Published online Nov 24, 2025. doi: 10.5306/wjco.v16.i11.111983
Revised: August 13, 2025
Accepted: September 30, 2025
Published online: November 24, 2025
Processing time: 128 Days and 23.4 Hours
Although chronic-phase chronic myeloid leukemia (CP-CML) is treatable and nearly curable in about 50% of patients, accelerated-phase chronic myeloid leukemia (AP-CML) shows concerning drug resistance, while blast crisis chronic myeloid leukemia (BC-CML) is highly lethal. Advances in whole exome sequencing (WES) reveal pan-cancer mutations in BC-CML, supporting mutation-guided therapies beyond Breakpoint cluster region-Abelson. Artificial intelligence (AI) and machine learning (ML) enable genomic stratification and drug repur
To stratify BC-CML into molecular subtypes using WES, ML, and AI for precision drug repurposing.
Included 123 CML patients (111 CP-CML, 5 AP-CML, 7 BC-CML). WES identified pan-cancer mutations. Variants annotated via Ensembl Variant Effect Predictor and Catalogue of Somatic Mutations in Cancer (COSMIC). ML (principal component analysis, K-means) stratified BC-CML. COSMIC signatures and PanDrugs prioritized drugs. Analysis of variance/Kruskal-Wallis validated differences (P < 0.05).
In this exploratory, hypothesis-generating study of BC-CML patients (n = 7), we detected over 2500 somatic mutations. ML identified three BC-CML clusters: (1) Cluster 1 [breast cancer susceptibility gene 2 (BRCA2), TP53]; (2) Cluster 2 [isocitrate dehydrogenase (IDH) 1/2, ten-eleven translocation 2]; and (3) Cluster 3 [Janus kinase (JAK) 2, colony-stimulating factor 3 receptor], with distinct COSMIC signatures. Therapies: (1) Polyadenosine-diphosphate-ribose polymerase inhibitors (olaparib); (2) IDH inhibitors (ivosidenib); and (3) JAK inhibitors (ruxolitinib). Mutational burden, signatures, and targets varied significantly across clusters, supporting precision stratification.
This WES-AI-ML framework provides mutation-guided therapies for BC-CML, enabling real-time stratification and Food and Drug Administration-approved drug repurposing. While this exploratory study is limited by its small sample size (n = 7), it establishes a methodological framework for precision oncology stratification that warrants validation in larger, multi-center cohorts.
Core Tip: This study integrates whole-exome sequencing, machine learning, and artificial intelligence-driven drug repurposing to stratify blast crisis chronic myeloid leukemia (BC-CML) into three molecular subtypes based on pan-cancer mutations and Catalogue of Somatic Mutations in Cancer signatures. By identifying cluster-specific therapies (e.g., polyadenosine-diphosphate-ribose polymerase, isocitrate dehydrogenase, and Janus kinase inhibitors), the framework enables precision oncology for BC-CML, offering a scalable model for real-time patient stratification and Food and Drug Administration/European Medicines Agency-approved drug repurposing in re-lapsed/refractory hematologic malignancies.
- Citation: AlGarni A, Alanazi N, AlMukhaylid S, Alqahtani S, Almasoudi H, Samir Taleb Y, Alkhamis N, Shaheen S, Haji Siyal A, Aleem A, Naeem R, Shammas MA, Saglio G, Alroweilly D, Hussain A, Iqbal Z. Omics and artificial intelligence integration for stratifying blast crisis CML using COSMIC signatures and pan-cancer precision drug repurposing. World J Clin Oncol 2025; 16(11): 111983
- URL: https://www.wjgnet.com/2218-4333/full/v16/i11/111983.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i11.111983
Blast crisis chronic myeloid leukemia (BC-CML) represents a terminal transformation of CML that remains challenging to treat, even with modern tyrosine kinase inhibitors (TKIs)[1]. Despite breakpoint cluster region-Abelson (BCR-ABL1) suppression, many patients eventually relapse with aggressive disease due to additional oncogenic mutations[2]. This underscores the need for mutation-guided, non-TKI therapeutic strategies that consider the broader genomic landscape. Advances in whole exome sequencing (WES) and next-generation sequencing technologies have revealed that BC-CML accumulates pan-cancer mutations affecting genes such as TP53, breast cancer susceptibility gene (BRCA) 1/2, epidermal growth factor receptor (EGFR), isocitrate dehydrogenase (IDH) 1, and regulatory associated protein of mechanistic target of rapamycin complex 1 (RPTOR), among others[3]. These observations support a model of disease progression whereby chronic myeloid leukemia (CML) increasingly adopts genetic features of solid tumors and high-grade myeloid mali
Artificial intelligence (AI) and machine learning (ML) have emerged as powerful tools for leveraging large-scale genomic data to guide drug repositioning in cancer[5]. ML models such as unsupervised clustering and principal co
Recent initiatives by regulatory bodies such as the United States Food and Drug Administration (FDA) have further validated the use of AI for mutation-based therapy discovery in orphan diseases and refractory malignancies (United States, 2024). Clinical adoption is also accelerating through integration with electronic health records and real-time sequencing data[10]. The potential for cross-indication therapeutic discovery was highlighted in our earlier study by Iqbal et al[11], which systematically mapped myeloid/Lymphoid gene mutations in advanced CML to repurposable drugs using computational drug discovery tools. That work laid the foundation for understanding how somatic mutation profiles could shape individualized therapy in BC-CML. More recently, Alanazi et al[12] presented evidence at American Association for Cancer Research that pan-cancer genes contribute to CML progression and transformation through diverse oncogenic mechanisms, including transcriptional deregulation, immune evasion, and chromatin remodeling.
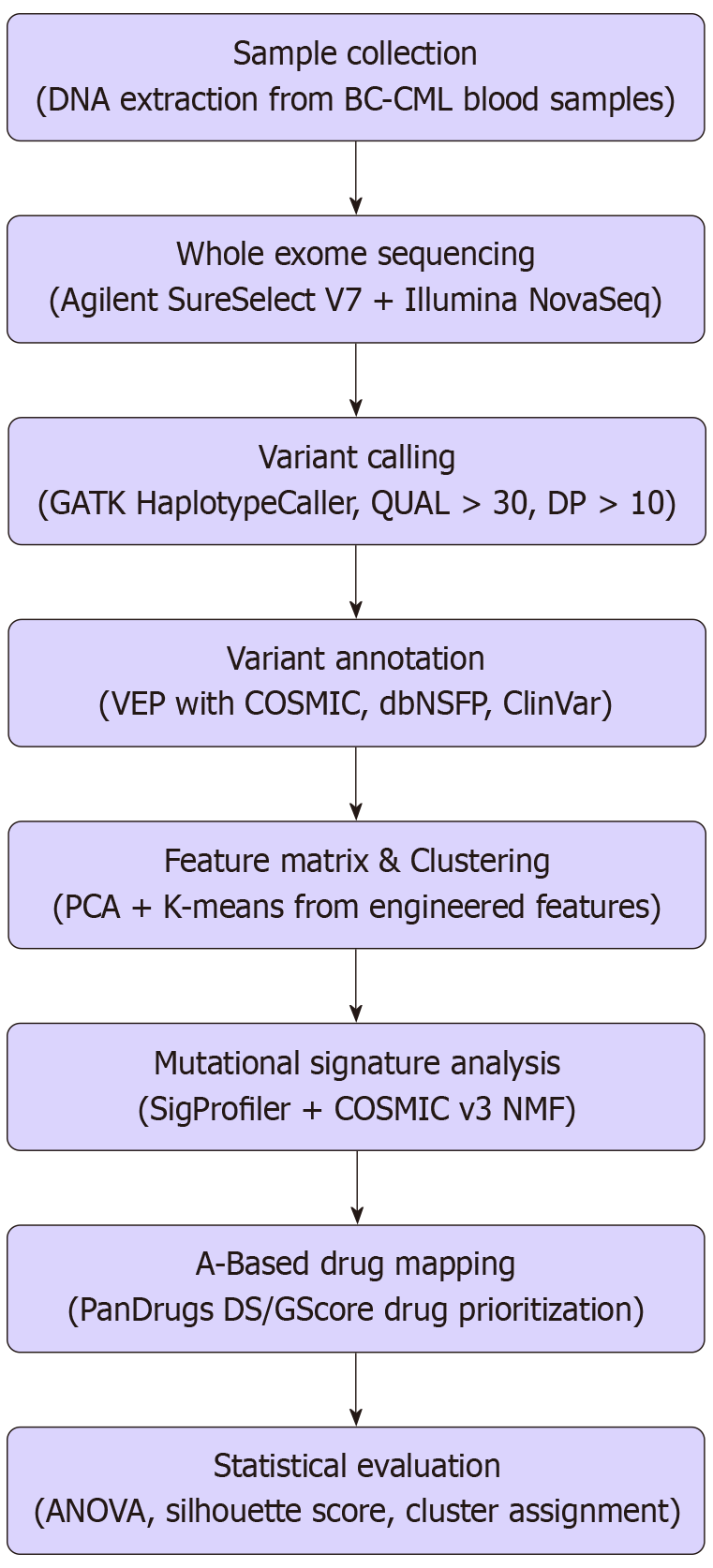
This current manuscript represents a direct extension of those studies. Here, we present an integrated pipeline combining WES, AI-based drug repurposing, and unsupervised ML clustering to identify therapeutic candidates for BC-CML patients using real genomic data (Figure 1). By evaluating mutation frequency, pathway involvement, and clinical drug accessibility, we aim to redefine the therapeutic roadmap for this lethal phase of CML, with implications extending to other relapsed/refractory and rare diseases.
This study employed an exploratory, hypothesis-generating design to investigate the feasibility of integrating omics and AI approaches for BC-CML stratification. Given the rarity of BC-CML cases and the pilot nature of this integrated pipeline, we analyzed genomic data from seven BC-CML patients. While this sample size limits statistical power and generalizability, it aligns with established frameworks for exploratory research in rare hematologic malignancies. The primary objective was not to provide definitive clinical conclusions, but rather to generate testable hypotheses regarding molecular subtypes and therapeutic targets that can guide future confirmatory studies. This approach follows FDA guidance supporting AI-assisted biomarker discovery in early-phase oncology research, particularly when integrated with high-confidence evidence platforms such as Catalogue of Somatic Mutations in Cancer (COSMIC) signatures and curated drug databases.
Clinical data and peripheral blood samples were obtained from twelve clinically diagnosed advanced phase CML, with 5 accelerated-phase CML (AP-CML) and 7 BC-CML patients (experimental group) and 123 chronic phase CML patients (control group) attending Hayatabad Medical Complex Peshawar Pakistan, following institutional ethics committee approval[11].
Mononuclear cells were isolated using Ficoll-Paque Plus density centrifugation. Genomic DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen), eluted in Tris-EDTA buffer, and quantified using Qubit fluorimeter, per manufacturer’s instructions[11]. DNA quality and integrity were assessed by agarose gel electrophoresis.
WES libraries were prepared using the Agilent SureSelect Human All Exon V7 kit. Sequencing was performed on an Illumina NovaSeq platform, producing paired-end 150 bp reads at an average depth of 100 ×. Quality control of raw reads was assessed using FastQC, and adapters were removed using Trimmomatic. Reads were aligned to the GRCh38 reference genome using BWA-MEM[13]. Polymerase chain reaction duplicates were removed using Picard tools. Variant calling was conducted using Genome Analysis Toolkit HaplotypeCaller with base recalibration and joint genotyping[14]. Variants were filtered to retain those with quality (QUAL > 30) and read depth (DP > 10). High-confidence variants were annotated using Ensembl Variant Effect Predictor (VEP) with integrated COSMIC, ClinVar, dbNSFP, SIFT, and PolyPhen annotations[14]. Mutations were categorized as missense, nonsense, frameshift, or splice site. Population allele fre
Mutational features including gene counts per chromosome, mutation types, and indel lengths were compiled into a matrix. Standardization was performed using StandardScaler from scikit-learn. PCA was applied for dimensionality reduction, and K-means clustering (k = 3) was conducted with optimal k determined by silhouette and elbow methods[15]. The workflow begins with patient recruitment and sample collection, followed by WES and variant annotation. Pan-cancer gene mutations were filtered, annotated, and subjected to AI-based drug mapping using PanDrugs. In parallel, mutation features were extracted from variant call format (VCF) files for ML analysis using PCA and K-means clustering. Identified mutation clusters and drug-target associations were evaluated to inform drug-repurposing strategies for BC-CML.
Single nucleotide variants were analyzed using SigProfilerExtractor to identify mutational signatures[16]. Extracted signatures were matched against COSMIC v3 reference profiles, and only those achieving a cosine similarity ≥ 0.85 were retained. Each sample was then assigned its predominant signature(s), and these dominant exposures were summarized within the three identified molecular clusters.
Druggable gene products were identified using PanDrugs.org, which integrates drug-target data from DrugBank, PubChem, and clinical evidence[8]. PanDrugs computes two key metrics: (1) DrugScore (DS), reflecting the strength of evidence linking a drug to the molecular target; and (2) GeneScore (GScore), indicating the target’s pathogenic relevance in the mutation profile. We prioritized drugs with DS ≥ 0.7 and GScore ≥ 0.5, focusing exclusively on FDA/European Medicines Agency (EMA) – approved or late-stage investigational agents. To mitigate potential biases from cluster fragility, we cross-checked top pan-drug predictions against the OncoKB knowledgebase to confirm clinical relevance of suggested therapies.
Cluster assignment was performed by minimizing Euclidean distance from each sample to the centroids determined through K-means clustering[15,17,18]. Statistical analyses included PCA coordinate extraction and silhouette scoring to validate cluster separation[19,20]. Analysis of variance (ANOVA) was used to compare variant types, counts, and indel sizes between clusters. COSMIC signature contributions were compared using Kruskal-Wallis tests, followed by Dunn’s post-hoc tests where applicable[21,22].
Our exploratory analysis of seven BC-CML patients revealed preliminary evidence of three biologically distinct molecular clusters, each characterized by specific mutational signatures and potential therapeutic vulnerabilities. While these findings require validation in larger cohorts, they demonstrate the feasibility of our integrated omics-AI framework for precision oncology applications.
A total of 141 CML patients were evaluated, including 123 in chronic phase (chronic-phase CML) and 18 in accelerated phase (AP-CML), out of which 12 (66.7%) progressed to blast crisis (BC-CML). The mean age was 36.4 ± 11.6 years (range: 9-67 years), with a male-to-female ratio of 1.6:1. Splenomegaly was noted in 83% of AP-CML and 100% of BC-CML cases (P = 0.0134), and hepatomegaly in advanced phases (P = 0.0014). Mortality was 8.5%, with 75% of deaths in BC-CML. Median survival in BC-CML was less than one year despite advanced therapies. These findings are consistent with prior clinical trends in high-risk hematologic transformations[9].
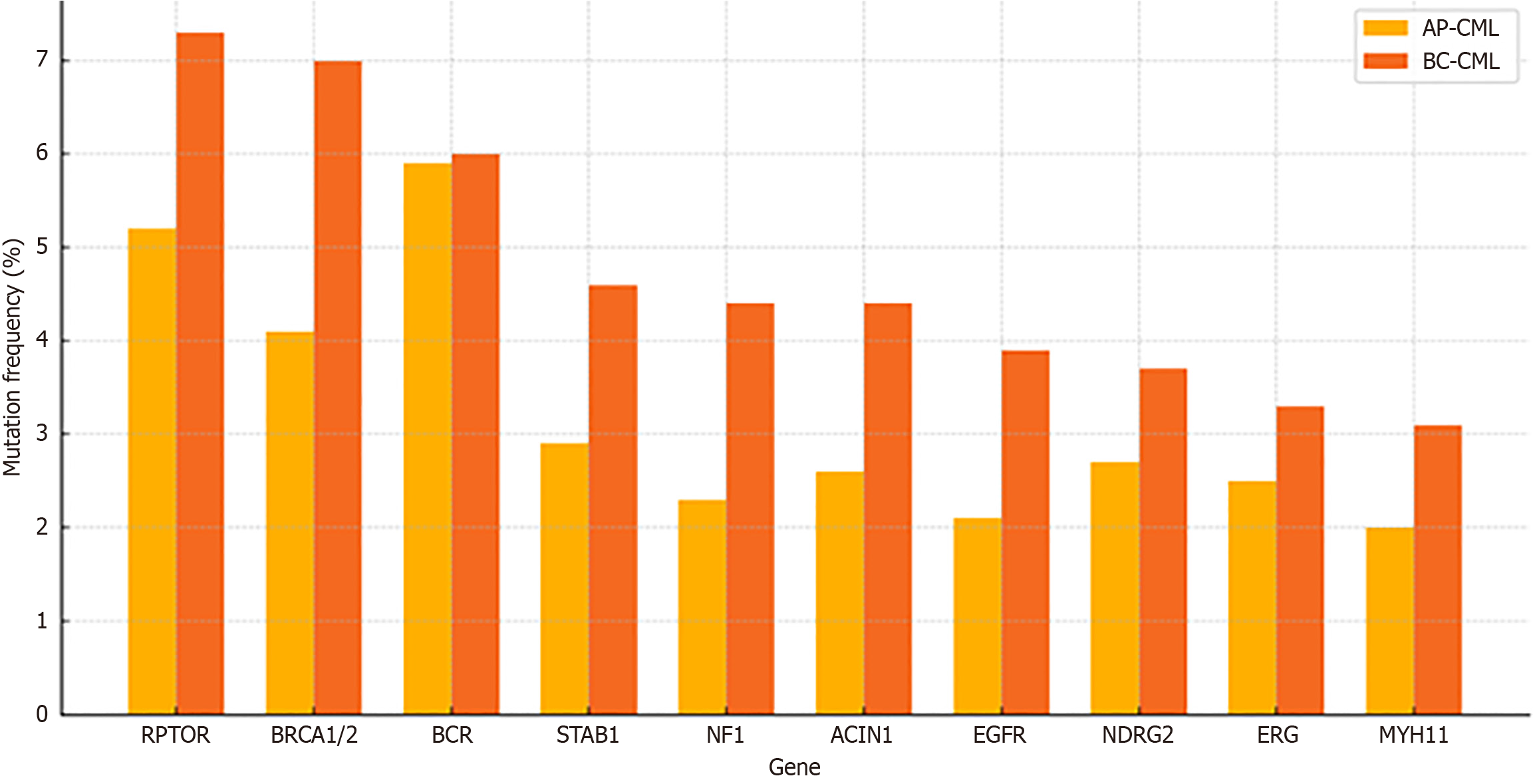
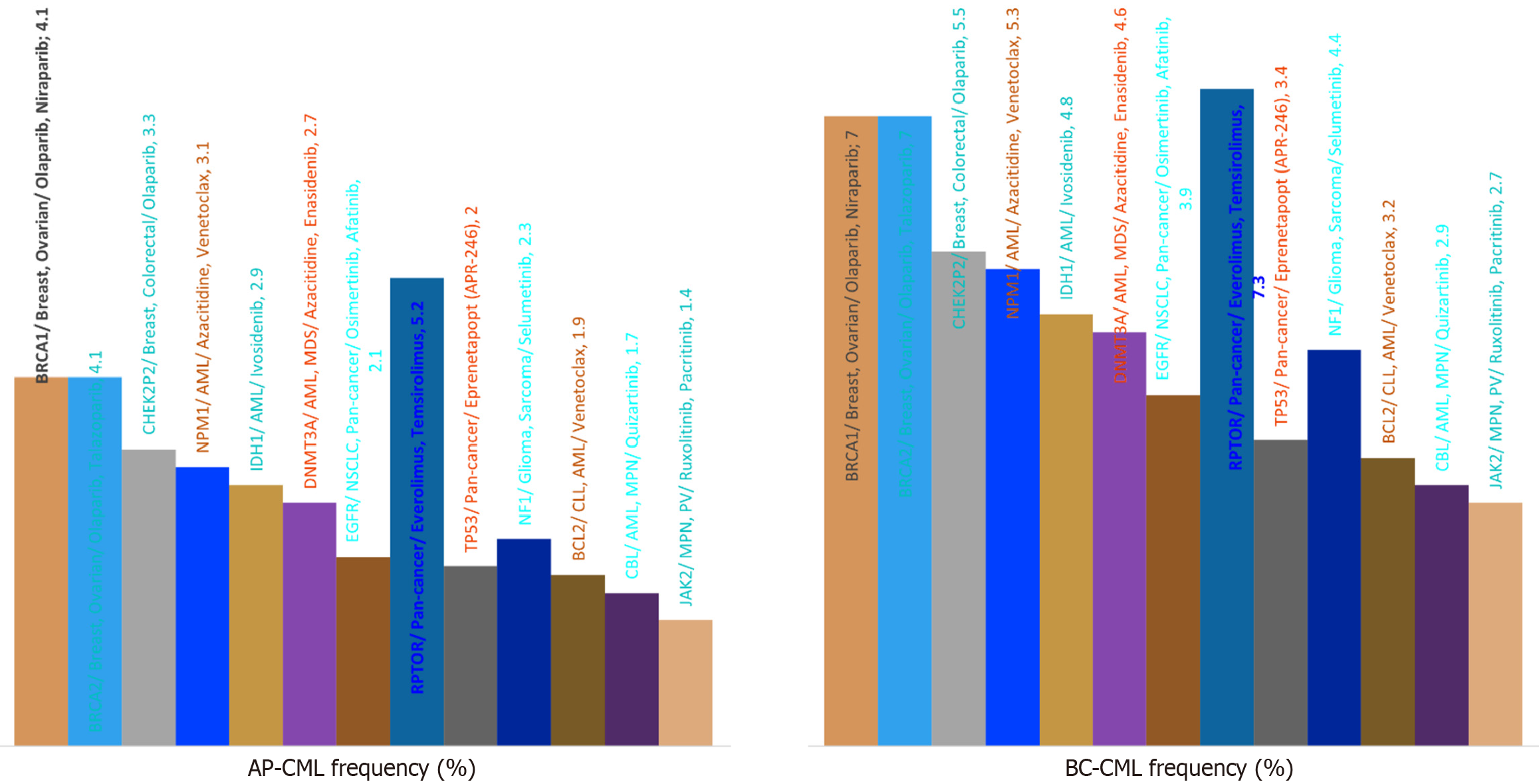
Whole-exome sequencing revealed over 2500 somatic mutations, with BC-CML samples showing a 54% higher mutational load than AP-CML (P < 0.000001). Comparative frequencies of the most commonly mutated genes in AP-CML and BC-CML included RPTOR, BRCA1/2, breakpoint cluster region, Stabilin 1, neurofibromatosis type 1, EGFR, and N-myc downstream-regulated gene 2 (Figure 2). These distributions aligned with known mutation accumulation patterns in blast-phase leukemias[3].
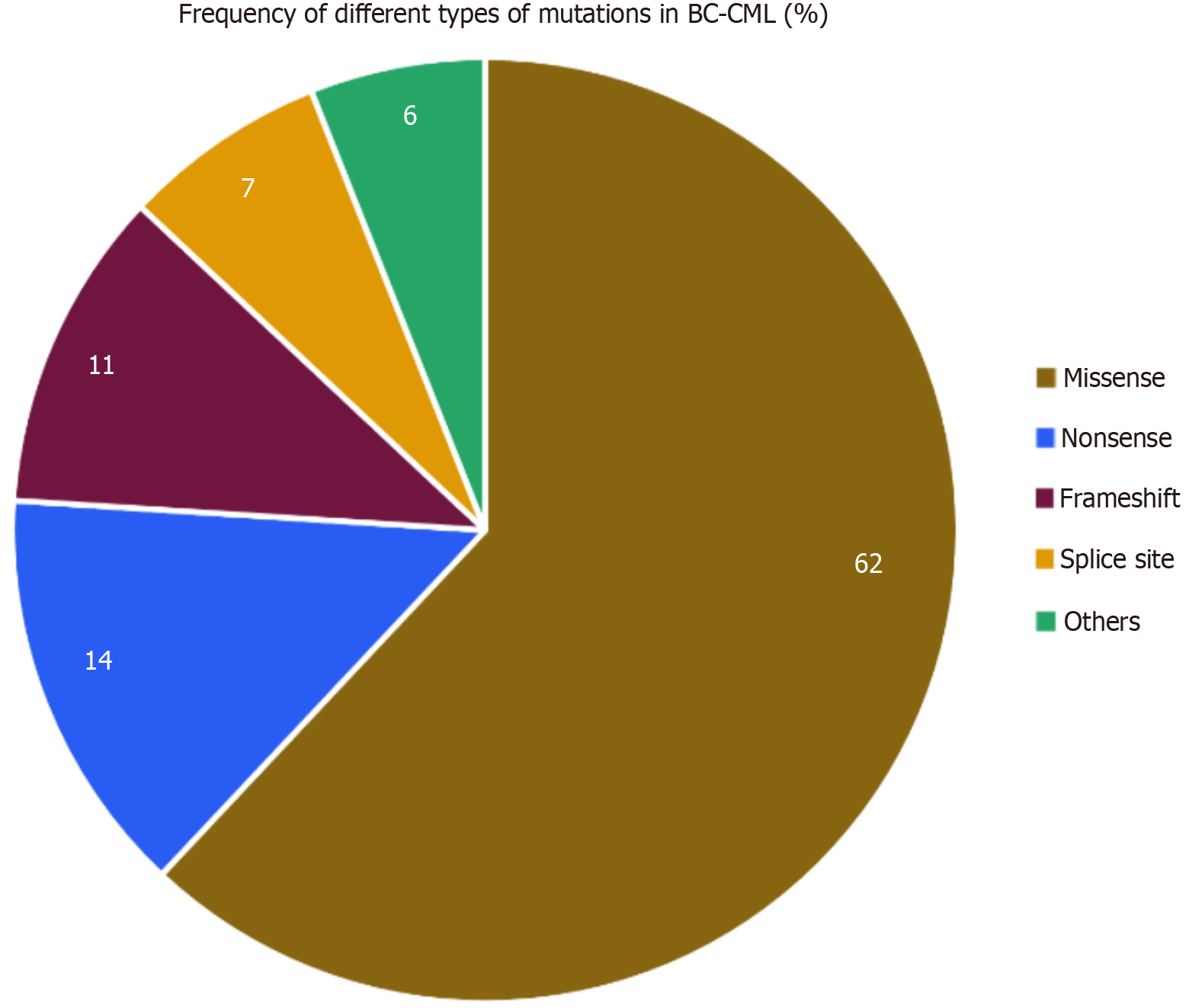
Comprehensive WES of 12 BC-CML samples resulted in the detection of 2963 high-confidence somatic variants. Variants were filtered using stringent quality criteria (QUAL > 30, DP > 10) and annotated using Ensembl VEP. The majority of variants were missense mutations, constituting approximately 62% of the dataset, followed by nonsense mutations (14%), frameshifts (11%), splice-site variants (7%), and others (6%). These distributions are illustrated in Table 1 and Figure 3[23]. The predominance of missense mutations suggests the presence of a broad range of protein function-modulating alte
| Cluster | Enriched genes | Pathogenic mechanism | Sample count |
| Cluster 1 | BRCA2, TP53, EGFR | Genomic instability | 2 |
| Cluster 2 | TET2, DNMT3A, IDH1/2 | Epigenetic deregulation | 2 |
| Cluster 3 | JAK2, CBL, CSF3R | Cytokine signaling | 3 |
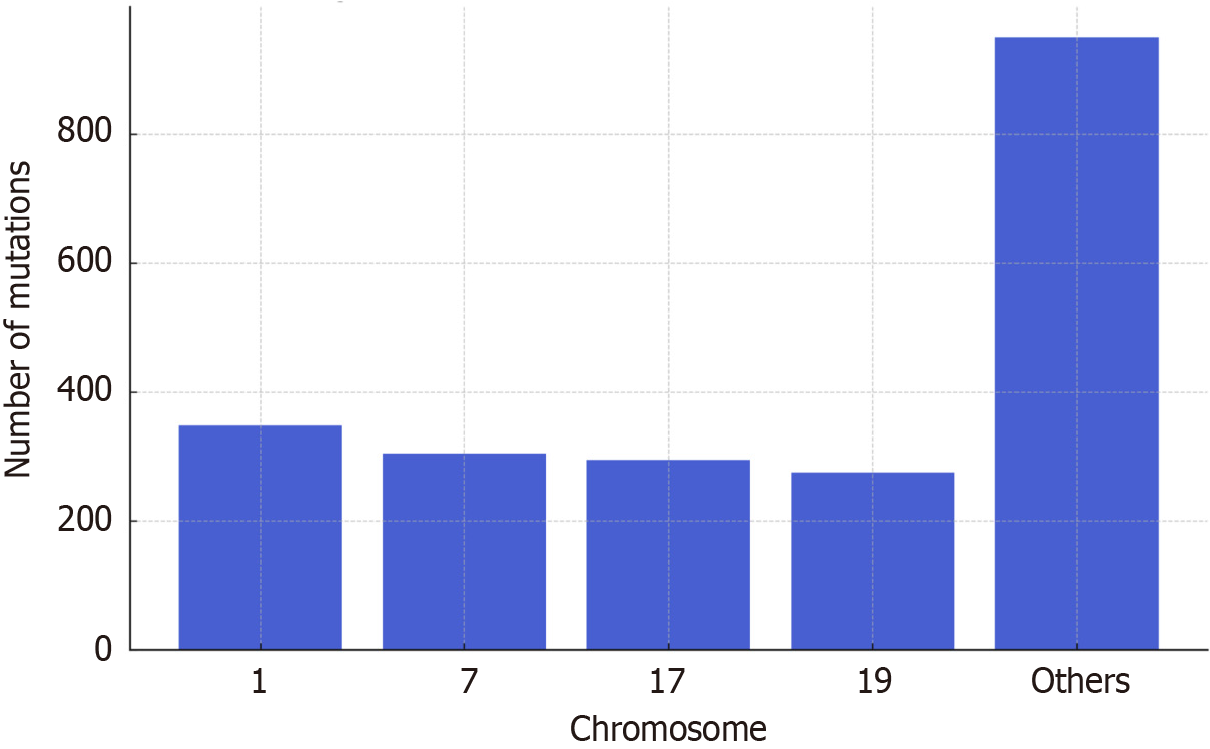
To evaluate the chromosomal distribution of variants, mutations were mapped across the genome. The majority of mutations clustered on chromosomes 1, 7, 17, and 19, highlighting these regions as potential hotspots in BC-CML pro
Unsupervised learning was applied to the filtered and annotated mutation data. Features including variant class, gene location, mutation impact, and allele length were used to construct a sample-wise feature matrix. PCA revealed distinct separation among samples. Subsequent K-means clustering (k = 3), guided by silhouette scoring, segregated samples into three biologically distinct clusters. Cluster 1 (n = 2) included samples enriched in BRCA2, TP53, and EGFR mutations, associated with DNA damage response and homologous recombination deficiency. Cluster 2 (n = 2) showed dominant mutations in epigenetic regulators and metabolic enzymes such as ten-eleven translocation 2 (TET2), DNA (cytosine-5)-methyltransferase 3 A (DNMT3A), IDH1, and IDH2. Cluster 3 (n = 3) was characterized by mutations in Janus kinase (JAK) 2, colony-stimulating factor (CSF) 3 receptor, and casitas B-cell lineage (CBL) – genes linked to cytokine signaling pathways. The cluster-specific characteristics are summarized in Table 1.
To further refine the biological relevance of each cluster, COSMIC mutational signature analysis was performed using SigProfilerExtractor. Distinct profiles emerged across the three clusters, emphasizing the unique mutagenic processes involved in blast crisis transformation. Cluster 1 showed high contribution of signature 3, indicative of BRCA-related homologous recombination deficiency, alongside signature 5, a flat signature associated with aging. Cluster 2 was enriched for signature 1, linked to spontaneous deamination of 5-methylcytosine, and signature 2, reflecting apolipoprotein B mRNA editing enzyme catalytic polypeptide (APOBEC) cytidine deaminase activity. Cluster 3 displayed signature 13 (replication slippage and polymerase error) and signature 18, a marker of reactive oxygen species (ROS)-driven DNA damage. These patterns highlight diverse DNA damage and repair contexts underpinning the three su
| Cluster | Dominant Catalogue of Somatic Mutations in Cancer signatures |
| Cluster 1 | Signature 3 (breast cancer susceptibility gene-deficiency), signature 5 (aging) |
| Cluster 2 | Signature 1 (5-methylcytosine deamination), signature 2 (apolipoprotein B mRNA editing enzyme catalytic polypeptide) |
| Cluster 3 | Signature 13 (polymerase error), signature 18 (reactive oxygen species-related) |
PanDrugs.org was used to identify actionable therapeutic candidates based on the mutational profiles of each cluster. Cluster 1 mutations in BRCA2 and TP53 were matched with PARP inhibitors (e.g., olaparib), EGFR inhibitors (e.g., erlotinib), and murine double minute 2 (MDM2) antagonists (e.g., nutlin-3), indicating sensitivity to agents targeting DNA repair and checkpoint response. Cluster 2, with dominant mutations in IDH1/2 and TET2, aligned with IDH inhibitors (e.g., ivosidenib) and hypomethylating agents (e.g., azacitidine, decitabine). Cluster 3, defined by JAK2 and CSF3R mutations, showed high prioritization scores for JAK inhibitors (e.g., ruxolitinib), CSF1R inhibitors, and agents targeting ROS. Table 3 summarizes these findings. Given the exploratory nature and small cohort size, the drug repurposing findings should be interpreted with caution. Although PanDrugs.org integrates high-quality drug–target relationships from DrugBank, PubChem, and curated clinical evidence platforms, the prioritization of agents (e.g., olaparib, ivosidenib, ruxolitinib) depends on cluster assignments that may be unstable. These results therefore represent preliminary, hy
| Cluster | Targeted therapies |
| Cluster 1 | Olaparib, erlotinib, nutlin-3 |
| Cluster 2 | Ivosidenib, azacitidine, decitabine |
| Cluster 3 | Ruxolitinib, colony-stimulating factor 1 receptor inhibitors, N-acetylcysteine |
ANOVA confirmed significant differences across clusters for key features, including average indel length, missense mutation frequency, and variant load per chromosome (P < 0.05). Kruskal-Wallis testing of mutational signature scores across clusters also yielded statistically significant differences (P < 0.05). These findings reinforce the stratification generated through unsupervised learning and validate the biological basis for the three-cluster model in BC-CML.
To visually reinforce this cluster-drug mapping strategy, a schematic table was constructed (Table 4), which recapitulates the relationships between key genetic features, mutational processes, and therapeutic options in a digestible format.
| Cluster | Key mutations | Catalogue of Somatic Mutations in Cancer signatures | Processes | Drugs |
| Cluster 1 | Breast cancer susceptibility gene 2, TP53 | S3, S5 | Homologous recombination deficiency | Olaparib, nutlin-3 |
| Cluster 2 | Isocitrate dehydrogenase 1/2, ten-eleven translocation 2 | S1, S2 | Methylation shift | Ivosidenib, azacitidine |
| Cluster 3 | Janus kinase 2, colony-stimulating factor 3 receptor | S13, S18 | Inflammation, reactive oxygen species | Ruxolitinib, N-acetylcysteine |
Overall, integrated WES, ML-guided and AI-guided (pandrugs) analyses led to discovery of common pan-cancer mutations in our AP-CML and BC-CML patient cohorts and corresponding drugs already approved and in active clinical practice for treatment of different cancers with these specific mutations (Figure 5).
WES of 12 BC-CML patients identified 2963 high-confidence somatic mutations, with an average of 247 mutations per sample. Missense variants predominated (62%), followed by nonsense (14%) and frameshift (11%) mutations. Ch
Despite significant advances in TKI therapy, BC-CML remains a biologically complex and clinically refractory stage with poor outcomes. Traditional diagnostic and therapeutic approaches fail to stratify patients based on transformation mechanisms[24]. To address this, we employed an integrated platform using WES, COSMIC mutational signatures, and unsupervised ML to define clinically relevant subgroups.
Our ML-based analysis revealed three biologically distinct clusters among BC-CML patients, each with specific mutational loads, variant classes, and signature patterns[25]. These clusters reflect divergent transformation routes and point toward tailored therapy strategies beyond standard TKI resistance profiling.
Cluster 1 (n = 2) exhibited BRCA2, TP53, and EGFR mutations, enriched with COSMIC signatures 3 (homologous recombination deficiency) and 5 (age-associated mutation accumulation). These genomic features are functionally linked to PARP inhibitor sensitivity in hematologic malignancies[26]. Importantly, TP53 loss is a marker of resistance and poor prognosis, but also suggests benefit from MDM2 inhibitors such as nutlin-3 and RG7388, which restore p53 function and are now being evaluated in advanced leukemia[27].
Cluster 2 (n = 2) was driven by IDH1/2, DNMT3A, and TET2 mutations and associated with COSMIC signatures 1 and 2, indicating spontaneous methylcytosine deamination (resulting in C mutations to T mutations) and APOBEC-mediated mutagenesis. APOBEC cytosine deaminases are prominent mutators in cancer, mediating mutations in over 50% of cancers. APOBEC mutagenesis has been linked to tumor heterogeneity, persistent cell evolution, and therapy responses. These events underpin the epigenetic instability seen in advanced-phase myeloid malignancies[28]. IDH inhibitors such as ivosidenib and hypomethylating agents like azacitidine have shown synergy in this context and are currently in clinical use for IDH1-mutated AML[29].
Cluster 3 (n = 3) revealed JAK2, CSF3R, and CBL mutations, co-occurring with signatures 13 and 18 – markers of po
The genomic stratification framework developed in this study enabled not only subtype identification but also drug prioritization through AI-assisted repurposing tools. By integrating COSMIC mutational signatures with PanDrugs scoring, we mapped each molecular clusters to pathway-informed, FDA/EMA-approved agents, bridging preclinical biology with immediate translational options[31]. Rather than focusing on isolated mutations, this model emphasized signature-guided drug sensitivity, which has proven more predictive of therapeutic response in high-grade hematologic malignancies. For example, in cluster 1, PARP inhibitors like olaparib were not only linked to BRCA2 mutations but prioritized for signature 3 exposure, a marker of homologous recombination repair deficiency observed across leukemias[32]. This reflects recent trends in AML trial design, which use functional signatures to define inclusion criteria rather than genotype alone[33].
Similarly, cluster 2's dominant epigenetic mutations and signatures 1/2 matched to therapies such as ivosidenib and azacitidine, already in use for relapsed IDH-mutated AML. These drugs exploit vulnerabilities in DNA methylation and metabolic rewiring, aligning with current epigenetically guided treatment strategies[34]. In cluster 3, PanDrugs high
Our use of PanDrugs reflects a broader shift toward AI-supported oncology tools that integrate mutational burden, pathway activation, and drug-gene networks. Comparable platforms like OncoKB are already being deployed in precision molecular tumor boards to guide off-label access and early-phase leukemia trials[36,37]. This approach demonstrates that ML-stratified COSMIC-based clusters, matched with AI-prioritized therapies, can operationalize signature-guided treatment models across hematologic cancers – paving the way for next-generation clinical guideline integration.
Our integrative stratification model – rooted in ML, mutational signature biology, and AI-based drug repurposing – provides a framework for reshaping clinical decision-making in BC-CML. This approach aligns with recent precision oncology trends emphasizing dynamic, pathway-informed patient classification over static genomic panels[37].
Each of the three ML-defined clusters reflected distinct mechanisms of leukemic transformation, validated by COSMIC signature exposure and supported by drug prioritization via PanDrugs. These clusters are immediately relevant for AI-powered clinical trial design, particularly in rare, aggressive hematologic malignancies where traditional randomized approaches are underpowered[38]. Recent pilot programs in AML have already implemented signature-guided inclusion criteria for PARP inhibitors and metabolic therapies, confirming the feasibility of this strategy[39].
Regulatory bodies are now supporting these initiatives. The United States FDA’s 2024 draft guidance promotes the integration of AI-assisted biomarker models into early-phase oncology trials, particularly when paired with high-confidence evidence such as COSMIC signatures and curated drug-pathway knowledgebases like PanDrugs and OncoKB[40]. Our approach satisfies these principles by directly aligning mutational processes to prioritized therapeutic options in an interpretable, reproducible manner. Furthermore, the pipeline’s modularity enables adaptation across diverse settings: From university-affiliated genomic programs to public cancer hospitals using standardized VCF/WES platforms. In
This workflow also supports a shift toward "N-of-few" or basket-style precision trials – an increasingly accepted strategy in hematologic oncology where genetically cohesive but numerically small subgroups require actionable trials. In the BC-CML context, these clusters offer trial frameworks for testing JAK inhibitors, PARP inhibitors, and hypome
Our results provide a translational blueprint for integrating AI-driven mutational clustering and COSMIC signature stratification into the clinical management of BC-CML. The three-cluster architecture uncovered here offers a practical scaffold for therapeutic decision-making, supporting FDA/EMA-approved agents such as PARP inhibitors, IDH in
Furthermore, this pipeline sets the foundation for future “real-time oncology practice”, where exome data can be directly processed via open-source AI platforms like PanDrugs and OncoKB to stratify patients and suggest targeted interventions[10]. The pipeline’s scalability makes it deployable across tertiary and resource-limited healthcare settings, especially when paired with cloud-based mutational signature analysis[44].
From a research standpoint, our work opens several high-impact avenues. Future clinical trials can adopt this three-cluster model as eligibility criteria, replacing lineage-based labels with mutational signature-defined enrollment[45]. The mutation-signature-treatment model may be adapted to high-risk AML, philadelphia chromosome-like acute lymphoblastic leukemia, and even solid tumors with overlapping epigenetic and cytokine pathways[46]. Incorporating tran
While our ML-based stratification reveals robust biological clusters, several limitations remain. The most significant limitation of this study is the small sample size (n = 7), which substantially reduces statistical power, increases the risk of overfitting, and limits the generalizability of our findings. We acknowledge that with such limited patient numbers, there is considerable risk of chance findings and that the observed clustering patterns may not be reproducible in larger cohorts. The statistical validations performed (ANOVA, Kruskal-Wallis testing) should be interpreted cautiously given the small sample sizes per cluster. The 7-patient cohort (BC-CML) limits statistical power for signature exclusivity and drug-sensitivity generalization[48]. WES alone may miss regulatory and non-coding variants contributing to trans
This exploratory study establishes preliminary evidence for an integrative precision oncology framework in BC-CML, demonstrating the feasibility of combining genomic clustering, COSMIC signatures, and AI-assisted drug repurposing. While our findings are limited by the small sample size (n = 7) and require validation in larger, independent cohorts, they provide a methodological foundation and generate specific hypotheses for future confirmatory research. The three molecular subtypes identified – characterized by distinct mutational signatures and therapeutic vulnerabilities – warrant investigation in multi-center studies with adequate statistical power. As precision oncology embraces algorithmic decision-making, exploratory frameworks like ours provide essential groundwork for developing evidence-based, scalable solutions for rare hematologic malignancies.
| 1. | Kwon HJ, Shin JE, Khan A, Park SY, Kim J, Lee JY, Lee D, Lee S, Im CY, Moon H, Han YR, Tamai M, Akahane K, Inukai T, Lee W, Kim H, Kim HN, Ahn SM, Park HW, Kim DW. KF1601, a dual inhibitor of BCR::ABL1 and FLT3, overcomes drug resistance in FLT3(+) blast phase chronic myeloid leukemia. Mol Cancer. 2025;24:114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Sun C, Xu X, Chen Z, Zhou F, Wang W, Chen J, Sun M, Wang F, Jiang L, Ji M, Liu S, Xu J, He M, Su B, Liu X, Gao Y, Wei H, Li J, Wang X, Zhao M, Yu J, Ma Y. Selective translational control by PABPC1 phase separation regulates blast crisis and therapy resistance in chronic myeloid leukaemia. Nat Cell Biol. 2025;27:683-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 3. | Cruz-Rodriguez N, Deininger MW. Novel treatment strategies for chronic myeloid leukemia. Blood. 2025;145:931-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 4. | Kamizela AE, Leongamornlert D, Williams N, Wang X, Nyamondo K, Dawson K, Spencer Chapman M, Guo J, Lee J, Mane K, Milne K, Green AR, Chevassut T, Campbell PJ, Ellinor PT, Huntly BJP, Baxter EJ, Nangalia J. Timing and trajectory of BCR::ABL1-driven chronic myeloid leukaemia. Nature. 2025;640:982-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 5. | Herráiz-gil S, Nygren-jiménez E, Acosta-alonso DN, León C, Guerrero-aspizua S. Artificial Intelligence-Based Methods for Drug Repurposing and Development in Cancer. Appl Sci. 2025;15:2798. [DOI] [Full Text] |
| 6. | Perillo T, de Giorgi M, Giorgio C, Frasca C, Cuocolo R, Pinto A. The Role of Machine Learning in the Most Common Hematological Malignancies: A Narrative Review. Hemato. 2024;5:380-387. [DOI] [Full Text] |
| 7. | Wang J, Zeng Z, Li Z, Liu G, Zhang S, Luo C, Hu S, Wan S, Zhao L. The clinical application of artificial intelligence in cancer precision treatment. J Transl Med. 2025;23:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 23] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 8. | Piñeiro-Yáñez E, Reboiro-Jato M, Gómez-López G, Perales-Patón J, Troulé K, Rodríguez JM, Tejero H, Shimamura T, López-Casas PP, Carretero J, Valencia A, Hidalgo M, Glez-Peña D, Al-Shahrour F. PanDrugs: a novel method to prioritize anticancer drug treatments according to individual genomic data. Genome Med. 2018;10:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 9. | Awada H, Gurnari C, Xie Z, Bewersdorf JP, Zeidan AM. What's Next after Hypomethylating Agents Failure in Myeloid Neoplasms? A Rational Approach. Cancers (Basel). 2023;15:2248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Wang SX, Huang ZF, Li J, Wu Y, Du J, Li T. Optimization of diagnosis and treatment of hematological diseases via artificial intelligence. Front Med (Lausanne). 2024;11:1487234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 11. | Iqbal Z, Alanazi N, Algarni A, Almukhaylid S, Adeel K, Sabar MF, Alabdullah F, Almaghlouth D, Alsalem GB, Alanazi S, Alfayez SF, Aljabr MAJ, Almasoudi HH, Taleb YS, Alsaab FM, Malik SR, Aleem A, Naeem R, Shamas MA, Saglio G. Exploring drug potential of myeloid/lymphoid lineage gene mutations in advanced-phase CML using drug discovery tools: Insights for precision oncology in blast crisis CML in the post-covid-19 ERA. J Popul Ther Clin Pharmacol. 2022;29:4994-5012. [DOI] [Full Text] |
| 12. | Alanazi S, Alfayez S, Butuwaybah M, Alanazi N, Algarni A, Adeel K, Taleb Y, Aleem A, Mohammed A, Iqbal Z. Abstract 4638: Investigations on association of prognostically important pan-cancer genes with disease progression and acute transformation in CML indicate involvement of diverse cellular mechanisms. Cancer Res. 2025;85:4638-4638. [DOI] [Full Text] |
| 13. | Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754-1760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29052] [Cited by in RCA: 36450] [Article Influence: 2144.1] [Reference Citation Analysis (0)] |
| 14. | DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491-498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9842] [Cited by in RCA: 8572] [Article Influence: 571.5] [Reference Citation Analysis (0)] |
| 15. | Qi X, Huang X. Machine learning-driven identification of key risk factors for predicting depression among nurses. BMC Nurs. 2025;24:368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 16. | Sondka Z, Dhir NB, Carvalho-Silva D, Jupe S, Madhumita, McLaren K, Starkey M, Ward S, Wilding J, Ahmed M, Argasinska J, Beare D, Chawla MS, Duke S, Fasanella I, Neogi AG, Haller S, Hetenyi B, Hodges L, Holmes A, Lyne R, Maurel T, Nair S, Pedro H, Sangrador-Vegas A, Schuilenburg H, Sheard Z, Yong SY, Teague J. COSMIC: a curated database of somatic variants and clinical data for cancer. Nucleic Acids Res. 2024;52:D1210-D1217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 321] [Article Influence: 160.5] [Reference Citation Analysis (0)] |
| 17. | Mosquera Orgueira A, González Pérez MS, Diaz Arias J, Rosiñol L, Oriol A, Teruel AI, Martinez Lopez J, Palomera L, Granell M, Blanchard MJ, de la Rubia J, López de la Guia A, Rios R, Sureda A, Hernandez MT, Bengoechea E, Calasanz MJ, Gutierrez N, Martin ML, Blade J, Lahuerta JJ, San Miguel J, Mateos MV; PETHEMA/GEM Cooperative Group. Unsupervised machine learning improves risk stratification in newly diagnosed multiple myeloma: an analysis of the Spanish Myeloma Group. Blood Cancer J. 2022;12:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Maxson JE, Ries RE, Wang YC, Gerbing RB, Kolb EA, Thompson SL, Guidry Auvil JM, Marra MA, Ma Y, Zong Z, Mungall AJ, Moore R, Long W, Gesuwan P, Davidsen TM, Hermida LC, Hughes SB, Farrar JE, Radich JP, Smith MA, Gerhard DS, Gamis AS, Alonzo TA, Meshinchi S. CSF3R mutations have a high degree of overlap with CEBPA mutations in pediatric AML. Blood. 2016;127:3094-3098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 19. | Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374:20150202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2837] [Cited by in RCA: 2423] [Article Influence: 242.3] [Reference Citation Analysis (1)] |
| 20. | Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53-65. [DOI] [Full Text] |
| 21. | Kruskal WH, Wallis WA. Use of Ranks in One-Criterion Variance Analysis. J Am Stat Assoc. 1952;47:583-621. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5360] [Cited by in RCA: 4366] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 22. | Dunn OJ. Multiple Comparisons Using Rank Sums. Technometrics. 1964;6:241-252. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2025] [Cited by in RCA: 1827] [Article Influence: 29.5] [Reference Citation Analysis (1)] |
| 23. | Fortin J, Chiang MF, Meydan C, Foox J, Ramachandran P, Leca J, Lemonnier F, Li WY, Gams MS, Sakamoto T, Chu M, Tobin C, Laugesen E, Robinson TM, You-Ten A, Butler DJ, Berger T, Minden MD, Levine RL, Guidos CJ, Melnick AM, Mason CE, Mak TW. Distinct and opposite effects of leukemogenic Idh and Tet2 mutations in hematopoietic stem and progenitor cells. Proc Natl Acad Sci U S A. 2023;120:e2208176120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 24. | Pamuk GE, Ehrlich LA. An Overview of Myeloid Blast-Phase Chronic Myeloid Leukemia. Cancers (Basel). 2024;16:3615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 25. | Mangiante L, Alcala N, Sexton-Oates A, Di Genova A, Gonzalez-Perez A, Khandekar A, Bergstrom EN, Kim J, Liu X, Blazquez-Encinas R, Giacobi C, Le Stang N, Boyault S, Cuenin C, Tabone-Eglinger S, Damiola F, Voegele C, Ardin M, Michallet MC, Soudade L, Delhomme TM, Poret A, Brevet M, Copin MC, Giusiano-Courcambeck S, Damotte D, Girard C, Hofman V, Hofman P, Mouroux J, Cohen C, Lacomme S, Mazieres J, de Montpreville VT, Perrin C, Planchard G, Rousseau N, Rouquette I, Sagan C, Scherpereel A, Thivolet F, Vignaud JM, Jean D, Ilg AGS, Olaso R, Meyer V, Boland-Auge A, Deleuze JF, Altmuller J, Nuernberg P, Ibáñez-Costa A, Castaño JP, Lantuejoul S, Ghantous A, Maussion C, Courtiol P, Hernandez-Vargas H, Caux C, Girard N, Lopez-Bigas N, Alexandrov LB, Galateau-Salle F, Foll M, Fernandez-Cuesta L. Multiomic analysis of malignant pleural mesothelioma identifies molecular axes and specialized tumor profiles driving intertumor heterogeneity. Nat Genet. 2023;55:607-618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 53] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 26. | Skelding KA, Lincz LF. PARP Inhibitors and Haematological Malignancies-Friend or Foe? Cancers (Basel). 2021;13:5328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Gungordu S, Aptullahoglu E. Targeting MDM2-mediated suppression of p53 with idasanutlin: a promising therapeutic approach for acute lymphoblastic leukemia. Invest New Drugs. 2024;42:603-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Amalinei C, Grigoraș A, Pricope D, Pricop B. Synthetic-Based Tumor-Infiltrating Lymphocytes (TILs) in Adoptive Cell Therapies. In: Rezaei N, editor. Handbook of Cancer and Immunology. Berlin: Springer, 2022. [DOI] [Full Text] |
| 29. | Montesinos P, Recher C, Vives S, Zarzycka E, Wang J, Bertani G, Heuser M, Calado RT, Schuh AC, Yeh SP, Daigle SR, Hui J, Pandya SS, Gianolio DA, de Botton S, Döhner H. Ivosidenib and Azacitidine in IDH1-Mutated Acute Myeloid Leukemia. N Engl J Med. 2022;386:1519-1531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 356] [Article Influence: 89.0] [Reference Citation Analysis (0)] |
| 30. | Kühn MWM, Pemmaraju N, Heidel FH. The evolving landscape of epigenetic target molecules and therapies in myeloid cancers: focus on acute myeloid leukemia and myeloproliferative neoplasms. Leukemia. 2025;39:1824-1837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 31. | Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z, Zhu Y, Wang Q, Lu L, Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther. 2020;5:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 238] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 32. | Habaka M, Daly GR, Shinyanbola D, Alabdulrahman M, McGrath J, Dowling GP, Hehir C, Huang HYR, Hill ADK, Varešlija D, Young LS. PARP Inhibitors in the Neoadjuvant Setting; A Comprehensive Overview of the Rationale for their Use, Past and Ongoing Clinical Trials. Curr Oncol Rep. 2025;27:533-551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 33. | Rutella S, Vadakekolathu J, Mazziotta F, Reeder S, Yau TO, Mukhopadhyay R, Dickins B, Altmann H, Kramer M, Knaus HA, Blazar BR, Radojcic V, Zeidner JF, Arruda A, Wang B, Abbas HA, Minden MD, Tasian SK, Bornhäuser M, Gojo I, Luznik L. Immune dysfunction signatures predict outcomes and define checkpoint blockade-unresponsive microenvironments in acute myeloid leukemia. J Clin Invest. 2022;132:e159579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 34. | Adan A. DNA Methylation Alterations in Acute Myeloid Leukemia: Therapeutic Potential. In: Rezaei N, editor. Hematological Cancer Diagnosis and Treatment: An Interdisciplinary Approach. Berlin: Springer, 2023. [DOI] [Full Text] |
| 35. | Bruedigam C, Porter AH, Song A, Vroeg In de Wei G, Stoll T, Straube J, Cooper L, Cheng G, Kahl VFS, Sobinoff AP, Ling VY, Jebaraj BMC, Janardhanan Y, Haldar R, Bray LJ, Bullinger L, Heidel FH, Kennedy GA, Hill MM, Pickett HA, Abdel-Wahab O, Hartel G, Lane SW. Imetelstat-mediated alterations in fatty acid metabolism to induce ferroptosis as a therapeutic strategy for acute myeloid leukemia. Nat Cancer. 2024;5:47-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 53] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 36. | Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, Traina TA, Paik PK, Ho AL, Hantash FM, Grupe A, Baxi SS, Callahan MK, Snyder A, Chi P, Danila D, Gounder M, Harding JJ, Hellmann MD, Iyer G, Janjigian Y, Kaley T, Levine DA, Lowery M, Omuro A, Postow MA, Rathkopf D, Shoushtari AN, Shukla N, Voss M, Paraiso E, Zehir A, Berger MF, Taylor BS, Saltz LB, Riely GJ, Ladanyi M, Hyman DM, Baselga J, Sabbatini P, Solit DB, Schultz N. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017:PO.17.00011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1079] [Cited by in RCA: 1372] [Article Influence: 152.4] [Reference Citation Analysis (0)] |
| 37. | Tsimberidou AM, Kahle M, Vo HH, Baysal MA, Johnson A, Meric-Bernstam F. Molecular tumour boards - current and future considerations for precision oncology. Nat Rev Clin Oncol. 2023;20:843-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 67] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 38. | Srivastava R. Advancing precision oncology with AI-powered genomic analysis. Front Pharmacol. 2025;16:1591696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 39. | Jain KK. Personalized Management of Cancers of Various Organs/Systems. Textbook of Personalized Medicine. Berlin: Springer, 2021. [DOI] [Full Text] |
| 40. | Jiménez-Santos MJ, García-Martín S, Fustero-Torre C, Di Domenico T, Gómez-López G, Al-Shahrour F. Bioinformatics roadmap for therapy selection in cancer genomics. Mol Oncol. 2022;16:3881-3908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 41. | Gazola AA, Lautert-Dutra W, Archangelo LF, Reis RBD, Squire JA. Precision oncology platforms: practical strategies for genomic database utilization in cancer treatment. Mol Cytogenet. 2024;17:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 42. | Gutiérrez-González A, Del Hierro I, Cariaga-Martínez AE. Advancements in Multiple Myeloma Research: High-Throughput Sequencing Technologies, Omics, and the Role of Artificial Intelligence. Biology (Basel). 2024;13:923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 43. | Mao Y, Shangguan D, Huang Q, Xiao L, Cao D, Zhou H, Wang YK. Emerging artificial intelligence-driven precision therapies in tumor drug resistance: recent advances, opportunities, and challenges. Mol Cancer. 2025;24:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 44. | Wang Z, Zhou G, Cao R, Zhang G, Zhang Y, Xiao M, Liu L, Zhang X. Harnessing multi-omics and artificial intelligence: revolutionizing prognosis and treatment in hepatocellular carcinoma. Front Immunol. 2025;16:1592259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 45. | Zullo A, De Francesco V, Gatta L, Scaccianoce G, Colombo M, Bringiotti R, Azzarone A, Rago A, Corti F, Repici A, Hassan C, Rossi RE. Small bowel lesions in patients with iron deficiency anaemia without overt bleeding: a multicentre study. Ann Hematol. 2024;103:1-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 46. | Wei L, Niraula D, Gates EDH, Fu J, Luo Y, Nyflot MJ, Bowen SR, El Naqa IM, Cui S. Artificial intelligence (AI) and machine learning (ML) in precision oncology: a review on enhancing discoverability through multiomics integration. Br J Radiol. 2023;96:20230211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 72] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 47. | Singh JP, Gaur V, Dubey P. Precision, Prediction and Progress: A New Era in Pharmacology. Int J Innov Sci Res Technol. 2025;10. [DOI] [Full Text] |
| 48. | Shen M, Jiang Z. Artificial Intelligence Applications in Lymphoma Diagnosis and Management: Opportunities, Challenges, and Future Directions. J Multidiscip Healthc. 2024;17:5329-5339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 49. | Post B, Klapaukh R, Brett SJ, Faisal AA. Harnessing temporal patterns in administrative patient data to predict risk of emergency hospital admission. Lancet Digit Health. 2025;7:e124-e135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/