Published online Sep 24, 2024. doi: 10.5306/wjco.v15.i9.1188
Revised: July 21, 2024
Accepted: August 2, 2024
Published online: September 24, 2024
Processing time: 113 Days and 17.5 Hours
Primary liver cancer is a prevalent and deadly cancer type. Despite treatment advances, prognosis remains poor, with high recurrence rates. Early detection is crucial but challenging due to the disease’s insidious nature. Myosin proteins play significant roles in cancer development, influencing cell migration, invasion, and tumor suppression. MYL6B, a myosin light chain, is involved in various cellular processes and has been associated with poor prognosis in colorectal adenocarcinoma and potential as a biomarker in breast cancer.
To investigate the expression of MYL6B in liver hepatocellular carcinoma (LIHC) and its impact on prognosis and potential mechanisms of action using bioin
The expression of MYL6B in pan-cancer and normal tissues was analyzed using the gene expression profiling interactive analysis 2 and tumor immune estimation resource databases. The expression level of MYL6B in LIHC tissues and its relationship with prognosis were analyzed, immunohistochemical analysis of MYL6B and its effect on immune cell infiltration, and the protein network were further studied.
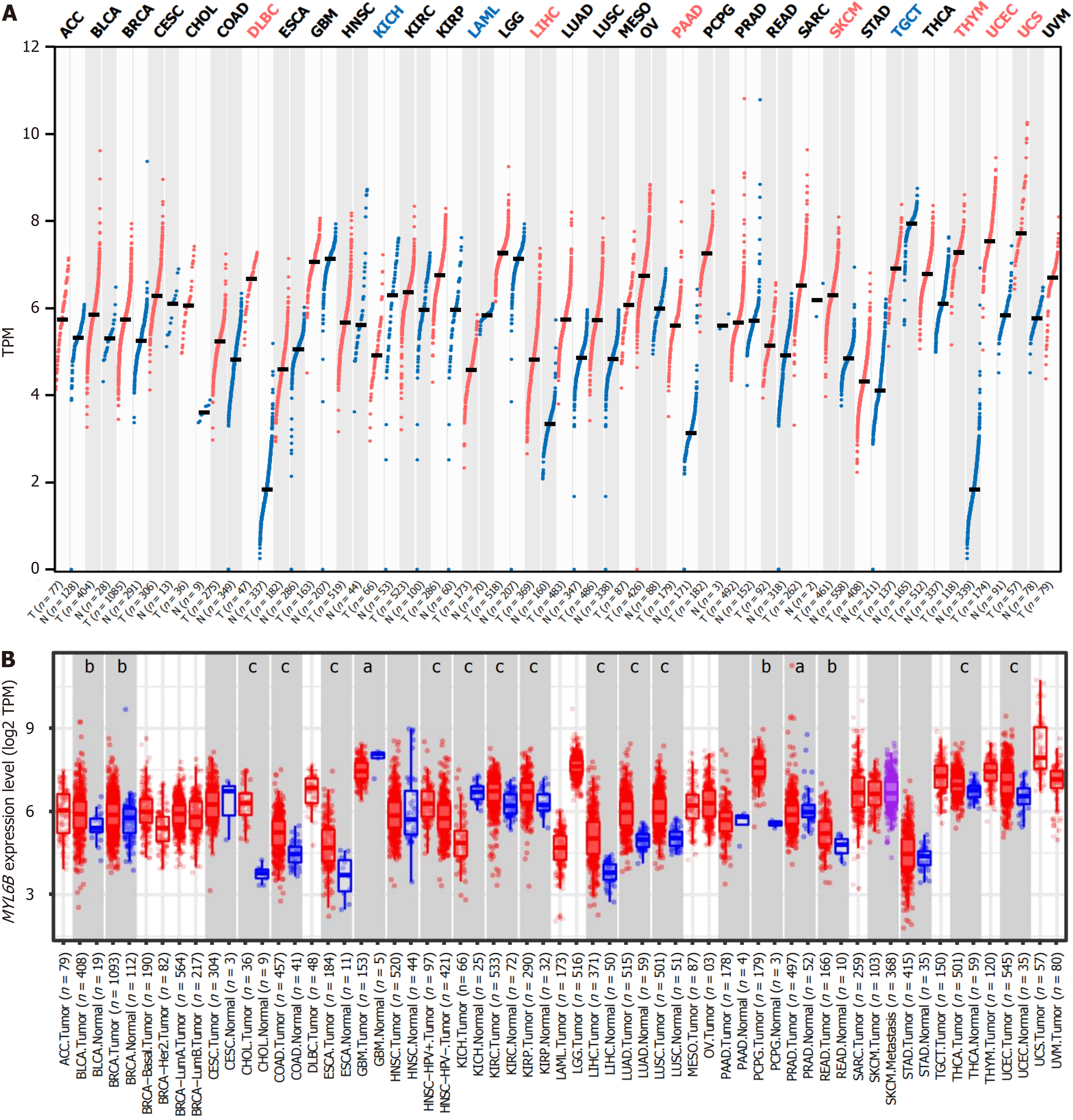
MYL6B was highly expressed in diffuse large b-cell lymphoma, LIHC, pancreatic adenocarcinoma, skin cutaneous melanoma, thymoma, uterine corpus endometrial carcinoma, uterine carcinosarcoma, and lowly expressed in kidney chromophobe, acute myeloid leukemia, testicular germ cell tumors. The expression level of MYL6B was sig
The expression level of MYL6B in LIHC was significantly higher than in normal liver tissues, and it was correlated with the degree of differentiation survival rate, and immune infiltration. MYL6B is a potential target for LIHC treatment.
Core Tip: In the study, we employed advanced bioinformatics methodologies to meticulously scrutinize the intricate relationship between MYL6B and liver hepatocellular carcinoma (LIHC). It entailed a comprehensive analysis encompassing the contrasting expression patterns observed between LIHC and normal tissue samples, coupled with an intricate exa
- Citation: Lv HB, Wu QY, Zhang YJ, Quan SW, Ma N, Dai YQ, Sun Y. Study on the expression and prognostic relationship of MYL6B in liver cancer based on bioinformatics. World J Clin Oncol 2024; 15(9): 1188-1197
- URL: https://www.wjgnet.com/2218-4333/full/v15/i9/1188.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i9.1188
Primary liver cancer is a common cancer type with a high mortality rate. In 2020, there were 905677 new cases (4.7%) and 830180 deaths (8.3%) from liver cancer worldwide[1]. Different treatment strategies are applied for liver cancer patients at different stages. Early-stage liver cancer patients should undergo curative treatments such as liver resection, trans
However, the high malignancy and insidious nature of liver hepatocellular carcinoma (LIHC) mean that most ad
Myosin plays an important role in cancer development. Myosin is involved in cancer progression through their roles in cell migration and invasion and their tumor suppressor functions. Genetic and epigenetic alterations of genes encoding myosin heavy chains have been found in many types of cancer. In some cases, changes in myosin expression can serve as a predictor of patient survival. Therefore, some members of the myosin superfamily have potential as cancer biomarkers[11]. For example, myosin 1e can promote the malignant progression of breast cancer by enhancing tumor cell proliferation and stimulating tumor cell dedifferentiation[12], while the activation of myosin II in cancer cells drives tumor progression through interaction with the secretions of the immune microenvironment[13]. Additionally, The elevated expression of myosin X in tumors contributes to the invasiveness and metastasis of breast cancer[14], myosin Vb can function as a tumor suppressor gene in colorectal cancer[15].
MYL6B is myosin light chain 6B, a protein encoded by the MYL6B gene. Myosins are superfamily of motor proteins that have a significant effect on the process of movement, and myosin light chains can regulate Ca2+ transduction[16,17]. As an important myosin light chain, MYL6B is involved in cell viability, adhesion, migration, and endocytosis, tissue structure, and transportation[18-20].
MYL6B is associated with various cancers. For instance, in colorectal adenocarcinoma, MYL6B is correlated with poor prognosis in patients. In vitro functional experiments have validated that knocking down MYL6B can inhibit the proliferation, migration, and invasion of colorectal adenocarcinoma cells, while promoting apoptosis[21]. In breast cancer, MYL6B is upregulated and serves as a potential biological marker. The expression level of MYL6B mRNA in breast tumor tissues is higher than in normal tissues, and in Luminal A type breast cancer, the expression level of MYL6B is signi
Therefore, this study used bioinformatics methods to analyze the expression level, staging, and prognostic significance of MYL6B in liver cancer. Based on these analyses, this study elucidates the role of MYL6B in liver cancer and provides potential therapeutic targets for its progression and diagnosis.
The investigation into MYL6B expression profiles involved a meticulous comparison between various cancer tissues and their corresponding normal counterparts, facilitated by the utilization of the gene expression profiling interactive analysis (GEPIA2) database (http://gepia2.cancer-pku.cn/). Furthermore, a comprehensive analysis of MYL6B expression across diverse tissue types was undertaken utilizing the tumor immune estimation resource (TIMER) database (http://timer.cistrome.org/). Notably, a rigorous statistical analysis was conducted to discern significant differences in MYL6B expression among various cancer types, thereby enriching our understanding of its potential roles in malignancy pro
Survival outcomes associated with MYL6B were meticulously evaluated through meticulous scrutiny of data obtained from the Kaplan-Meier plotter database (https://kmplot.com/). This encompassed an in-depth analysis of both overall survival (OS) among a cohort of 364 subjects and recurrence-free survival (RFS) within a subgroup comprising 316 individuals. These survival analyses provided invaluable insights into the prognostic implications of MYL6B expression levels in cancer.
Immune infiltration analysis using TIMER database. The exploration of MYL6B expression extended beyond mere quantitative assessments, delving into qualitative aspects through immunohistochemical staining analyses. Leveraging the resources provided by the human protein atlas (HPA) database (https://www.proteinatlas.org/), this endeavor involved the meticulous examination of MYL6B expression in both high-expression cancer tissues and normal tissue counterparts. The antibody HPA063034 (Sigma-Aldrich) was utilized for immunohistochemical analyses.
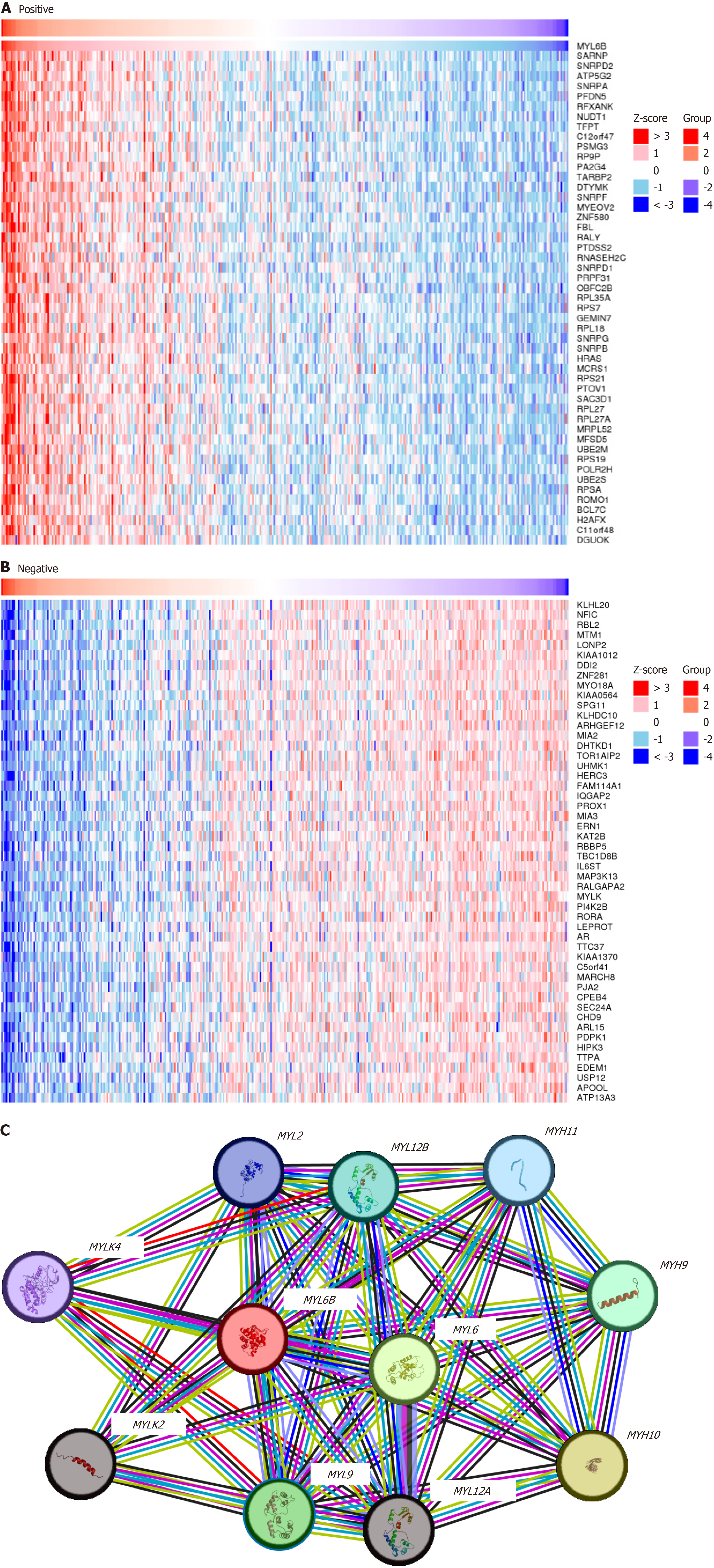
Furthermore, a deeper understanding of MYL6B’s functional implications was sought through an exploration of its co-expression network. This endeavor was facilitated by the utilization of the LinkedOmic database (https://www.linkedomics.org/), enabling the identification and analysis of genes that exhibit coordinated expression patterns with MYL6B. Subsequently, comprehensive analyses of both upregulated and downregulated genes were undertaken, with a focus on the top 50 significantly altered genes. The visualization of these findings was facilitated through the generation of heat maps, enhancing the interpretability and utility of the results.
Additionally, the construction of protein-protein interaction networks surrounding MYL6B was undertaken utilizing the resources provided by the STRING database (https://string-db.org/).
The statistical analysis was automatically computed based on the above online databases. aP < 0.05 indicated a statistically significant difference.
In multiple cancer tissues, the expression level of MYL6B showed variability compared to normal tissues, MYL6B expression levels differed significantly between cancer and normal tissues (Figure 1A). GEPIA2 analysis revealed MYL6B high expression in diffuse large B-cell lymphoma, pancreatic adenocarcinoma, skin cutaneous melanoma, thymoma, uterine corpus endometrial carcinoma, and uterine carcinosarcoma cancer tissues, while low expression was observed in kidney chromophobe, acute myeloid leukemia, testicular germ cell tumors. Furthermore, in LIHC, MYL6B showed high expression in GEPIA2 analysis. Similarly, TIMER analysis showed high expression in LIHC with significant differences (Tumor n = 371, Normal n = 50) (Figure 1B).
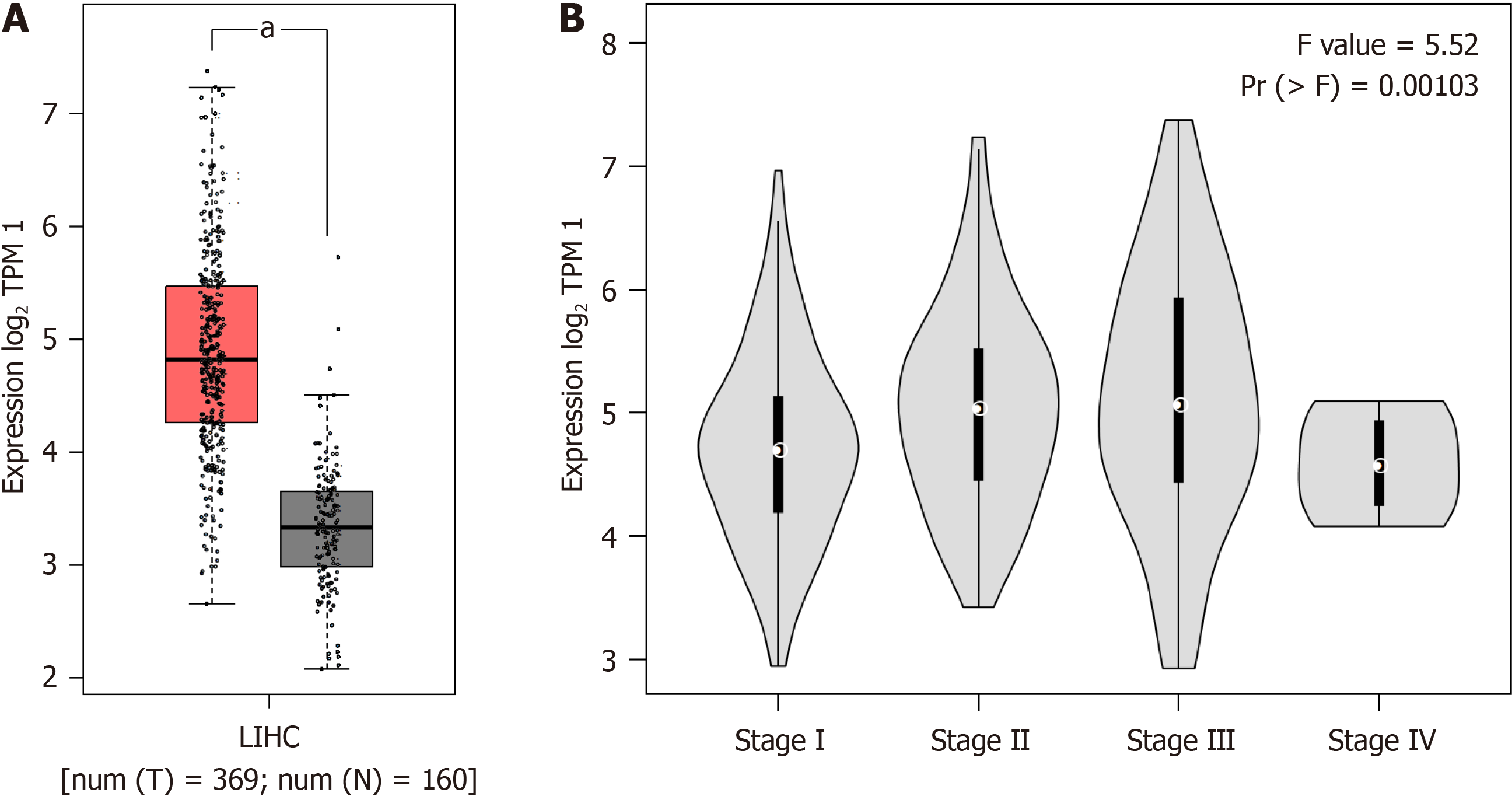
In the context of hepatocellular carcinoma (HCC), MYL6B demonstrated significant differences when subjected to comparative analysis (Tumor n = 369, Normal n = 160) (P < 0.05), revealing markedly elevated expression levels spe
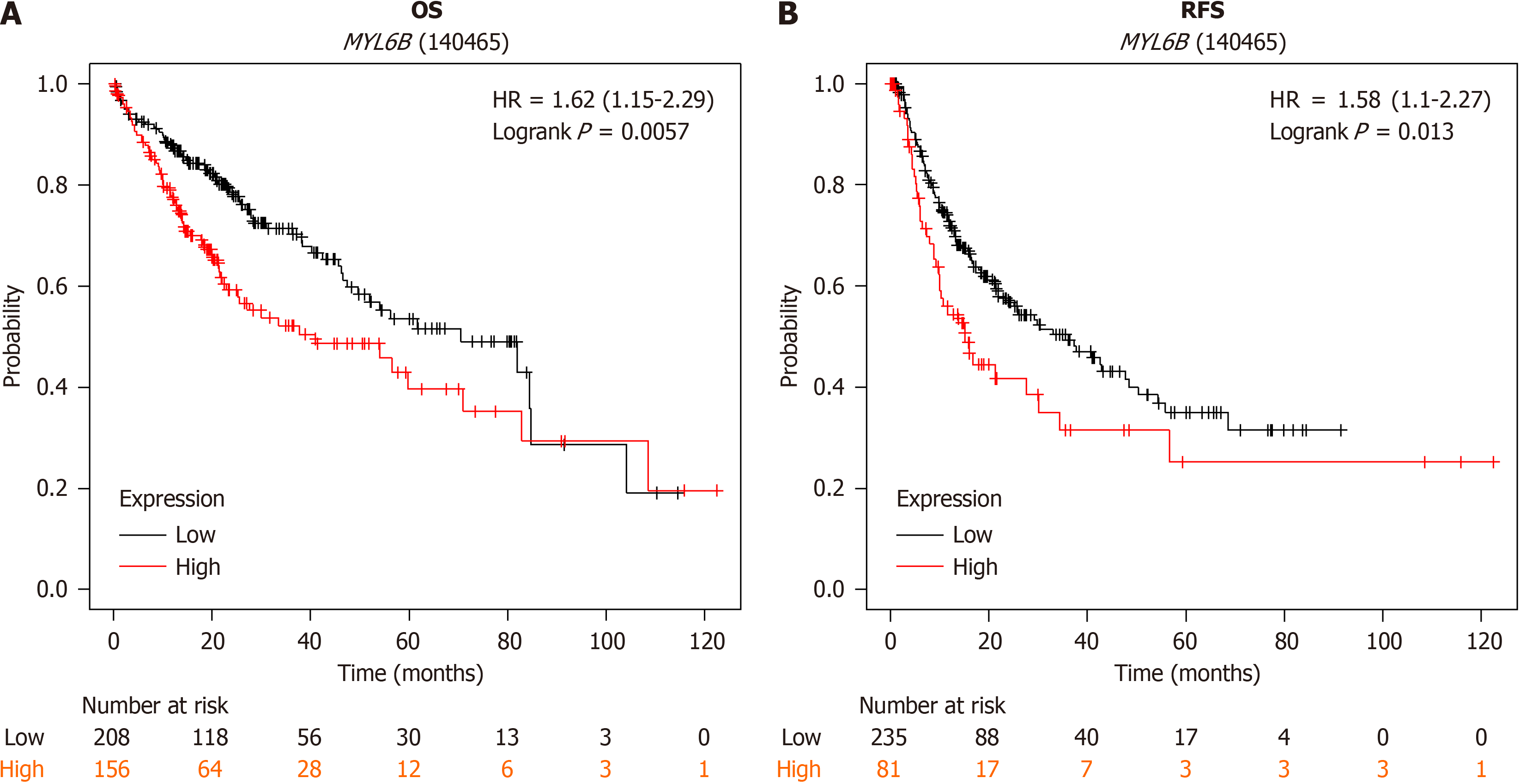
To investigate the correlation between MYL6B and LIHC prognosis, Kaplan-Meier plotter database was used to explore the association between high and low expression groups of MYL6B with survival rates, patient prognosis, and recurrence risk. Results indicated a close association between high gene expression and shorter OS and progression-free survival. Analysis of OS (n = 364), with low expression group (n = 208) and high expression group (n = 156), showed higher OS in the low expression group compared to the high expression group (logrank P = 0.0057), with median survival times of 70.5 months and 41 months, respectively (Figure 3A). For RFS, with low expression group (n = 235) and high expression group (n = 81), median survival times were 36.1 months and 15.17 months, respectively (Figure 3B).
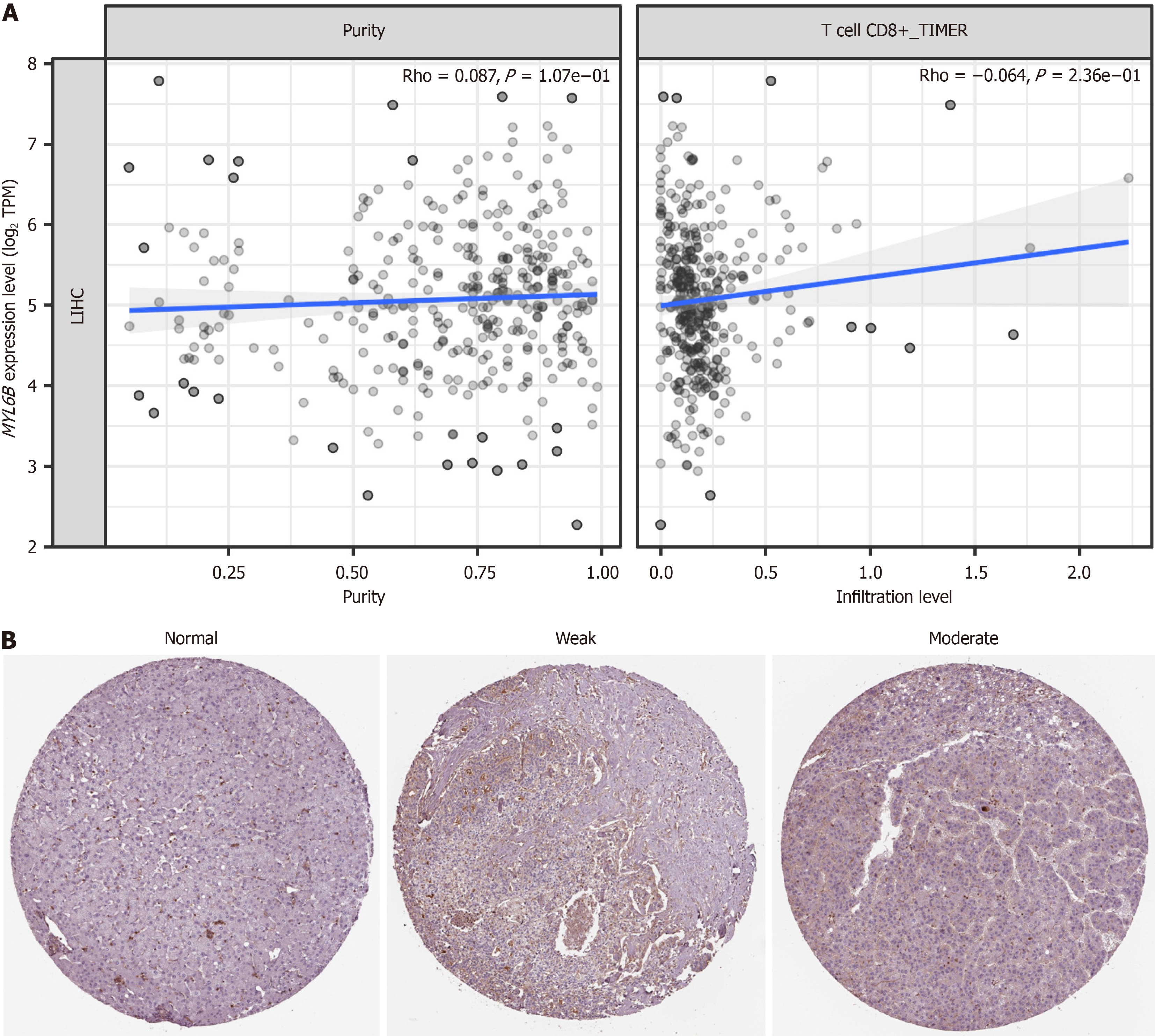
Through comprehensive immune infiltration analyses, we uncover a compelling association between elevated MYL6B expression and heightened immune cell infiltration, particularly characterized by increased activation of CD8 + T cells. This observation, depicted graphically in Figure 4A, underscores the immunomodulatory roles of MYL6B within the tumor milieu, potentially shaping the host immune response to cancer. Additionally, immunohistochemical staining images obtained from the HPA database further corroborate these findings, revealing differential MYL6B expression patterns between normal and cancer tissues (Figure 4B). Notably, high MYL6B expression in cancer tissues correlates with the severity of malignancy, underscoring its potential as a biomarker of tumor aggressiveness and immune modulation.
Furthermore, the LinkedOmic database was harnessed to delve into the repertoire of co-expressed genes associated with MYL6B. This comprehensive analysis unveiled a diverse array of genes that exhibited either positive or negative corre
Analysis of the protein interaction network of MYL6B using the STRING database revealed correlations with MYL2, MYL6, MYL9, MYLK4, MYLK2, MYL12A, MYL12B, MYH11, MYH9, and MYH10 (Figure 5B). This facilitated the eluci
This study employed advanced bioinformatics methodologies to meticulously scrutinize the intricate relationship between MYL6B and LIHC. It entailed a comprehensive analysis encompassing the contrasting expression patterns observed between LIHC and normal tissue samples, coupled with an intricate examination of how MYL6B expression levels correlate with the staging of cancer progression and the rates of survival. Furthermore, an exhaustive immunohistochemical investigation was conducted to elucidate the nuances of inflammation across varying levels of MYL6B ex
MYL6B, as an exosomal gene, has been studied in relation to HCC. A study focusing on the prognostic, recurrence risk, and immune infiltration prediction models based on two exosomal genes, MYL6B and THOC2, in HCC revealed that the prognostic and recurrence risk prediction model built upon these two exosomal genes (MYL6B and THOC2) was con
In this study, bioinformatics techniques alongside other research methodologies were employed to evaluate the expression pattern of MYL6B across a spectrum of cancers, with subsequent exploration of its impact on the progression and prognosis of HCC. The findings unveiled a conspicuous upregulation of MYL6B expression in HCC tissues, with substantially elevated levels of MYL6B expression demonstrating a strong correlation with unfavorable prognosis and advanced tumor staging.
Tumor occurrence is a complex and multi-step process primarily caused by the accumulation of gene mutations associated with organismal growth and development. It is widely believed that genetic and chromosomal instability are the main reasons for gene mutations and potential tumor progression. Subsequently, malignant cells detach from primary tumors and enter the stages of metastasis and invasion. To facilitate this, a series of physiological and metabolic processes need to be altered. These include loss of cell polarity and tissue disintegration, formation of cell protrusions, damage to cell adhesion, and inhibition of apoptosis. The seven classes of myosin superfamily, including myosin I, II, V, VI, VII, IX, and X, have been shown to participate in these processes during tumor occurrence. Additionally, myosins are involved in various factors, pathways, and mechanisms related to tumor progression, and they have specific functions in nuclear division and myosin superfamily optimization processes[25].
MYL6B, as a myosin, is correlated with cancer development. The modification of the actin cytoskeleton’s structure is a fundamental mechanism in the progression of cancer, enabling the proliferation and dissemination of cancerous cells. This intricate process involves a myriad of contributors, with myosin motors standing out as pivotal regulators that oversee numerous stages of tumorigenesis. From orchestrating nuclear transcriptional programs to orchestrating the reshaping of the cellular cortex during cancer cell migration and division, myosin motors exert indispensable control over the intricate dance of cancer progression[11,26].
Hence, MYL6B emerges as a versatile prognostic marker across a spectrum of cancer types. Its multifaceted roles encompassing diverse biological functions, modulation of immune infiltration, and orchestration of protein interaction networks position MYL6B as a potent facilitator of tumor progression. Delving into its protein interactions unveils a nexus with MYL2, MYL6, and MYL9, underscoring MYL6B’s potential as an appealing therapeutic target for addressing LIHC. Within the realm of HCC, MYL6B assumes the role of a predictive risk factor of LIHC, offering valuable insights into disease prognosis and management strategies. However, there are several limitations in our study: (1) Some im
The expression level of MYL6B in LIHC was significantly higher than in normal liver tissues, and it was correlated with the degree of differentiation survival rate, and immune infiltration. MYL6B is a potential target for LIHC treatment.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68827] [Article Influence: 13765.4] [Reference Citation Analysis (202)] |
| 2. | Lencioni R, Crocetti L. Local-regional treatment of hepatocellular carcinoma. Radiology. 2012;262:43-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 267] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 3. | Wedd JP, Nordstrom E, Nydam T, Durham J, Zimmerman M, Johnson T, Thomas Purcell W, Biggins SW. Hepatocellular carcinoma in patients listed for liver transplantation: Current and future allocation policy and management strategies for the individual patient. Liver Transpl. 2015;21:1543-1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Doyle MB, Vachharajani N, Maynard E, Shenoy S, Anderson C, Wellen JR, Lowell JA, Chapman WC. Liver transplantation for hepatocellular carcinoma: long-term results suggest excellent outcomes. J Am Coll Surg. 2012;215:19-28; discussion 28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 5. | Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873:188314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 762] [Cited by in RCA: 983] [Article Influence: 163.8] [Reference Citation Analysis (2)] |
| 6. | Guglielmi A, Ruzzenente A, Conci S, Valdegamberi A, Vitali M, Bertuzzo F, De Angelis M, Mantovani G, Iacono C. Hepatocellular carcinoma: surgical perspectives beyond the barcelona clinic liver cancer recommendations. World J Gastroenterol. 2014;20:7525-7533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 47] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 7. | Ochiai T, Ikoma H, Okamoto K, Kokuba Y, Sonoyama T, Otsuji E. Clinicopathologic features and risk factors for extrahepatic recurrences of hepatocellular carcinoma after curative resection. World J Surg. 2012;36:136-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Poon RT, Fan ST, O'Suilleabhain CB, Wong J. Aggressive management of patients with extrahepatic and intrahepatic recurrences of hepatocellular carcinoma by combined resection and locoregional therapy. J Am Coll Surg. 2002;195:311-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 91] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Chen J, Wang H, Zhou L, Liu Z, Chen H, Tan X. A necroptosis-related gene signature for predicting prognosis, immune landscape, and drug sensitivity in hepatocellular carcinoma. Cancer Med. 2022;11:5079-5096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 10. | Balogh J, Victor D 3rd, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour HP Jr. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 755] [Cited by in RCA: 835] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 11. | Ouderkirk JL, Krendel M. Non-muscle myosins in tumor progression, cancer cell invasion, and metastasis. Cytoskeleton (Hoboken). 2014;71:447-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 12. | Ouderkirk-Pecone JL, Goreczny GJ, Chase SE, Tatum AH, Turner CE, Krendel M. Myosin 1e promotes breast cancer malignancy by enhancing tumor cell proliferation and stimulating tumor cell de-differentiation. Oncotarget. 2016;7:46419-46432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Georgouli M, Herraiz C, Crosas-Molist E, Fanshawe B, Maiques O, Perdrix A, Pandya P, Rodriguez-Hernandez I, Ilieva KM, Cantelli G, Karagiannis P, Mele S, Lam H, Josephs DH, Matias-Guiu X, Marti RM, Nestle FO, Orgaz JL, Malanchi I, Fruhwirth GO, Karagiannis SN, Sanz-Moreno V. Regional Activation of Myosin II in Cancer Cells Drives Tumor Progression via a Secretory Cross-Talk with the Immune Microenvironment. Cell. 2019;176:757-774.e23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 14. | Cao R, Chen J, Zhang X, Zhai Y, Qing X, Xing W, Zhang L, Malik YS, Yu H, Zhu X. Elevated expression of myosin X in tumours contributes to breast cancer aggressiveness and metastasis. Br J Cancer. 2014;111:539-550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 15. | Cartón-García F, Brotons B, Anguita E, Dopeso H, Tarragona J, Nieto R, García-Vidal E, Macaya I, Zagyva Z, Dalmau M, Sánchez-Martín M, van Ijzendoorn SCD, Landolfi S, Hernandez-Losa J, Schwartz S Jr, Matias-Guiu X, Ramón Y Cajal S, Martínez-Barriocanal Á, Arango D. Myosin Vb as a tumor suppressor gene in intestinal cancer. Oncogene. 2022;41:5279-5288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 16. | Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 582] [Cited by in RCA: 601] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 17. | England J, Loughna S. Heavy and light roles: myosin in the morphogenesis of the heart. Cell Mol Life Sci. 2013;70:1221-1239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 167] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 18. | Maliga Z, Junqueira M, Toyoda Y, Ettinger A, Mora-Bermúdez F, Klemm RW, Vasilj A, Guhr E, Ibarlucea-Benitez I, Poser I, Bonifacio E, Huttner WB, Shevchenko A, Hyman AA. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat Cell Biol. 2013;15:325-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 19. | Hartman MA, Finan D, Sivaramakrishnan S, Spudich JA. Principles of unconventional myosin function and targeting. Annu Rev Cell Dev Biol. 2011;27:133-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 136] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 20. | Heissler SM, Sellers JR. Myosin light chains: Teaching old dogs new tricks. Bioarchitecture. 2014;4:169-188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 21. | Li JL, Wang ZQ, Sun XL. MYL6B drives the capabilities of proliferation, invasion, and migration in rectal adenocarcinoma through the EMT process. Open Life Sci. 2020;15:522-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 22. | Feser R, Opperman RM, Nault B, Maiti S, Chen VC, Majumder M. Breast cancer cell secretome analysis to decipher miRNA regulating the tumor microenvironment and discover potential biomarkers. Heliyon. 2023;9:e15421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 23. | Zhu J, Tang B, Gao Y, Xu S, Tu J, Wang Y, Yang W, Fang S, Weng Q, Zhao Z, Xu M, Yang Y, Chen M, Lu C, Ji J. Predictive Models for HCC Prognosis, Recurrence Risk, and Immune Infiltration Based on Two Exosomal Genes: MYL6B and THOC2. J Inflamm Res. 2021;14:4089-4109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 24. | Xie X, Wang X, Liao W, Fei R, Wu N, Cong X, Chen Q, Wei L, Wang Y, Chen H. MYL6B, a myosin light chain, promotes MDM2-mediated p53 degradation and drives HCC development. J Exp Clin Cancer Res. 2018;37:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 25. | Li YR, Yang WX. Myosins as fundamental components during tumorigenesis: diverse and indispensable. Oncotarget. 2016;7:46785-46812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 26. | Naydenov NG, Lechuga S, Huang EH, Ivanov AI. Myosin Motors: Novel Regulators and Therapeutic Targets in Colorectal Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/