronos scores < -1). Functional enrichment implicated ribosome biogenesis and RNA splicing. Significant enrichment of signalling pathways such as RNA degradation, ribosomes, and chemical carcinogenesis—reactive oxygen species. Drug sensitivity analysis and a molecular docking model showed that high expression of PSMD6 was associated with the tolerance of HCC cells to drugs such as ML323, sepantronium bromide, and GDC0810. Overexpressed PSMD6 effectively distinguished HCC tissues (AUC = 0.75, 95%CI: 0.71-0.79).
This study was the first to discover that PSMD6 was overexpressed in HCC tissues. PSMD6 is essential for the growth of HCC cells and may be involved in ribosome biogenesis and RNA splicing.
Core Tip: This study integrated 8339 hepatocellular carcinoma (HCC) tissue mRNA expression profiles from 41 platforms, 383 pairs of hepatocellular tissue samples from inhouse immunohistochemistry experiments, and HCC cell lines using the clustered regularly interspaced short palindromic repeats system for proteasome 26s subunit non-ATPase 6 (PSMD6) knockout data to evaluate the abnormal expression pattern and clinical significance of PSMD6 in HCC tissues. This study was the first to discover that PSMD6 was overexpressed in HCC tissues. The promotion of PSMD6 in HCC progression is closely related to ribosome biogenesis and RNA splicing, suggesting that PSMD6 may enhance the proliferation of HCC cells.
- Citation: Zhou SS, Ye YP, Chen Y, Zeng DT, Zheng GC, He RQ, Chi BT, Wang L, Lin Q, Su QY, Dang YW, Chen G, Wei JL. Overexpression pattern, function, and clinical value of proteasome 26S subunit non-ATPase 6 in hepatocellular carcinoma. World J Clin Oncol 2025; 16(2): 99839
- URL: https://www.wjgnet.com/2218-4333/full/v16/i2/99839.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i2.99839
Hepatocellular carcinoma (HCC) is one of the most common malignancies and is strongly linked with several key risk factors: Hepatitis virus infection, aflatoxin exposure, metabolic disorders, heavy alcohol consumption and smoking[1]. The high infection rates of hepatitis B virus, hepatitis C virus (HCV), and aflatoxin are important reasons for the long-term high morbidity and mortality of HCC[2-5]. Due to the lack of clinical symptoms in the early stages of HCC, many patients are diagnosed in the late stages[6]. For early HCC, common treatments are liver transplantation and surgical resection; for advanced HCC, common treatments are radiofrequency ablation, transarterial chemoembolization, targeted therapy, and immunotherapy[6]. Although there are many treatment options that have had a positive impact on patients with advanced HCC, drug resistance and recurrence still exist[7]. Timeliness and accuracy of diagnosis are still the keys to improving the prognosis of HCC[8]. Therefore, further understanding of the molecular biological mechanisms related to HCC is crucial to improving the timeliness of HCC diagnosis, treatment efficacy, and patient prognosis.
Proteasome 26S subunit non-ATPase 6 (PSMD6) is one of the important components of the ubiquitin-proteasome system. In recent years, many studies have shown that PSMD6 plays an important role in the occurrence and deve
To this end, this study is the first to comprehensively evaluate the abnormal expression pattern, potential molecular mechanism, and clinical significance of PSMD6 in HCC tissues based on multicentre large-sample bulk RNA-Seq, inhouse immunohistochemistry, and clustered regularly interspaced short palindromic repeats (CRISPR) knockout screen technology.
In this study, 284 and 99 HCC tissues and their corresponding adjacent tissues were collected from The First Affiliated Hospital of Guangxi Medical University (hereinafter referred to as Institution A) and Redcross Hospital of Yulin (he
This study widely collected bulk RNA-Seq data and corresponding clinical data of CRC tissues available worldwide from databases such as the Cancer Genome Atlas (TCGA), Gene Expression Omnibus, Sequence Read Archive, ArrayExpress, and International Cancer Genome Consortium. The specific process of retrieving and screening datasets is detailed in Figure 1. We finally included PSMD6 mRNA expression profiles of 4672 HCC tissues and 3667 non-HCC tissues from 41 platforms. Then, we merged, de-batched, standardised, corrected, and removed abnormal data samples based on plat
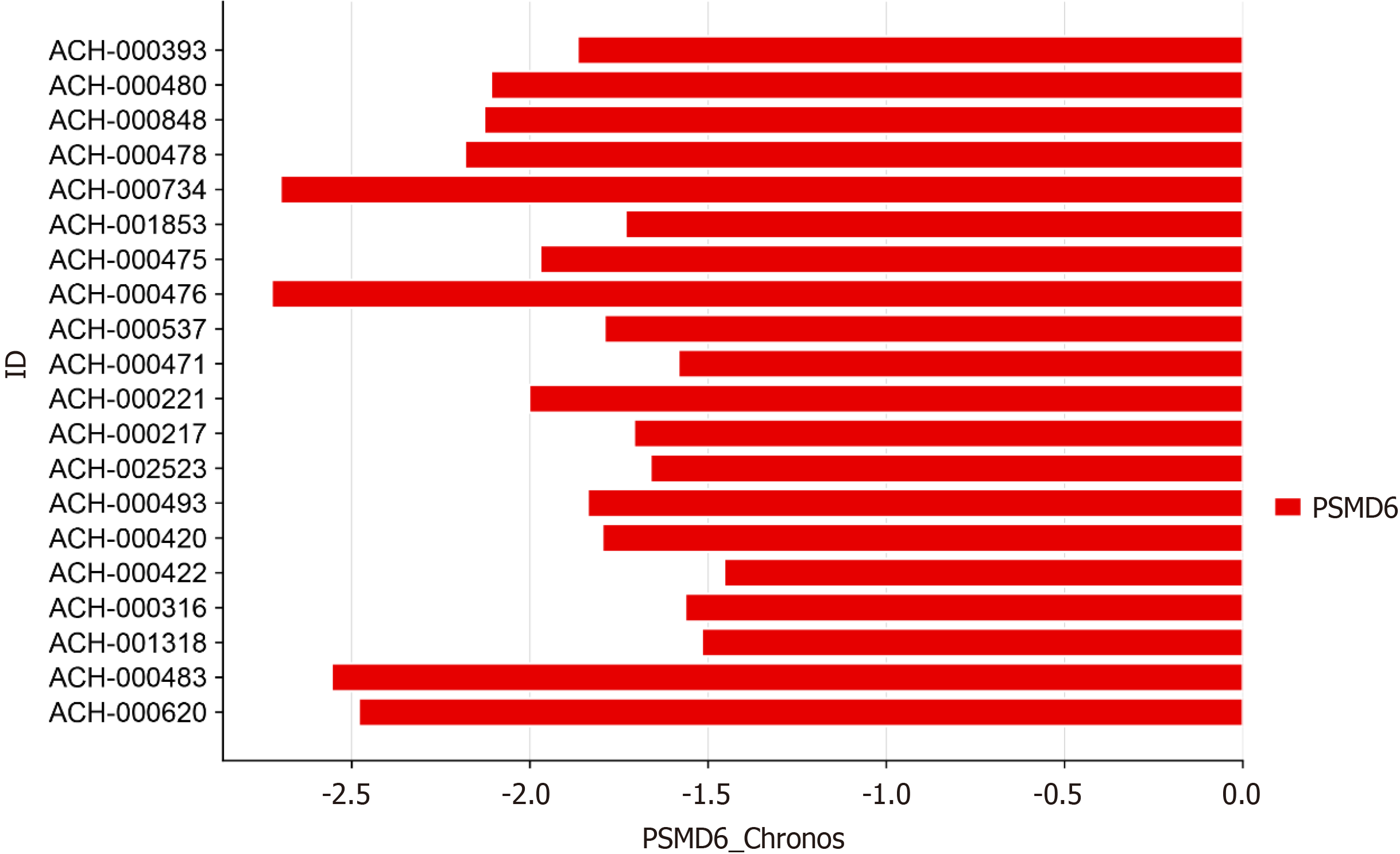
This study used CRISPR knockout screen technology from the DepMap database to analyse the effect of PSMD6 on the growth of 20 primary HCC cell lines[12]. After the PSMD6 gene was knocked out in the HCC cell lines, the chronos algorithm was used to evaluate the PSMD6 chronos score to evaluate the effect of PSMD6 knockout on the growth of HCC cell lines[13]. In this study, a chronos score less than -1 indicated that PSMD6 was highly necessary for the growth and proliferation of the cell line.
Based on the included multi-centre mRNA expression profiles, this study used the meta and metafor packages of R v4.2.2 to batch calculate the SMD of each gene to screen out differentially expressed genes (DEGs) (the 95%CI of SMD does not include 0, P < 0.05). Next, we used cBioPortal to analyse the HCC mRNA data from TCGA to identify genes co-expressed with PSMD6 (|R| > 0.25, P < 0.05)[14]. By taking the intersection of DEGs and PSMD6 co-expressed genes, we obtained the intersection genes. Subsequently, we performed Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis on these intersection genes to explore the possible molecular biological mechanism of PSMD6 in HCC.
Based on the HCC tissue mRNA expression profile in the TCGA database and the GDSC 2.0 database, the correlation between PSMD6 mRNA expression level and targeted drug sensitivity score (as measured by IC50 value) was calculated using the oncopdict package R v 4.2.2. The effect of PSMD6 expression on IC50 was evaluated by regression analysis. In this study, if there is a strong correlation between PSMD6 mRNA expression level and drug sensitivity (|R|> 0.35, P < 0.05), it is believed that PSMD6 has a significant effect on the drug sensitivity of targeted therapeutic drugs in HCC cells.
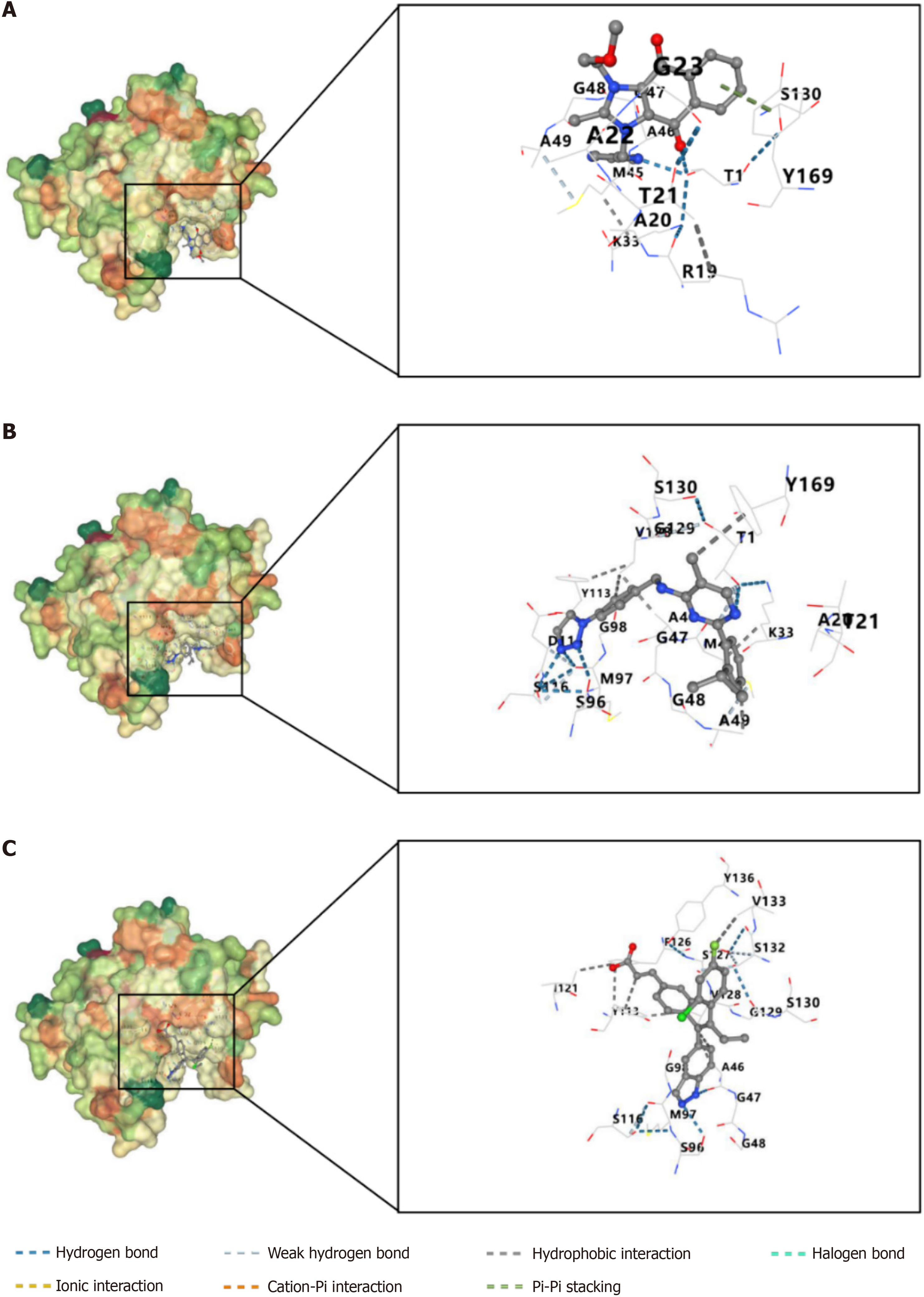
This study obtained the three-dimensional structure of PSMD6 protein (PDB format) from the PDB database and the three-dimensional structure (sdf format) of small molecule targeted therapeutic drugs from PubChem. This study also used CB-Dock2 to repair missing atoms and hydrogen atoms, remove water molecules and other heteroatoms, detect protein pockets, estimate spatial parameters, and perform Vina scoring on the three-dimensional structure of PSMD6 protein and small molecule targeted therapeutic drugs for protein drug blind docking analysis[15]. The molecular docking model with the best Vina score is visually displayed. In this study, Vina scores less than -6 kcal/mol are considered to have good binding ability; scores greater than -6 and less than -4 kcal/mol are considered to have average binding ability; scores greater than -4 kcal/mol are considered to have poor binding ability.
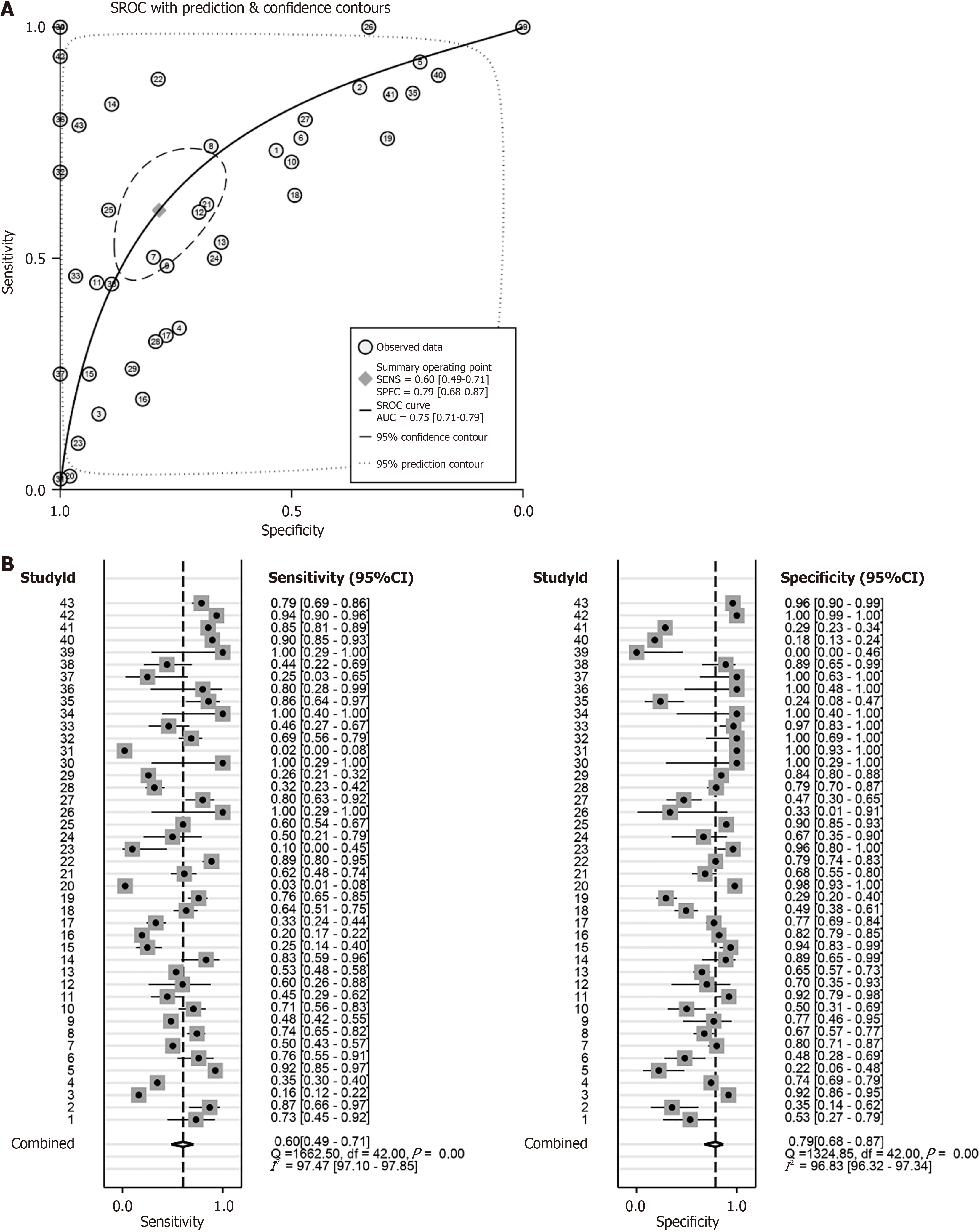
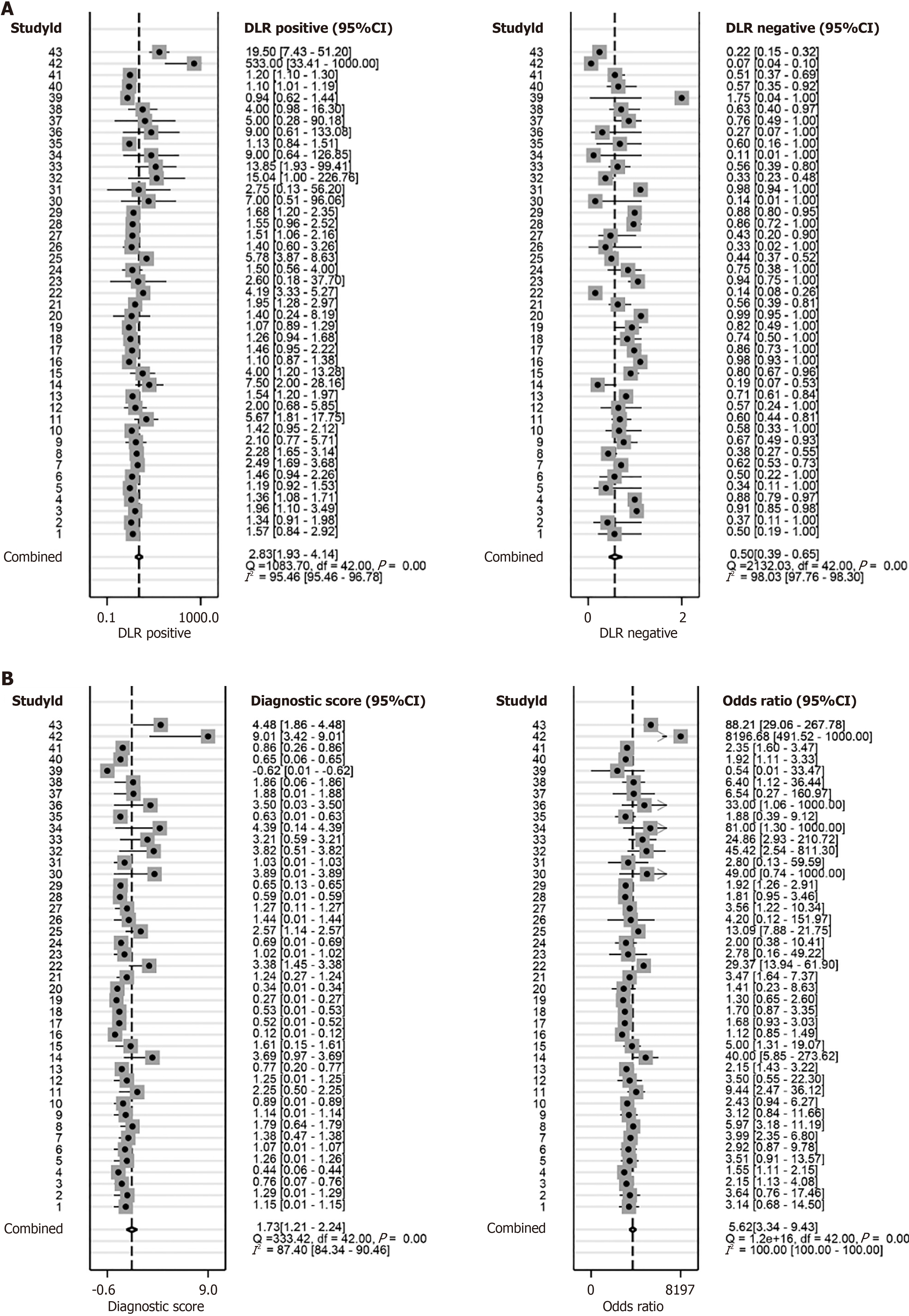
This study used receiver operating characteristic curve (ROC), summary ROC (sROC), positive likelihood ratio, and negative likelihood ratio forest plots to analyse the above-mentioned collected HCC PSMD6 mRNA expression profiles, HCC tissue internal protein expression levels, and corresponding clinical information. The ability of PSMD6 expression levels to identify HCC tissues was evaluated. Since most of the HCC patients in Institution B's internal samples have relatively complete clinical information, this study also evaluated the relationship between PSMD6 protein expression levels and the clinicopathological characteristics of corresponding HCC patients to further analyse the influencing factors of PSMD6 expression levels.
All statistical analyses involved in this study were completed on R v4.2.2, Stata v12, and SPSS v23. Q test and I2 statistical analysis were used to select the calculation model to be used to calculate the SMD of PSMD6. If I2 > 50%, it indicates that there is large heterogeneity, and the random effects model is used to calculate SMD. Otherwise, the fixed effects model was used. When comparing the differences in the means of two or more groups, choose the t test, one-way analysis of variance, or rank sum test according to the type and characteristics of the data. In this study, P < 0.05 was considered statistically significant.
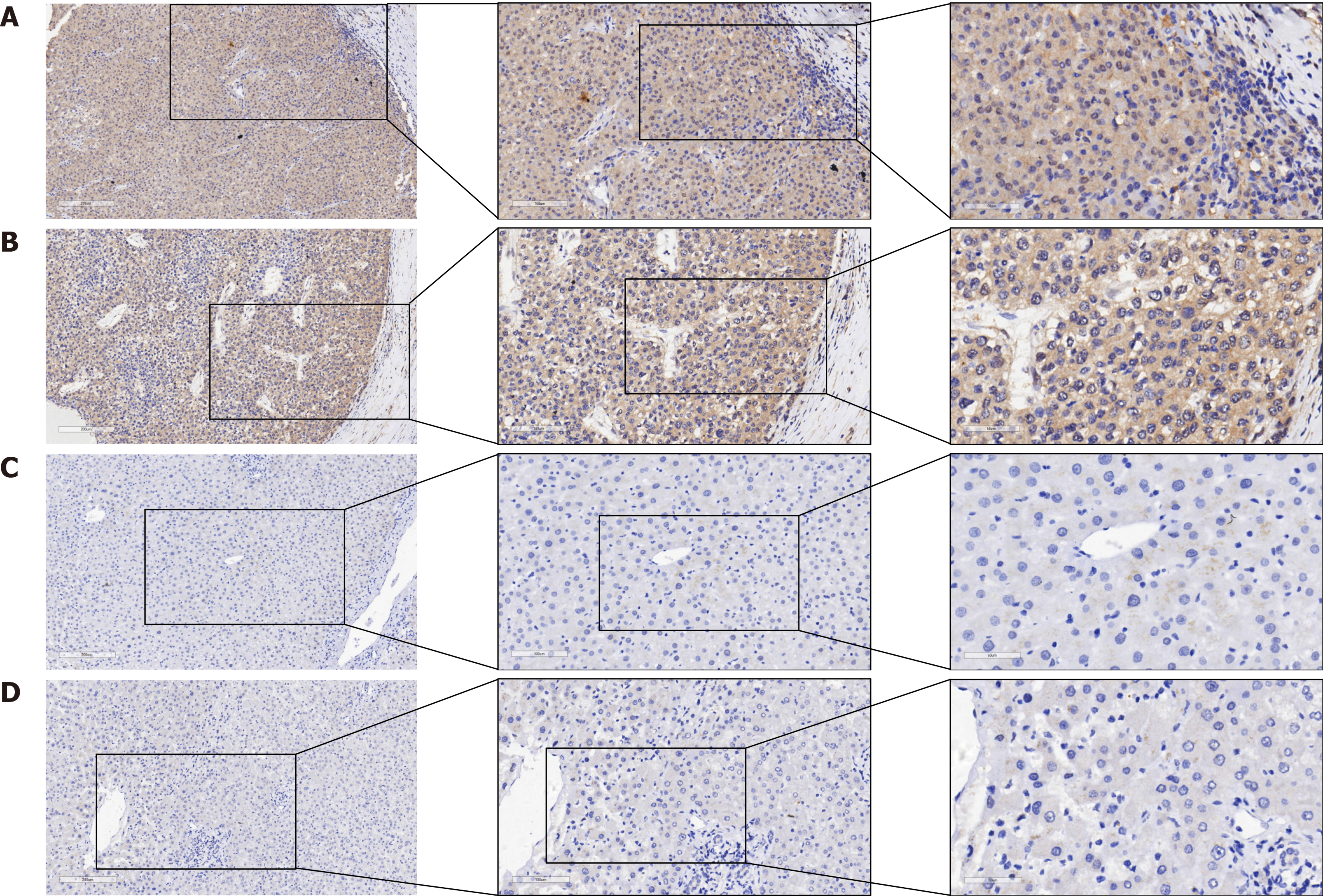
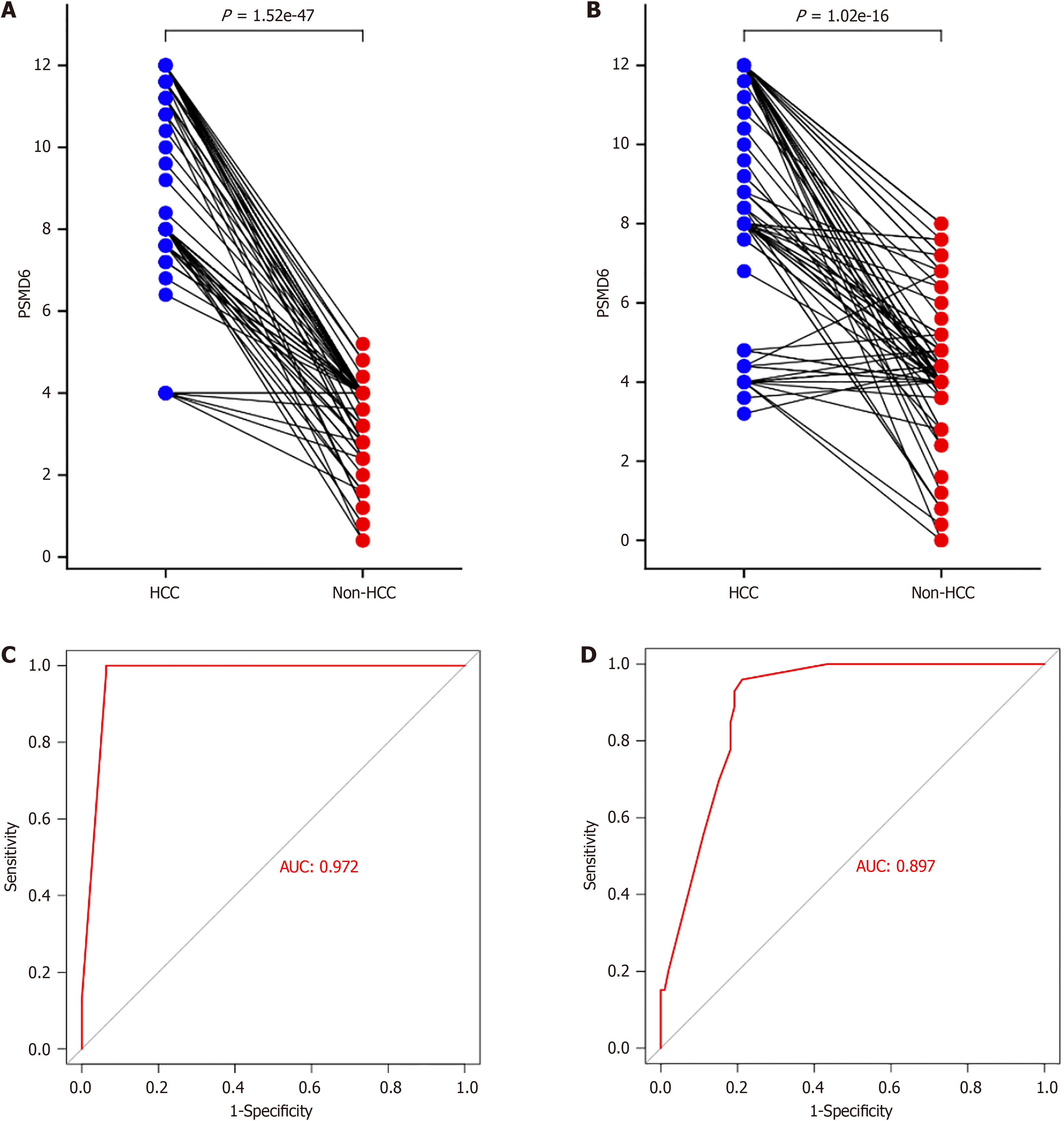
In the study, 383 HCC tissues and their corresponding paracancerous tissues were collected for in-house immunohistochemistry to evaluate the overexpression pattern of PSMD6 protein in HCC tissues. From a qualitative perspective, PSMD6 showed positive or strong positive staining in HCC tissues (Figure 2A and B). PSMD6 showed weak positive or negative staining in the corresponding paracancerous tissues of HCC (Figure 2C and D). To more objectively evaluate the overexpression pattern of PSMD6, we also scored the inhouse immunohistochemistry results for quantitative analysis. The results showed that whether in institution A samples (Figure 3A) or in institution B samples (Figure 3B), the PSMD6 protein expression level in HCC tissues was significantly higher (P < 0.05) than the corresponding paracancerous tissues.
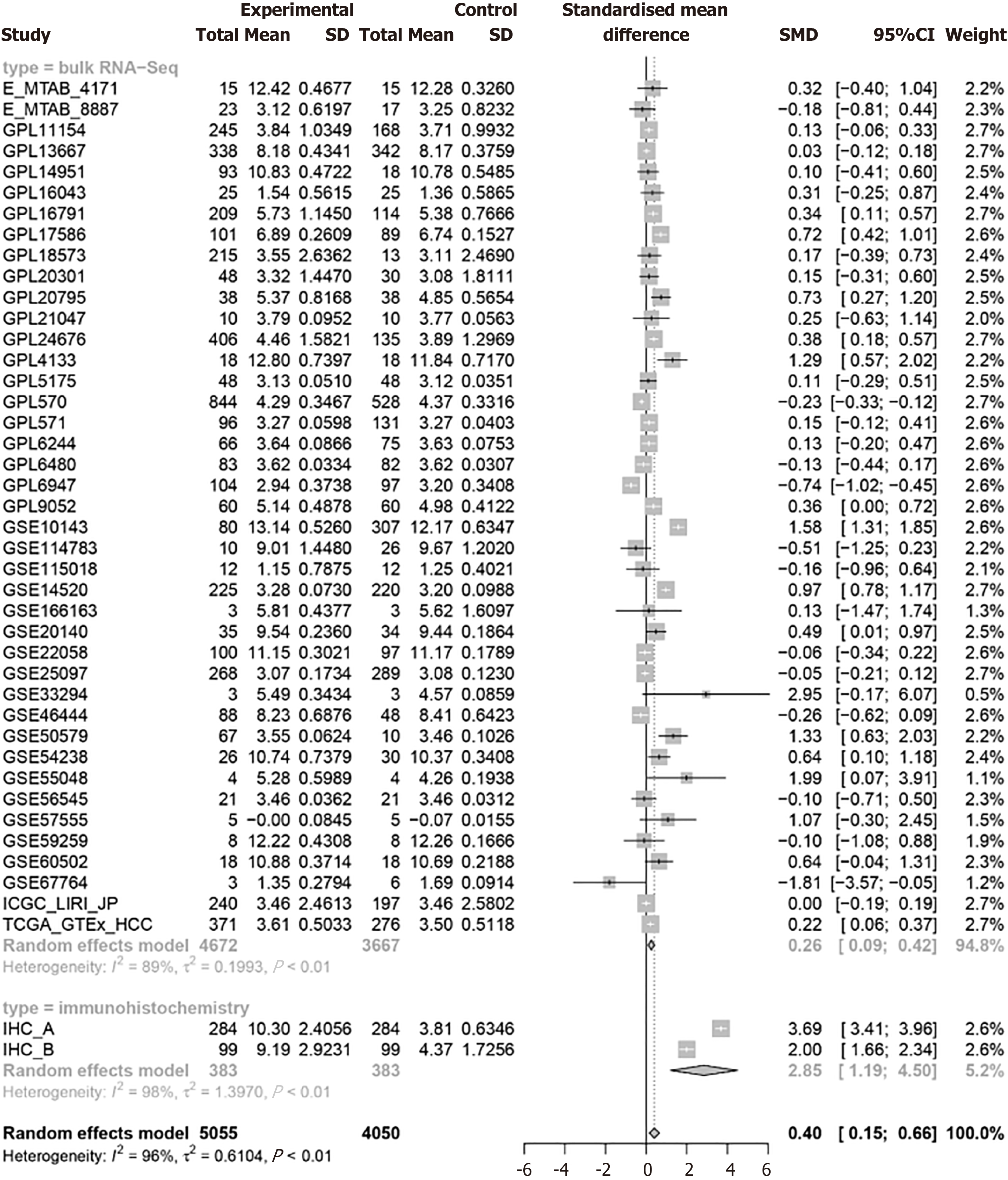
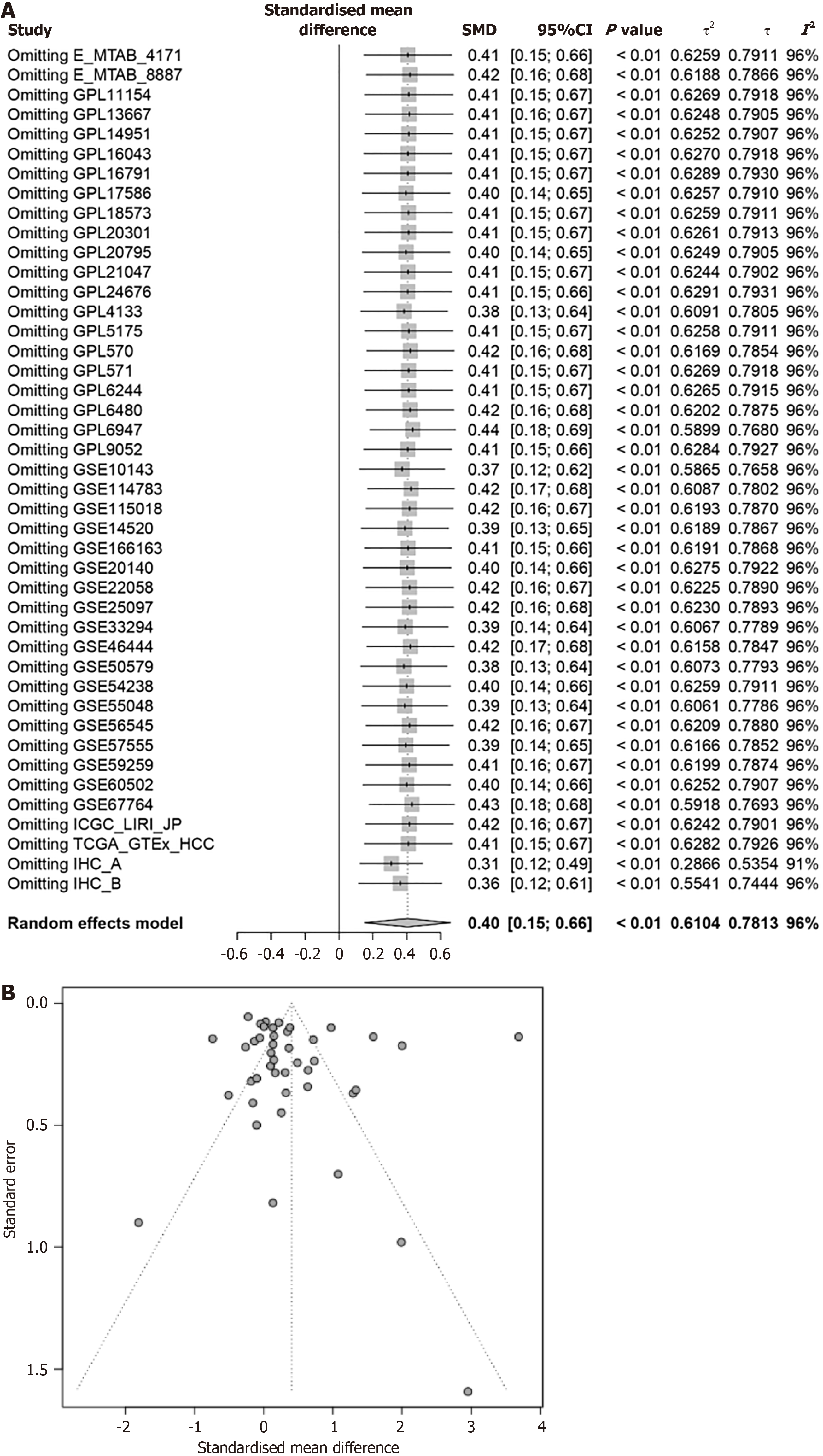
This approach aims to the findings of high PSMD6 in HCC tissues by accessing and analyzing a large, diverse dataset of RNA data from multiple sources. Since I2 is all greater than 50%, the random effects model is used to calculate PSMD6 SMD (Figure 4). Results from 41 external datasets and 2 internal datasets showed that PSMD6 mRNA (SMD = 0.26, 95%CI: 0.09-0.42, P < 0.05) and protein (SMD = 2.85, 95%CI: 1.19-4.50, P < 0.05) are significantly highly expressed in HCC tissues (Figure 4). The integration of external and internal data further showed that PSMD6 has a significant overexpression pattern in HCC tissues (SMD = 0.40, 95%CI: 0.15-0.66, P < 0.05; Figure 4). Sensitivity analysis of PSMD6 SMD showed that no significant changes were found when PSMD6 SMD was re-evaluated by discarding individual HCC tissue data sets one by one (Figure 5A). In addition, Begg’s test (P > 0.05) and Egger’s test (P > 0.05) did not detect significant publication bias (Figure 5B). These results illustrate that external and internal datasets from multiple centres can be combined for analysis without appreciable impact on the results.
This study used CRISPR knockout screen technology to analyse the effect of PSMD6 on the growth of 20 HCC cell lines. The gene chronos score of PSMD6 in 20 types of HCC cells showed that after knocking out the PSMD6 gene, all 20 types of HCC cells showed significant growth inhibition (chronos scores were all less than -1; Figure 6). This result suggests that the PSMD6 gene is an essential gene for HCC cell growth.
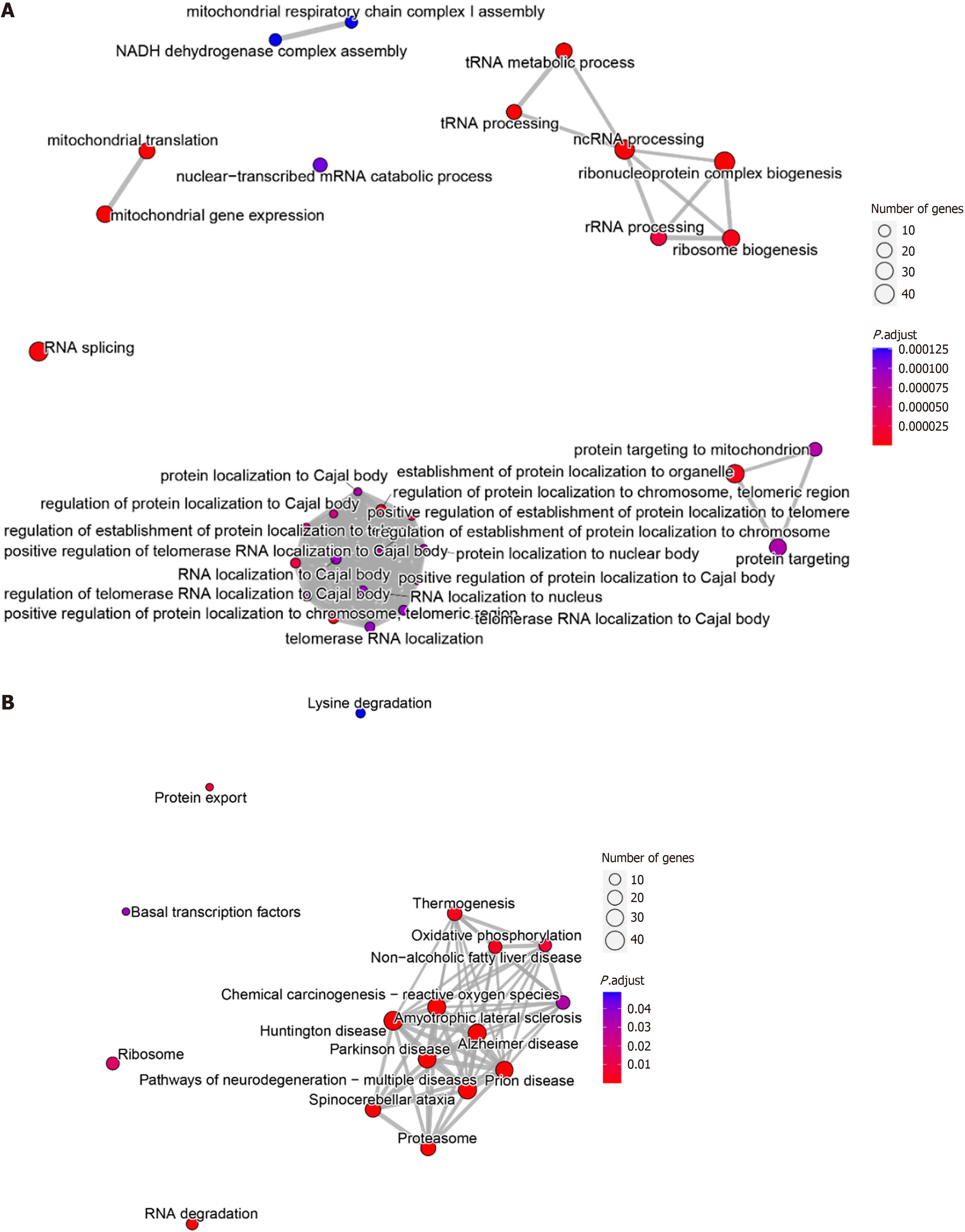
This study obtained 600 intersection genes through differential analysis and co-expression analysis. GO enrichment analysis showed that PSMD6 abnormally expressed in HCC might influence or disrupt signalling pathways such as the tRNA metabolic process, ncRNA processing, ribosome biogenesis, RNA splicing, and RNA localization to the nucleus. In addition, PSMD6 is involved in the biogenesis of ribosomes, suggesting that it may contribute to dysregulation of protein synthesis. Its effect on RNA splicing may lead to the production of oncogene subtypes that drive HCC progression. Alterations in ncRNA processing may also lead to dysregulation of tumor suppressor rna or accumulation of carcinogenic mirnas (Figure 7A). KEGG enrichment analysis showed significant enrichment of signalling pathways such as RNA degradation, ribosomes, and chemical carcinogenesis—reactive oxygen species (Figure 7B). These molecular pathways highlight the potential role of PSMD6 as a key regulator in the development and progression of HCC.
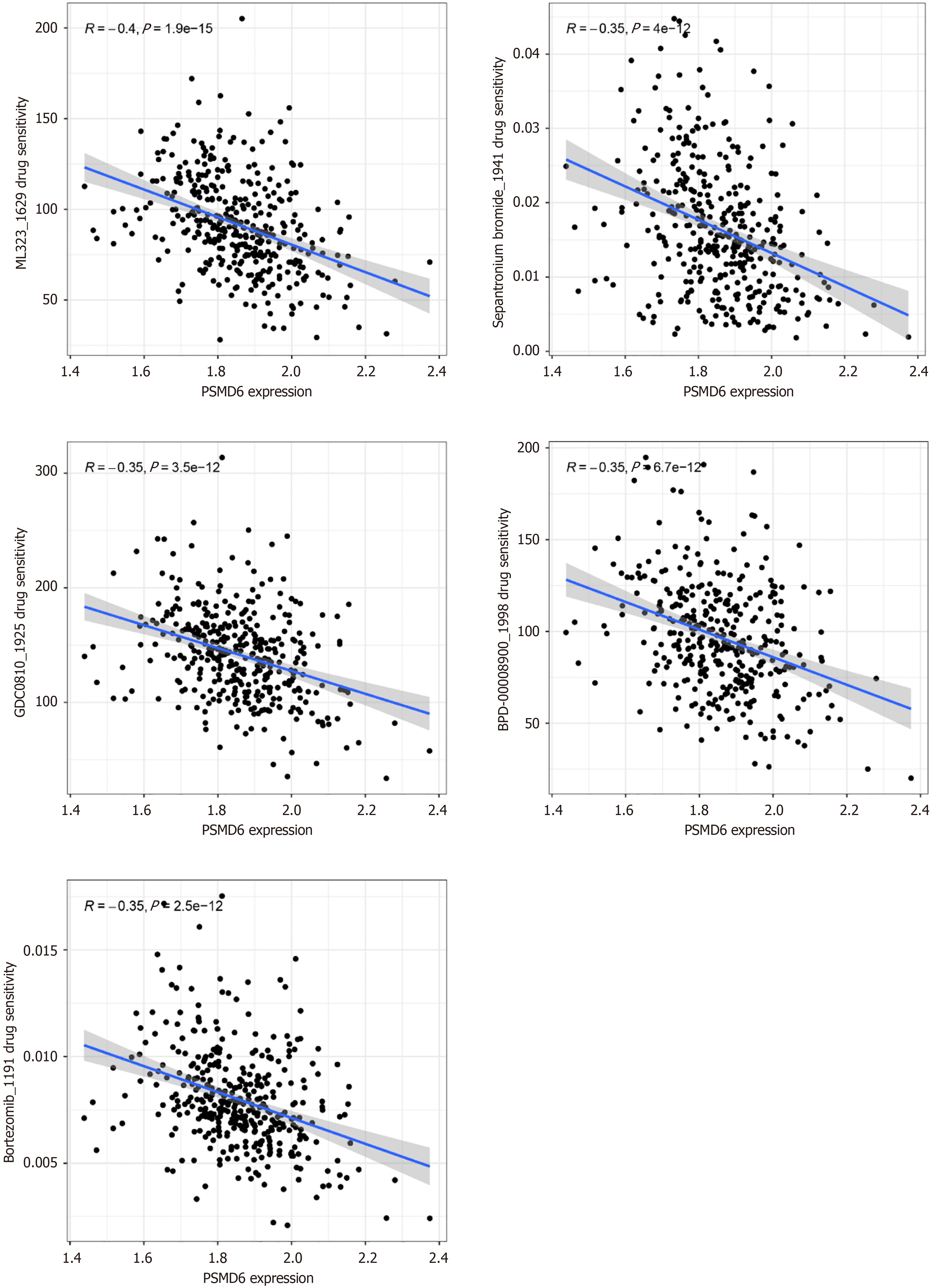
Drug sensitivity analysis showed that the drug sensitivity of ML323 (R = -0.40, P < 0.05), Sepantronium bromide (R = -0.35, P < 0.05), GDC0810 (R = -0.35, P < 0.05), BPD-00008900 (R = -0.35, P < 0.05), and Bortezomib (R = -0.35, P < 0.05) in HCC cells was negatively correlated with the expression level of PSMD6 mRNA (Figure 8). This suggests that HCC cells overexpressing PSMD6 have a certain tolerance to these targeted therapeutic drugs. In addition, this study also constructed a molecular docking model to further evaluate this drug-sensitivity relationship. Given that BPD-00008900 and Bortezomib do not have 3D structures suitable for this analysis process, only the docking models of the other three small molecules with PSMD6 are shown here. The results showed that ML323 (Vina score = -7.5 kcal/mol), Sepantronium bromide (Vina score = -6.1 kcal/mol), and GDC0810 (Vina score = -6.6 kcal/mol) all had strong binding ability with PSMD6 protein (Figure 9). And there were strong intermolecular forces in these three molecular docking models (Figure 9). For example, hydrogen bond, Pi-Pi stacking, and hydrophobic interaction. This further illustrates the stability of these molecular docking models.
First, in the immunohistochemical samples of institution A (AUC = 0.972; Figure 3C) and institution B (AUC = 0.897; Figure 3D), the highly expressed PSMD6 protein had a strong recognition ability for HCC tissue. The combination of external multicentre PSMD6 mRNA and internal protein analysis further demonstrated that PSMD6 had a strong ability to discriminate HCC tissues (AUC = 0.75, 95%CI: 0.71-0.79; Sensitivity = 0.60, 95%CI: 0.49-0.71; Specificity = 0.79, 95%CI: 0.68-0.87; Positive likelihood ratio = 2.83, 95%CI: 1.93-4.14; Negative likelihood ratio= 0.50, 95%CI 0.39-0.65; OR = 5.62, 95%CI: 3.34-9.43; Figure 10 and Figure 11). In addition, Table 1 suggests that the expression level of PSMD6 in HCC tissues of HCV-positive patients was higher than that of HCV-negative patients (P < 0.05). The remaining clinicopathological characteristics (e.g., age, gender, vascular invasion, tumour necrosis, hepatitis grade, satellite nodules, and Alpha-fetoprotein) were not statistically significant (P > 0.05).
| Clinicopathological Features | PSMD6 protein expression | |||||
| n | Mean | SD | t (t-test) or F (ANOVA) | P value | ||
| Gender | Male | 82 | 9.06 | 2.94 | -0.93 | 0.36 |
| Female | 17 | 9.79 | 2.91 | |||
| Age | > 60 years | 28 | 8.77 | 2.99 | -0.89 | 0.38 |
| ≤ 60 years | 71 | 9.35 | 2.92 | |||
| Tumour necrosis | Yes | 34 | 9.08 | 2.88 | -0.26 | 0.80 |
| No | 65 | 9.24 | 2.99 | |||
| Tumour recurrence | Yes | 4 | 9.00 | 3.60 | -0.13 | 0.90 |
| No | 95 | 9.20 | 2.93 | |||
| Number of lesions | 1 | 91 | 9.20 | 2.88 | 0.09 | 0.93 |
| > 1 | 8 | 9.10 | 3.73 | |||
| Satellite nodules | Yes | 9 | 9.29 | 2.04 | 0.11 | 0.92 |
| No | 90 | 9.18 | 3.02 | |||
| Vascular invasion | M0 | 50 | 8.79 | 3.07 | 1.34 | 0.27 |
| M1 | 35 | 9.38 | 2.82 | |||
| M2 | 13 | 10.22 | 2.60 | |||
| Lymph node invasion | Yes | 10 | 9.88 | 1.92 | 1.12 | 0.29 |
| No | 89 | 9.11 | 3.03 | |||
| Hepatitis grade | G0 | 6 | 8.07 | 3.49 | 0.58 | 0.63 |
| G1 | 36 | 8.93 | 2.86 | |||
| G2 | 54 | 9.44 | 3.00 | |||
| G3 | 3 | 10.00 | 1.74 | |||
| Liver fibrosis grading | S1 | 9 | 9.87 | 2.71 | 0.23 | 0.88 |
| S2 | 19 | 8.88 | 3.72 | |||
| S3 | 12 | 9.03 | 3.41 | |||
| S4 | 52 | 9.15 | 2.59 | |||
| T stage | T1 | 45 | 8.75 | 3.04 | 1.10 | 0.34 |
| T2 | 44 | 9.65 | 2.87 | |||
| T3 | 9 | 8.84 | 2.69 | |||
| N stage | N0 | 96 | 9.18 | 2.97 | -0.09 | 0.93 |
| N1 | 3 | 9.33 | 2.31 | |||
| HBV | Positive | 86 | 9.02 | 2.96 | -1.44 | 0.15 |
| Negative | 13 | 10.28 | 2.65 | |||
| HCV | Positive | 5 | 6.32 | 3.65 | -2.29 | 0.02 |
| Negative | 94 | 9.34 | 2.84 | |||
| AFP | > 400 ng/mL | 41 | 9.35 | 2.91 | 0.45 | 0.65 |
| < 400 ng/mL | 58 | 9.08 | 2.98 | |||
| BCLC staging | A | 81 | 9.27 | 2.93 | 0.15 | 0.86 |
| B | 13 | 8.80 | 3.04 | |||
| C | 3 | 9.33 | 2.31 | |||
Currently, HCC patients have not only a large base number but also a very poor prognosis. HCC accounts for 90% of liver cancer cases[16]. In 2022, the numbers of new cases of and deaths from liver cancer worldwide were 865269 and 757948, respectively, accounting for 4.3% and 7.8% of the total number of new cancer cases and total tumour-related deaths, respectively[17]. The five-year survival rate of HCC is less than 20%[16,18]. This situation is closely related to the ongoing challenges in achieving an early diagnosis of HCC[18]. Therefore, further research on molecular biomarkers related to HCC remains of great significance. At present, no studies have reported the abnormal expression pattern, molecular biological behaviour, and clinical pathological significance of PSMD6 in HCC. Consequently, this study integrated mul
Immunohistochemistry can intuitively evaluate the expression and spatial distribution characteristics of target proteins at the histological level, which sheds light on the potential role of target proteins in the occurrence and development of tumours[19]. In this study, a total of 383 pairs of HCC tissues and corresponding paracancerous tissues was collected from institutions A and B for in-house immunohistochemistry experiments, and the results showed that PSMD6 was significantly overexpressed in HCC tissues compared with paracancerous tissues. Given that results based on single-centre datasets can lack reliability, this study also collected 8,339 Liver tissue mRNA expression profiles from multiple external dataset libraries. This study then integrated and analysed these 41 external datasets and two internal datasets, and it was concluded that PSMD6 has an overexpression pattern in HCC tissues. Therefore, the results of this study, which was based on the design of a large external multicentre sample and internal sample experiments, have strong generalisability and reliability. In addition, the results of the CRISPR knockout screen technology analysis showed that the growth of HCC cells is dependent on PSMD6. Therefore, abnormal PSMD6 may play a certain role in the occurrence and development of HCC.
This study therefore conducted functional enrichment analysis to further explore the potential molecular biological behaviour of PSMD6 in HCC tissues. The above results show that the overexpression pattern of PSMD6 in HCC involves abnormal pathways, such as ribosome biogenesis and RNA splicing. Although this study is the first to reveal the con
In addition, given the clinical potential of the overexpression pattern of PSMD6 in HCC tissues, this study evaluated the drug sensitivity and clinical pathological significance of targeted therapeutic drugs. In terms of drug sensitivity, ML323, sepantronium bromide, GDC0810, BPD-00008900, and bortezomib have great potential anti-HCC effects. Previous studies have shown that bortezomib can inhibit the 26S proteasome in which PSMD6 protein participates and thus reduce the growth of HCC cells[27], confirming PSMD6 as a potential therapeutic target. In-vivo experiments have shown that ML323 can exert its anti-HCC function by inhibiting USP1[28], and targeting PSMD6 may increase the sensitivity of HCC cells to ML323 and similar drugs. Sepantronium bromide (YM155) can inhibit the proliferation of HCC cells by acting on genes related to cell cycle checkpoints[29]. HCC with a high m5C score is more sensitive to the effects of sepantronium bromide[30]. The latest study shows that GDC0810 has a better therapeutic effect on HCC with high-risk disulfidptosis-related prognostic model scores[31]. Other studies have shown that BPD-00008900 and ML323 have better therapeutic effects on HCC patients with high cholesterol metabolism[32], further illustrating the complex interaction between metabolic pathways and drug sensitivity. In this study, we revealed a negative correlation between the sen
Although this study makes many new findings, the following limitations are worth noting. First, the clinical in
This study used the mRNA expression profile from external multicentre and inhouse immunohistochemistry experiments and found for the first time that PSMD6 has an overexpression pattern in HCC tissues. PSMD6 is very necessary for the growth of HCC cells. This growth necessity may be related to pathways such as ribosome biogenesis and RNA splicing. They hold promise for improving the prognosis of HCC through the targeting of PSMD6.
We thank the Guangxi Zhuang Autonomous Region Clinical Medicine Research Center for Molecular Pathology and Intelligent Pathology Precision Diagnosis for providing technical support in computational pathology and experimental pathology.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68770] [Article Influence: 13754.0] [Reference Citation Analysis (202)] |
| 2. | Maomao C, He L, Dianqin S, Siyi H, Xinxin Y, Fan Y, Shaoli Z, Changfa X, Lin L, Ji P, Wanqing C. Current cancer burden in China: epidemiology, etiology, and prevention. Cancer Biol Med. 2022;19:1121-1138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 202] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 3. | Tourkochristou E, Kalafateli M, Triantos C, Aggeletopoulou I. Evaluation of PAGE-B Score for Hepatocellular Carcinoma Development in Chronic Hepatitis B Patients: Reliability, Validity, and Responsiveness. Biomedicines. 2024;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 4. | Mezzacappa C, Kim NJ, Vutien P, Kaplan DE, Ioannou GN, Taddei TH. Screening for Hepatocellular Carcinoma and Survival in Patients With Cirrhosis After Hepatitis C Virus Cure. JAMA Netw Open. 2024;7:e2420963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 5. | Szilveszter RM, Muntean M, Florea A. Molecular Mechanisms in Tumorigenesis of Hepatocellular Carcinoma and in Target Treatments-An Overview. Biomolecules. 2024;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 6. | Chakraborty E, Sarkar D. Emerging Therapies for Hepatocellular Carcinoma (HCC). Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 240] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 7. | Ceballos MP, Quiroga AD, Palma NF. Role of sirtuins in hepatocellular carcinoma progression and multidrug resistance: Mechanistical and pharmacological perspectives. Biochem Pharmacol. 2023;212:115573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 8. | Wang Z, Qin H, Liu S, Sheng J, Zhang X. Precision diagnosis of hepatocellular carcinoma. Chin Med J (Engl). 2023;136:1155-1165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 49] [Reference Citation Analysis (0)] |
| 9. | Zhou C, Li H, Han X, Pang H, Wu M, Tang Y, Luo X. Prognostic Value and Molecular Mechanisms of Proteasome 26S Subunit, Non-ATPase Family Genes for Pancreatic Ductal Adenocarcinoma Patients after Pancreaticoduodenectomy. J Invest Surg. 2022;35:330-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 10. | Li L, Cheng H, Gong L, Huang Y, Yang J, Yan Q, Dai S, Wang J. Cuproptosis/OXPHOS tendency prediction of prognosis and immune microenvironment of esophageal squamous cell carcinoma: Bioinformatics analysis and experimental validation. Gene. 2024;902:148156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 11. | Zhang JY, Shi KZ, Liao XY, Li SJ, Bao D, Qian Y, Li DJ. The Silence of PSMC6 Inhibits Cell Growth and Metastasis in Lung Adenocarcinoma. Biomed Res Int. 2021;2021:9922185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Lin Y, Zhong W, Lin Q, Ye Y, Li S, Chen H, Liu H, Xu L, Zhuang W, Chen S, Lin Y, Wang Q. SFPQ promotes the proliferation, migration and invasion of hepatocellular carcinoma cells and is associated with poor prognosis. Am J Cancer Res. 2023;13:2269-2284. [PubMed] |
| 13. | Dempster JM, Boyle I, Vazquez F, Root DE, Boehm JS, Hahn WC, Tsherniak A, McFarland JM. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol. 2021;22:343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 262] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 14. | Du Z, Zhang Z, Han X, Xie H, Yan W, Tian D, Liu M, Rao C. Comprehensive Analysis of Sideroflexin 4 in Hepatocellular Carcinoma by Bioinformatics and Experiments. Int J Med Sci. 2023;20:1300-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 15. | Liu Y, Yang X, Gan J, Chen S, Xiao ZX, Cao Y. CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022;50:W159-W164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 968] [Article Influence: 242.0] [Reference Citation Analysis (0)] |
| 16. | Fite EL, Makary MS. Transarterial Chemoembolization Treatment Paradigms for Hepatocellular Carcinoma. Cancers (Basel). 2024;16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 17. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 12862] [Article Influence: 6431.0] [Reference Citation Analysis (8)] |
| 18. | Zhang S, Tuo P, Ji Y, Huang Z, Xiong Z, Li H, Ruan C. Identification of 1-Methylnicotinamide as a specific biomarker for the progression of cirrhosis to hepatocellular carcinoma. J Cancer Res Clin Oncol. 2024;150:310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 19. | Qin Y, Liu Y, Xiang X, Long X, Chen Z, Huang X, Yang J, Li W. Cuproptosis correlates with immunosuppressive tumor microenvironment based on pan-cancer multiomics and single-cell sequencing analysis. Mol Cancer. 2023;22:59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 110] [Reference Citation Analysis (1)] |
| 20. | Yang XM, Wang XQ, Hu LP, Feng MX, Zhou YQ, Li DX, Li J, Miao XC, Zhang YL, Yao LL, Nie HZ, Huang S, Xia Q, Zhang XL, Jiang SH, Zhang ZG. Nucleolar HEAT Repeat Containing 1 Up-regulated by the Mechanistic Target of Rapamycin Complex 1 Signaling Promotes Hepatocellular Carcinoma Growth by Dominating Ribosome Biogenesis and Proteome Homeostasis. Gastroenterology. 2023;165:629-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 21. | Xue M, Dong L, Zhang H, Li Y, Qiu K, Zhao Z, Gao M, Han L, Chan AKN, Li W, Leung K, Wang K, Pokharel SP, Qing Y, Liu W, Wang X, Ren L, Bi H, Yang L, Shen C, Chen Z, Melstrom L, Li H, Timchenko N, Deng X, Huang W, Rosen ST, Tian J, Xu L, Diao J, Chen CW, Chen J, Shen B, Chen H, Su R. METTL16 promotes liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation. J Hematol Oncol. 2024;17:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 56] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 22. | Chen MY, Hsu CH, Setiawan SA, Tzeng DTW, Ma HP, Ong JR, Chu YC, Hsieh MS, Wu ATH, Tzeng YM, Yeh CT. Ovatodiolide and antrocin synergistically inhibit the stemness and metastatic potential of hepatocellular carcinoma via impairing ribosome biogenesis and modulating ERK/Akt-mTOR signaling axis. Phytomedicine. 2023;108:154478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 23. | Õunap K, Leetsi L, Matsoo M, Kurg R. The Stability of Ribosome Biogenesis Factor WBSCR22 Is Regulated by Interaction with TRMT112 via Ubiquitin-Proteasome Pathway. PLoS One. 2015;10:e0133841. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Yan Q, Fang X, Liu X, Guo S, Chen S, Luo M, Lan P, Guan XY. Loss of ESRP2 Activates TAK1-MAPK Signaling through the Fetal RNA-Splicing Program to Promote Hepatocellular Carcinoma Progression. Adv Sci (Weinh). 2024;11:e2305653. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 25. | Zhan YT, Li L, Zeng TT, Zhou NN, Guan XY, Li Y. SNRPB-mediated RNA splicing drives tumor cell proliferation and stemness in hepatocellular carcinoma. Aging (Albany NY). 2020;13:537-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 26. | Ka HI, Lee S, Han S, Jeong AL, Park JY, Joo HJ, Soh SJ, Park D, Yang Y. Deubiquitinase USP47-stabilized splicing factor IK regulates the splicing of ATM pre-mRNA. Cell Death Discov. 2020;6:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Baiz D, Dapas B, Farra R, Scaggiante B, Pozzato G, Zanconati F, Fiotti N, Consoloni L, Chiaretti S, Grassi G. Bortezomib effect on E2F and cyclin family members in human hepatocellular carcinoma cell lines. World J Gastroenterol. 2014;20:795-803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 30] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Liu Y, Kong WY, Yu CF, Shao ZL, Lei QC, Deng YF, Cai GX, Zhuang XF, Sun WS, Wu SG, Wang R, Chen X, Chen GX, Huang HB, Liao YN. SNS-023 sensitizes hepatocellular carcinoma to sorafenib by inducing degradation of cancer drivers SIX1 and RPS16. Acta Pharmacol Sin. 2023;44:853-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 29. | Xia H, Chen J, Shi M, Deivasigamani A, Ooi LL, Hui KM. The over-expression of survivin enhances the chemotherapeutic efficacy of YM155 in human hepatocellular carcinoma. Oncotarget. 2015;6:5990-6000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Liu P, Zhu Z, Ma J, Wei L, Han Y, Shen E, Tan X, Chen Y, Cai C, Guo C, Peng Y, Gao Y, Liu Y, Huang Q, Gao L, Li Y, Jiang Z, Wu W, Liu Y, Zeng S, Li W, Feng Z, Shen H. Prognostic stratification based on m(5)C regulators acts as a novel biomarker for immunotherapy in hepatocellular carcinoma. Front Immunol. 2022;13:951529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 31. | Wang H, Wang W. Unlocking the future of hepatocellular carcinoma treatment: A comprehensive analysis of disulfidptosis-related lncRNAs for prognosis and drug screening. Open Med (Wars). 2024;19:20240919. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 32. | Bai W. The combined characteristics of cholesterol metabolism and the immune microenvironment may serve as valuable biomarkers for both the prognosis and treatment of hepatocellular carcinoma. Heliyon. 2023;9:e22885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/