Published online Jan 24, 2024. doi: 10.5306/wjco.v15.i1.145
Peer-review started: October 22, 2023
First decision: November 23, 2023
Revised: December 8, 2023
Accepted: December 28, 2023
Article in press: December 28, 2023
Published online: January 24, 2024
Processing time: 92 Days and 11.8 Hours
Emerging research suggests that microRNAs (miRNAs) play an important role in the development of hepatocellular carcinoma (HCC). A comprehensive analysis of recent research concerning miRNAs in HCC development could provide researchers with a valuable reference for further studies.
To make a comprehensive analysis of recent studies concerning miRNAs in HCC.
All relevant publications were retrieved from the Web of Science Core Collection database. Bibliometrix software, VOSviewer software and CiteSpace software were used to visually analyze the distribution by time, countries, institutions, journals, and authors, as well as the keywords, burst keywords and thematic map.
A total of 9426 publications on this topic were found worldwide. According to the keywords analysis, we found that the studies of miRNAs focused on their expression level, effects, and mechanisms on the biological behaviour of HCC. Keywords bursting analysis showed that in the early years (2013–2017), “micro
This study provides a comprehensive overview of the role of miRNAs in HCC development based on bibliometric analysis. The hotspots in this field focus on miRNAs expression level, effects, and mechanisms on the biological behavior of HCC. The frontiers turned to sorafenib resistance, tumor microenvironment and so on.
Core Tip: The main objective of this work is to systematically analyse the recent research concerning microRNAs (miRNAs) in hepatocellular carcinoma (HCC). Annual publications provided an overview of the trends in this area, while distribution by countries, institutions, journals, and authors analysis were outstandingly representative and provided some cooperation situation information. According to the keywords analysis, the studies of miRNAs focused on their expression level, effects, and mechanisms on the biological behaviour of HCC. Keywords bursting analysis showed that in the latest phase (2018–2022), the hot topics turned to “sorafenib resistance”, “tumor microenvironment” and so on.
- Citation: Zhang L, Chen ZY, Wei XX, Li JD, Chen G. What are the changes in the hotspots and frontiers of microRNAs in hepatocellular carcinoma over the past decade? World J Clin Oncol 2024; 15(1): 145-158
- URL: https://www.wjgnet.com/2218-4333/full/v15/i1/145.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i1.145
It is estimated that in 2020, 905700 people were diagnosed with liver cancer worldwide and 830200 died from liver cancer, with China accounting for 45.3% of the world's liver cancer cases and 47.1% of the world's liver cancer deaths[1]. In southern China, where liver cancer death rates are high (22.3 per 100000), especially in rural areas (26.6 per 100000), Fusui County in Guangxi Province is one of the nine regions with the highest incidence of liver cancer in the country[2]. As the understanding of the carcinogenic factors that cause liver cancer has improved worldwide, researchers have developed new treatment strategies, such as immunotherapy and targeted therapies. The continuous development of new technologies and drugs provides hope for further diagnosis and treatment of liver cancer[3]. Hepatocellular carcinoma (HCC) accounts for 85%-90% of primary liver cancers and is highly drug-resistant, making it difficult to treat. Most patients are diagnosed at an advanced stage, greatly affecting the prognosis and five-year survival rate[4]. Therefore, new therapies for HCC remain an unmet medical need. In this regard, important insights into the biology of the disease have been obtained through genomics, transcriptomics and epigenomics studies[5]. In recent years, exploration in the field of noncoding genes has provided a new understanding of the pathogenesis and development of HCC, early diagnostic markers, and novel therapeutic targets.
MicroRNA (miRNA), a small noncoding RNA molecule approximately 22 nucleotides in length, is estimated to regulate almost 60% of human protein-coding genes[6]. miRNA is closely associated with the pathogenesis of various types of tumors[7]. Previous research has shown that a great quantity of miRNAs are aberrantly expressed in various cancer cell lines and tissues, and they participate in the biological processes such as the tumor invasion, metastasis, autophagy, regulation of the tumor microenvironment, chemotherapy resistance, and immunotherapy modulation[8-12]. As competitive endogenous RNAs, long noncoding RNAs (lncRNAs) and circRNAs contain miRNA binding sites and promote miRNA target gene expression through competition with miRNAs[13-17]. For example, Wang et al[18] identified and verified a novel MAGI2-AS3/miRNA-374-5p/FOXO1 network that was associated with HBV-related HCC. Therefore, the identification of miRNAs and their binding sites on tumor will help to explore new mechanisms of tumor occurrence and development, as well as to discover new tumor diagnostic markers and therapeutic targets[19,20].
In our research, we used bibliometrics to explore the research relationship between miRNAs and HCC. Bibliometrics are able to study the distribution focus, number relationship and change pattern of literature in related fields using mathematical, statistical and other econometric research methods[21]. Potential relationships between large amounts of literature were presented in bibliometric plots, and large bibliometric plots were visualized and analysed with VOSviewer and CiteSpace. Related software was used to summarize and display the relationships between countries, institutions, journals, authors, and keywords. Then, the topic was examined at a macro level to discuss future directions and the extent of progress, providing a better overview and new ideas for the researchers involved.
All relevant publications were retrieved from the Web of Science Core Collection (WOSCC) database with the following search strategies: ((“hepatocellular carcinoma” OR “hepatocarcinoma” OR “HCC” OR “liver cancer” OR “liver carcinoma” OR “hepatic cancer” OR “hepatic carcinoma”) AND (“miRNA” OR “microRNA” OR “miR”)) AND Document types = (Article OR Review) AND Language = English, index = Science Citation Index Expanded (SCI-EXPANDED), with a limited time frame from 2013 to 2022. In total, 9,426 publications were ultimately included.
The bibliographic information was downloaded from the WOSCC database, including title, authors, year of publications, country, institution, keywords, citations, abstract and reference. The Bibliometrix software was used to extract all data eligible for inclusion in this study by two investigators alone and then imported into Microsoft Excel. Where there was any disagreement between the two investigators, a third investigator resolved it.
Microsoft Excel was used to calculate descriptive statistical analysis, including institutions, journals, and authors. Visualization analysis was based on the Bibliometrix software, VOSviewer software and CiteSpace software. Visual network analysis consisted of the node size, distance, and colour. Nodes represent specific elements, and the size of the node indicates the quantity or frequency of publication. The larger the node, the more often the element presented. A line between nodes meant that these nodes appeared together in the same article in the 9426 publications. The thicker the line, the more often they appeared together.
A total of 9426 records were initially found based on the search criteria. Annual publications provided an overview of the research field of miRNA in HCC. Figure 1 shows the distribution of publications related to the field of miRNAs in HCC development from 2013 to 2022. Noticeably, the annual output of papers was nearly tripled compared to 2013 (380 publications) in 2017 (1067 publications). A turning point occurred in 2020, and the number of publications began to decline. In 2021 and 2022, the number of publications was 1132 and 931, respectively. The change in the average annual number of citations is also shown in Figure 1. The result reflects that the average yearly citations decreased yearly.
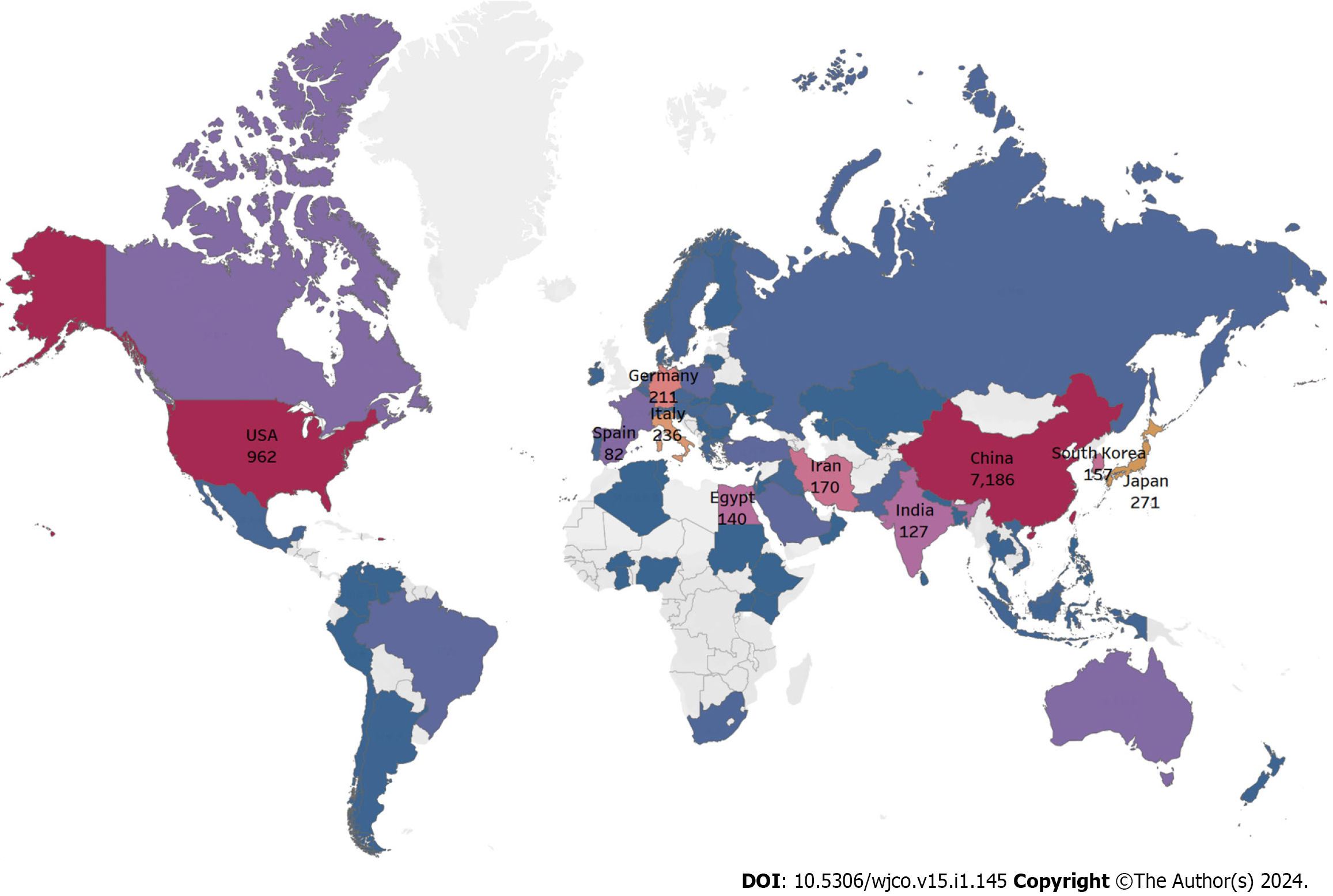
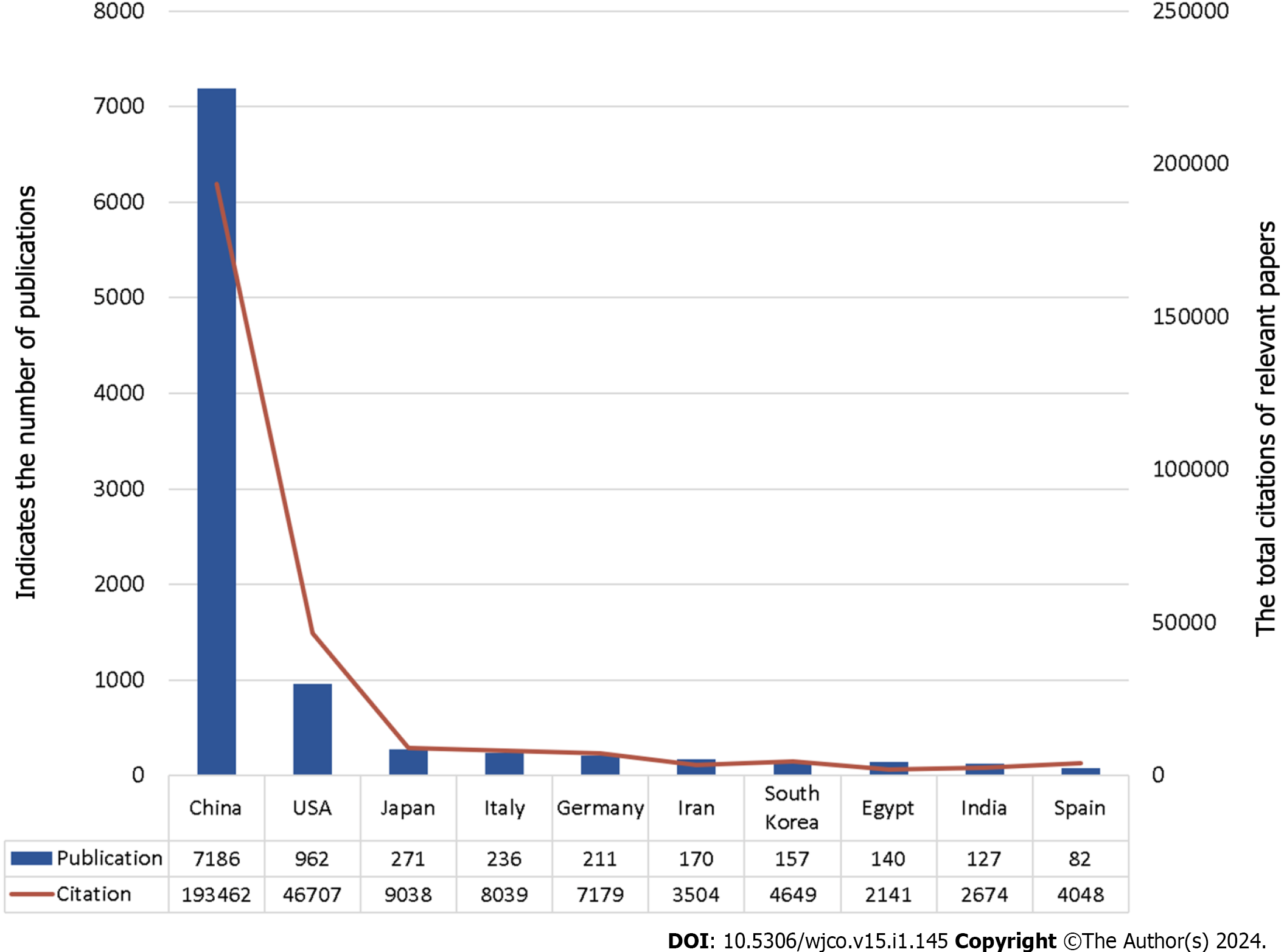
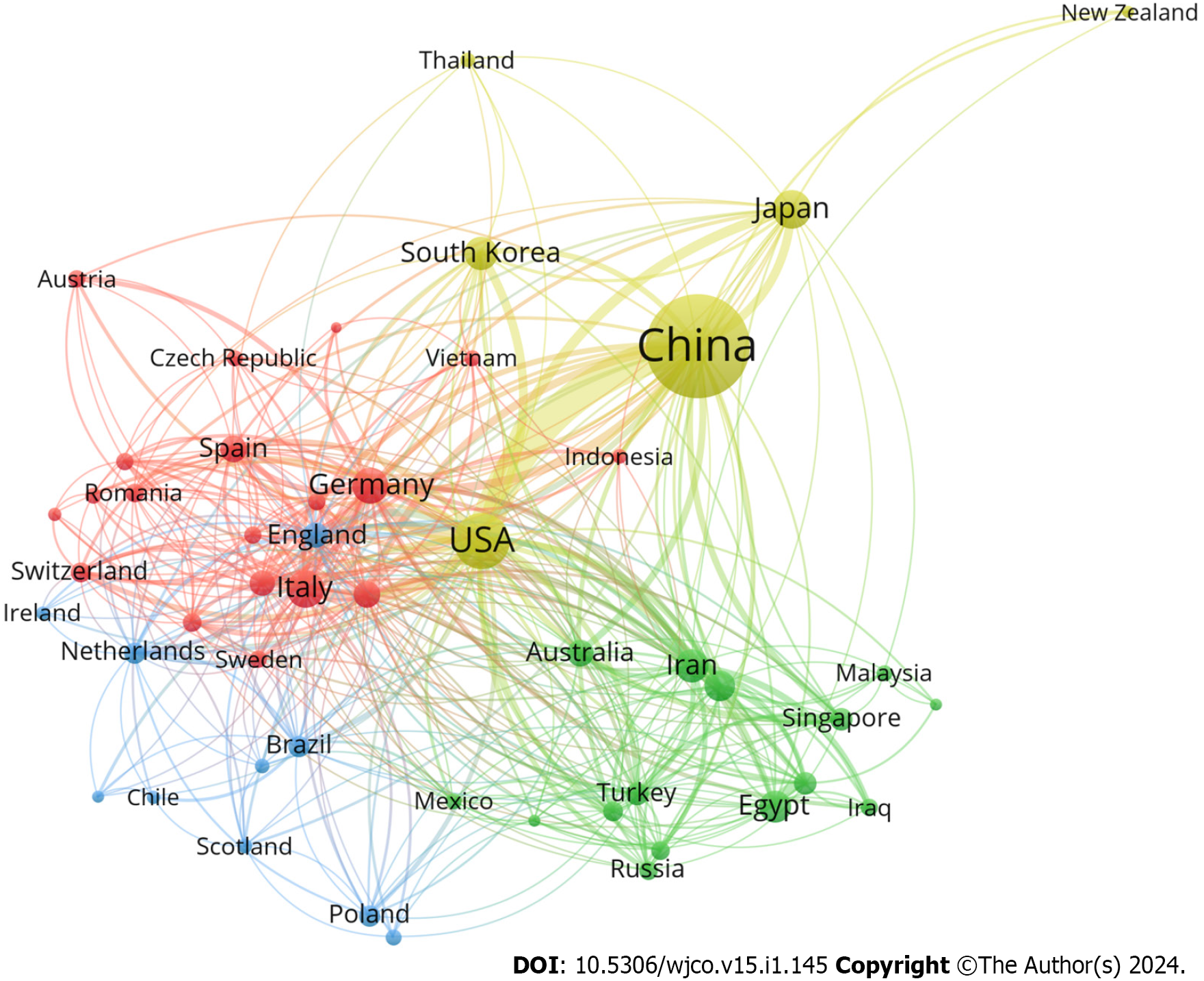
Figure 2 shows the number of articles in each country. Figure 3 shows the top 10 productive countries based on total citations in the research field of miRNA in HCC. China, the United States, Japan, and Italy contributed to the majority of scientific publications. China was the country with the most publications (7186 articles) and had the highest number of total citations (193462), followed by the United States (962 publications and 46707 total citations) and Japan (271 publications and 9038 total citations). According to the network map, the larger a node was, the more articles a country produced, while the thickness of lines between nodes indicated the strength of collaboration (Figure 4). The results show that China was the most prolific country and collaborated substantially with other countries. Table 1 shows the top 10 productive institutions based on citations concerning miRNA research in HCC. Sun Yat Sen University published 346 articles and had the most total citations (12488), followed by Nanjing Med University (337 publications and 11397 total citations) and Fudan University (311 publications and 11612 total citations).
| Institutions | Publications | Citations |
| Sun Yat Sen University | 346 | 12488 |
| Nanjing Med University | 337 | 11397 |
| Fudan University | 311 | 11612 |
| Shanghai Jiao Tong University | 267 | 11300 |
| Xi’an Jiao Tong University | 262 | 7411 |
| Zhejiang University | 261 | 8164 |
| Zhengzhou University | 230 | 5638 |
| Huazhong University Sci & Technol | 209 | 5223 |
| Wuhan University | 189 | 5180 |
| Shandong University | 346 | 4749 |
Table 2 shows the top 10 productive journals, their impact factor and their JCR ranking. Oncotarget (335) has the largest number of publications, followed by Oncology Letters (248) and PLoS One (200). Among the top 10 productive journals, Biomedicine and Pharmacotherapy has an impact factor greater than 5, and two journals (Biomedicine and Pharmacotherapy, Scientific Reports) belong to JCR quartile 1 (Q1).
| Rank | Journal | Publications | 2021 impact factor | 2022 JCR quartile |
| 1 | Oncotarget | 335 | Removed | - |
| 2 | Oncology Letters | 248 | 3.111 | Q4 |
| 3 | PLoS One | 200 | 3.752 | Q2 |
| 4 | Oncology Reports | 198 | 4.136 | Q3 |
| 5 | Molecular Medicine Reports | 189 | 3.432 | Q3 |
| 6 | Oncotargets and Therapy | 188 | 4.345 | Q2 |
| 7 | European Review for Medical and Pharmacological Sciences | 162 | 3.784 | Q2 |
| 8 | Biomedicine and Pharmacotherapy | 160 | 7.419 | Q1 |
| 9 | Tumor Biology | 156 | Removed | - |
| 10 | Scientific Reports | 154 | 4.996 | Q1 |
The author distribution was examined to identify the author with the strongest influence. Supplementary Table 1 shows the top 10 most prolific researchers. The most prolific author was Masaki Tsutomu from Kagawa University (42 publications), followed by Fan Jia from Fudan University (40 publications) and Morishita Asahiro from Kagawa University (40 publications).
We analysed the high-frequency keywords mentioned most frequently in the research articles related to miRNAs in HCC to help us understand the main research hot spots in this field. The top 20 keywords with the highest frequency are shown in Figure 5. “Expression”, “cancer”, “metastasis”, “proliferation”, “invasion”, “growth”, “down-regulation”, “apoptosis”, “progression”, and “cells” comprised the top 10, suggesting that the research on miRNAs focuses on their expression level in HCC and their impact on the biological behaviour of HCC. “Migration”, “breast-cancer”, “cell-proliferation”, “tumor-suppressor”, “promotes”, “epithelial-mesenchymal transition”, “gene”, “colorectal-cancer”, “identification”, and “gastric-cancer” are the next 10 keywords, indicating that the research focuses on the effects and mechanisms of miRNAs on the biological behaviour of HCC, and applies to other tumors.
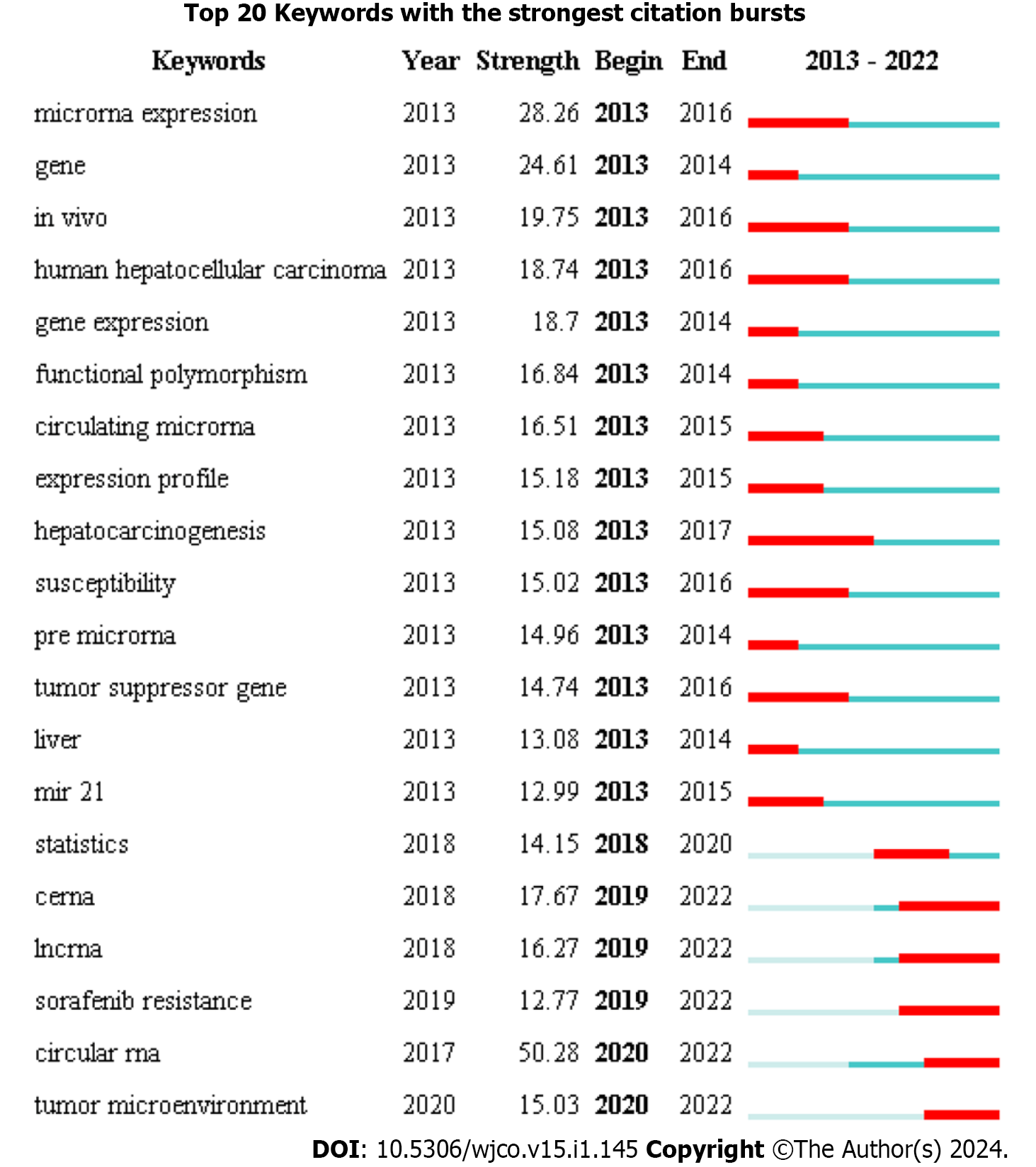
Burst keywords were regarded as an indicator for the frontiers of the specific field during a period of time. We used CiteSpace to explore the 20 keywords with the strongest citation bursts of the last 10 years (Figure 6). From 2013 to 2022, “microRNA expression”, “gene”, “in vivo”, “human hepatocellular carcinoma”, “gene expression”, “functional polymorphism”, “circulating microRNA”, “expression profile”, “hepatocarcinogenesis”, “susceptibility”, “pre microRNA”, “tumor suppressor gene”, “liver” and “mir 21” started to attract attention in the early years (2013–2017). Regarding the latest phase (2018–2022), “statistics”, “ceRNA”, “lncRNA”, “sorafenib resistance”, “circular RNA” and “tumor microenvironment” became trend topics.
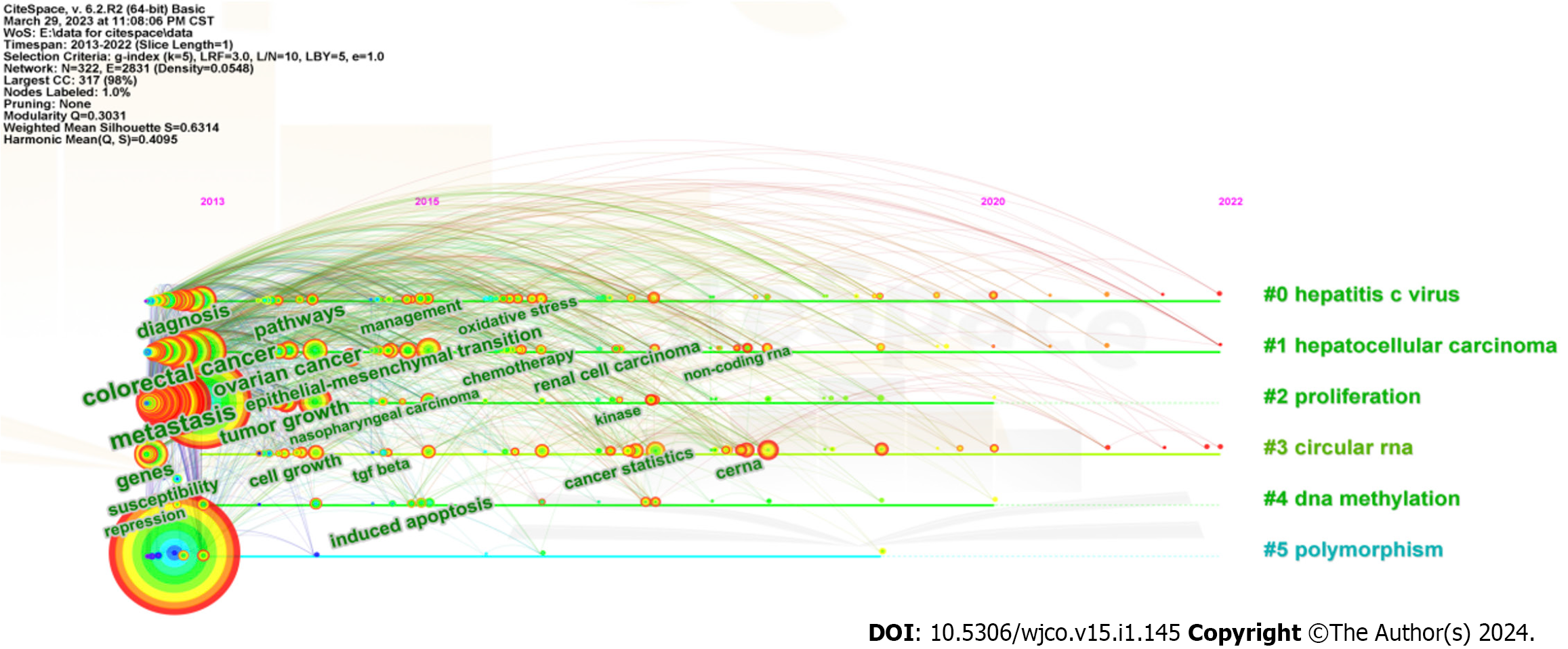
The timeline spectrum of keywords can clearly represent the time span of each cluster and the development trend of a specific cluster, explore the time characteristics of the research field reflected by each cluster, and thereby verify the evolutionary trend of hotspots. According to Figure 7, the articles on miRNA research in the HCC field published from 2013 to 2022 could be categorized into 6 main clusters. The first cluster was “#0 hepatitis c virus”, and followed by “#1 hepatocellular carcinoma”, “#2 proliferation”, “#3 circular RNA”, “#4 DNA methylation” and “#5 polymorphism”. The tree-rings with different sizes on the timeline indicate which keywords have a higher frequency.
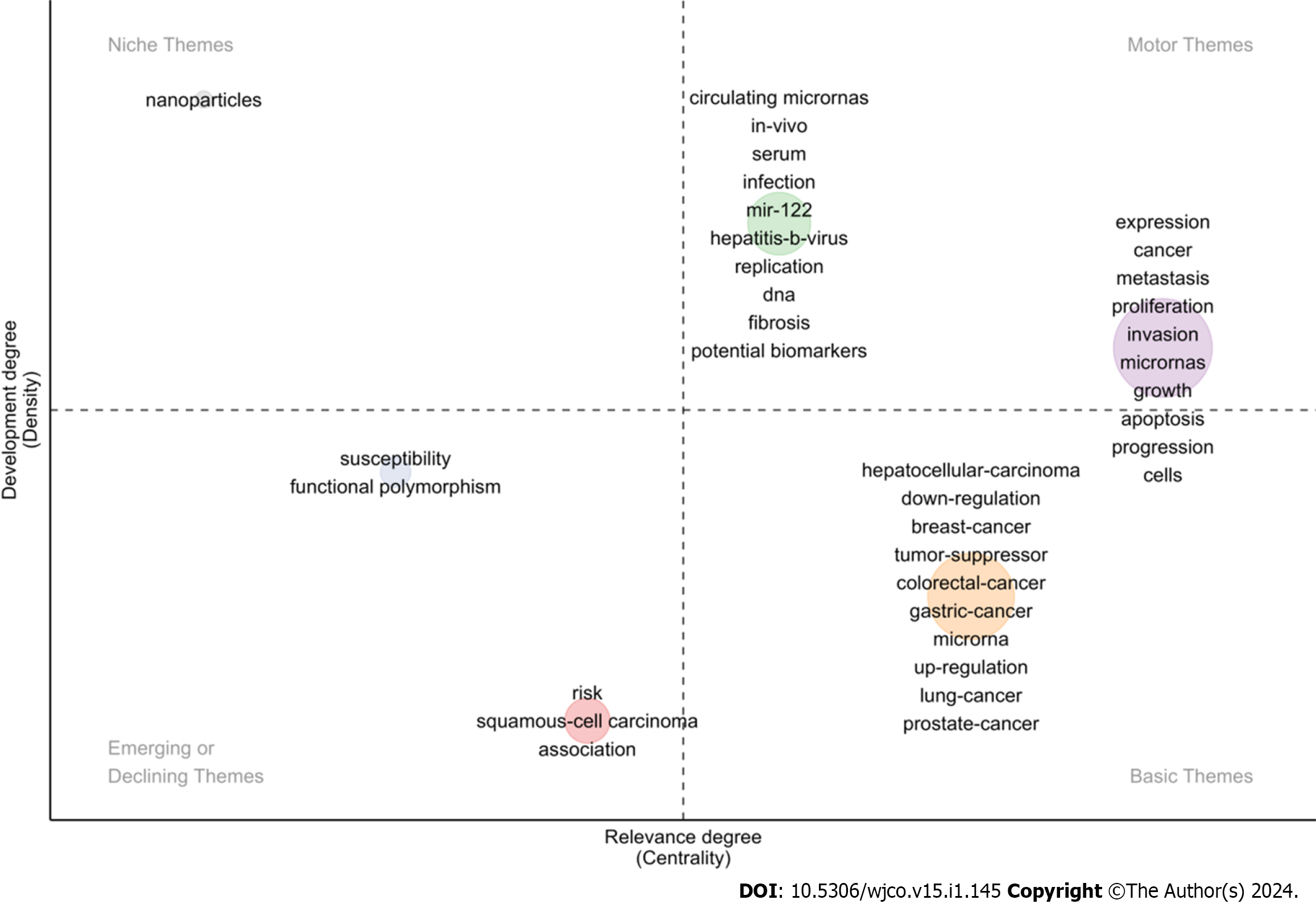
We used Bibliometrix software to draw a thematic map of miRNA in the field of HCC research, and the results indicate that topics such as “expression”, “cancer”, “metastasis”, “proliferation”, “invasion”, “mircoRNAs”, “growth”, “apoptosis”, “progression” and “cells” were in the Motor Themes, as well as the topics such as “circulating microRNAs”, “in-vivo”, “serum”, “infection”, “mir-122”, “hepatitis-b-virus”, “replication”, “DNA”, “fibrosis” and “potential biomarkers”; the topic “nanoparticles” was in the Niche Themes; and the topics such as “susceptibility”, “functional polymorphism” were in the Emerging or Declining Themes, as well as the topics such as “risk”, “squamous-cell carcinoma” and “association”; “hepatocellular-carcinoma”, “down-regulation”, “breast-cancer”, “tumor-suppressor”, “colorectal-cancer”, “gastric-cancer”, “microRNA”, “up-regulation”, “lung-cancer” and “prostate-cancer” were in the Basic Themes (Figure 8).
In this study, we conducted a bibliometric and visualized analysis of the last 10 years focusing on studies based on miRNAs in HCC. We achieve insight into the role of miRNAs in HCC. Annual publications provided an overview of the trends in this area, while distribution by countries, institutions, journals, and authors analysis were outstandingly representative and provided some cooperation situation information.
Keywords analysis could discover emerging trends or study frontiers in the field of miRNAs in HCC. In our study, we used Bibliometrix and CiteSpace software to analyze the active keywords mentioned most frequently in the research papers related to miRNA in HCC. The top 20 keywords with the highest frequency were “expression”, “cancer”, “metastasis”, “proliferation”, “invasion”, “growth”, “down-regulation”, “apoptosis”, “progression”, “cells”, “migration”, “breast-cancer”, “cell-proliferation”, “tumor-suppressor”, “promotes”, “epithelial-mesenchymal transition”, “gene”, “colorectal-cancer”, “identification” and “gastric-cancer”. Based on this result, we conclude that the studies of miRNAs focus on their expression level and impact on the biological behaviour in HCC, as well as the mechanism exploration. Interestingly, other tumor types also appeared in our results. After carefully reviewing the included literature, we found studies on tumors having different miRNA, and the miRNA in one tumor can also be applied to other tumors. This suggests that searching for specific miRNAs in one type of tumor and finding miRNAs with similar effects in multiple types of tumors is routine in tumor research. For example, we found that miR-21-3p plays an important role in the progression of multiple tumors, such as HCC, lung cancer, colorectal cancer, breast cancer and ovarian cancer[22-26].
Keywords bursting analysis shows that in the early years (2013–2017), “microRNA expression”, “gene”, “in vivo”, “human hepatocellular carcinoma”, “gene expression”, “functional polymorphism”, “circulating microRNA”, “expression profile”, “hepatocarcinogenesis”, “susceptibility”, “pre microRNA”, “tumor suppressor gene”, “liver” and “mir 21” started to attract attention. In the latest phase (2018–2022), the hot topics turned to “statistics”, “ceRNA”, “lncRNA”, “sorafenib resistance”, “circular RNA” and “tumor microenvironment”. At the early exploration stage, laboratory techniques such as Northern blotting and quantitative polymerase chain reaction were the main methods for studying miRNA expression. Further exploration of miRNAs was limited to some extent by the limitations of these techniques. The development of sequencing technology has greatly promoted the research of miRNAs in various diseases[27]. For example, Murakami et al[28] used next-generation sequencing and microarrays to evaluate the miRNA expression profile of HCC and detected novel miRNAs that could not be detected by microarrays, providing diagnostic insights for the study of miRNAs in HCC.
Polymorphism is mainly concentrated in the early stages of relevant research in the timeline spectrum, and as a major keyword, it shows significant significance. The same gene or protein exhibits different functions in different environments or cells, indicating polymorphism. Therefore, the high-frequency occurrence periods of polymorphism, gene, and gene expression are basically coincident. This polymorphism is closely related to the risk of HCC occurrence and progression[29-32]. For example, after long-term alcohol exposure, liver cells lacking ALDH2 produce a large amount of harmful oxidative mitochondrial DNA, which can be received by adjacent HCC cells. Subsequently, multiple carcinogenic pathways are activated, thereby promoting alcohol-related liver cancer[33]. With the growing understanding of the mechanism of miRNA action, research on polymorphism in HCC has shifted towards clinical drug therapy research, resulting in a decrease in the frequency of keyword occurrences. The deepening of miRNA research is often accompanied by the synchronization of circulating miRNA research, partly because circulating miRNAs exist in body fluids and have the characteristics of simple collection and detection for cancer prediction, showing high clinical application value and can be used as biomarkers for cancer[34-37].
The high-frequency occurrence period of the keyword susceptibility was almost in the early and middle stages of the entire related research. This is because studying the sensitivity of miRNAs can help determine which miRNAs have therapeutic potential in HCC and provide a basis for individualized treatment of HCC. From the selection of miRNAs related to HCC to the predictive role of miRNAs in HCC, it has important research significance. In the study by Zhang et al[38], it was found that miR-196a2 can be used as a predictive indicator of HCC sensitivity, especially for HBV-related HCC. Previous and mid-term studies have focused on understanding the occurrence and development of HCC; therefore, the keyword hepatocarcinogenesis contains the high-frequency occurrence period of relevant keywords. Existing preclinical studies and clinical trials have discovered different miRNA profiles in hepatocarcinogenesis. For example, miR-122 and miR-34a play important roles in hepatic lipid metabolism, which is associated with HCC[39,40]. miR-122 also has important roles in hepatic inflammation, as do miR-132 and miR-155[41-43]. miR-21 can mediate the activation of hepatic stellate cells via the PTEN/AKT pathway during hepatic fibrosis[44]. Hepatitis B virus (HBV) is the most common cause of HCC in China, and the miR-99 family can promote HBV replication, while miR-199-3p and miR-201 can suppress HBV replication[45,46]. A cohort study suggested that miR-221 and miR-222 play pivotal roles in the progression of liver fibrosis due to persistent hepatitis C virus (HCV) infection[47]. Another study demonstrated that miR-182 is associated with alcoholic hepatitis[48].
In 2013, Liu et al[49] used TargetScan, PicTar, and miRanda prediction algorithms based on the discovery of a new class of small molecule regulatory RNAs closely related to HCC and found that pre-microRNAs play important roles in cell differentiation and mitosis. Subsequent studies have proven that pre-microRNAs can not only mediate RNA splicing and cell signal transduction but also induce G1/S stagnation and may play a role in cancer. Meanwhile, Giacomo Diaz detected a complete set of 2226 human miRNAs, including 1121 pre-miRNAs, 1105 mature miRNAs, and several tumor suppressors, by comparing tumor tissue with a wide range of liver specimens, further confirming that inactivation of tumor suppressors is the cause of most human cancer development[50]. In the following years, a large amount of research was conducted on tumor suppressor factors. Hishida et al[51] invented the triple array analysis method, which identified genes with altered expression through gene expression profiles, single nucleotide polymorphisms, and methylation arrays and subsequently identified tumor suppressor genes. Subsequently, a large number of tumor suppressors were confirmed.
From 2013 to 2015, studies were conducted on miR-21 (one of the most frequently overexpressed small RNAs in cancer). Wagenaar et al[52] used cell culture and found that miR-21 is related to the regulation of cell metabolism and is cell environment dependent. In 2019-2022, Wang, Antonio, and others identified the carcinogenic lncRNA lncUCID and its role in the BRAF pathway. They found that upregulation of lncUCID can enhance CDK6 expression and promote HCC growth. The lncRNA of BRAF is associated with cancer proliferation and tyrosine kinase inhibitor escape in HCC, and inhibiting the use of lncRNA in the BRAF pathway may become a possible therapeutic strategy for HCC. These findings provide insights into the biological function of lncRNAs, the regulatory network of cell cycle control, and the development mechanism of HCC[53,54].
The keyword “sorafenib resistance” appeared frequently in 2019-2022 because sorafenib became the first-line treatment drug for HCC and has been shown to effectively improve the prognosis of patients with advanced HCC by strong evidence and clinical experience[55-60]. However, according to the research of Tang Weiwei and others, the number of patients who can benefit from sorafenib is small (30%), and the population usually develops drug resistance within 6 months. The adverse events found in patients receiving sorafenib treatment mainly include digestive and skin diseases and may even cause hypertension and abdominal pain, leading to treatment interruption. Therefore, it is urgent to study the drug resistance mechanism of sorafenib and more reasonable clinical treatment methods[61]. Studies have explored various relationships between miRNAs and sorafenib. For example, miR-23a-3p contributes to sorafenib resistance in HCC by regulating ferroptosis[62]. miR-10b-3p can be a biomarker for predicting sorafenib efficacy[63]. miR-494-3p promotes sorafenib resistance in HCC cells by targeting PTEN[64]. Wang et al[65] found that compared with patients with HCC treated with sorafenib, patients treated with lenvatinib developed 3 differentially expressed miRNAs, including miR-548ah, miR-888 and miR-196a-1. Wang et al[65] further investigated the adverse events of sorafenib and lenvatinib and found that the patients in the sorafenib group and lenvatinib group developed different frequent symptoms, such as hypertension, diarrhoea and hand-foot skin reactions. At present, systemic therapy has become the standard therapy for unresectable HCC in the middle and late stages. Small molecule antiangiogenic targeted drugs and immune checkpoint inhibitors are the main systemic therapy options. The combination of the two often has a synergistic effect and improves the prognosis of HCC patients. During the use of these systems for treatment, changes in miRNA expression profiles and levels in HCC patients may have important value in predicting drug efficacy, drug-related adverse events and prognosis[66].
From 2020 to 2022, research in the field of miRNAs in HCC began to focus on the “tumor microenvironment”. The tumor microenvironment is a complex environment in which tumor cells survive and develop, including surrounding blood vessels, immune cells, fibroblasts, bone marrow-derived inflammatory cells, various signaling molecules and the extracellular matrix. As the location of tumor cells survival, the tumor microenvironment plays a vital role in the occurrence, development, and metastasis of tumors. The amount of research on the tumor microenvironment in HCC has gradually increased, and some studies have focused on the mechanism, which involves some miRNAs. For example, Zhou et al[67] found that exosomal miR-761 modulated the tumor microenvironment via SOCS2/JAK2/STAT3 pathway-dependent activation of cancer-associated fibroblasts. In the research of Yugawa et al[68], the findings suggested that the loss of antitumoral miR-150-3p in cancer-associated fibroblast-derived exosomes greatly promotes HCC progression. Research on the effect of miRNAs on the tumor microenvironment can provide more therapeutic strategies for HCC.
The thematic map is a two-dimensional map constructed with the density index as the vertical axis and the centrality index as the horizontal axis. Density represents the strength of the connections between basic knowledge units within a single topic. If the density value of a certain topic is higher, it indicates that the maturity of the topic is higher. Centrality represents the strength of the connection between a certain topic and other topics. If the centrality value of a certain topic is higher, it indicates that the topic is closely related to other topics and is at the core of all research topics. According to the density and centrality value, the rectangular coordinate system is divided into four regions: Motor Themes, a core theme with high maturity; Niche Themes, a highly mature isolated theme; Emerging or Declining Themes, new or disappearing themes; Basic Themes, a basic theme with low maturity and some influence but has not been well developed and should continue to be strengthened in the future. According to our results, themes such as “susceptibility”, “functional polymorphism”, “risk”, “squamous-cell carcinoma” and “association” are the new or disappearing themes, while “nanoparticles” is a highly mature but isolated theme. “Expression”, “cancer”, “metastasis”, “proliferation”, “invasion”, “microRNAs”, “growth”, “apoptosis”, “progression”, “cells”, “circulating microRNAs”, “in-vivo”, “serum”, “infection”, “mir-122”, “hepatitis-b-virus”, “replication”, “DNA”, “fibrosis” and “potential biomarkers” are the mainstream themes. “Hepatocellular-carcinoma”, “down-regulation”, “breast-cancer”, “tumor-suppressor”, “colorectal-cancer”, “gastric-cancer”, “microRNA”, “up-regulation”, “lung-cancer” and “prostate-cancer” are the basic themes with low maturity, which have some influence but have not been well developed and should continue to be strengthened in the future.
This study is novel for its comprehensive extraction and evaluation of research outputs from the available global data to explore the relationship between miRNA and HCC. However, there are also some limitations in this study. For example, it did not include all relevant literature and was limited to the Web of Science Core Collection database, which may lead to selection bias. Meanwhile, some recently published high-quality papers might not be highlighted due to low citation frequency because of the short time since their publication.
Our study provides a comprehensive overview of the role of miRNAs in HCC development from 2013 to 2022 based on bibliometric analysis. According to the keywords analysis, we found that the studies of miRNAs focused on their expression level, effects, and mechanisms on the biological behaviour of HCC. Keywords bursting analysis showed that in the early years (2013–2017), “microRNA expression”, “gene expression”, “expression profile”, “functional polymorphism”, “circulating microRNA”, “susceptibility” and “mir 21” started to attract attention. In the latest phase (2018–2022), the hot topics turned to “sorafenib resistance”, “tumor microenvironment” and so on.
Over the past 10 years, studies have increasingly focused on the role of microRNAs (miRNAs) in hepatocellular carcinoma (HCC), generating significant scientific output. The accumulated knowledge needs to be systematically organized in order to improve research efficiency.
The purpose of this article is to make a comprehensive analysis of recent studies concerning miRNAs in HCC.
We aim to systematically analyze the research on miRNAs in HCC in the past decade. A comprehensive analysis of recent research concerning miRNAs in HCC development could provide researchers with a valuable reference for further studies.
This study collected relevant publications from the Web of Science Core Collection database (https://www.webofscience.com/) with a specific search strategy and the limited time frame was from 2013 to 2022. Bibliometrix software, VOSviewer software and CiteSpace software were used to visually analyze the distribution by time, countries, institutions, journals, and authors, as well as the keywords, burst keywords and thematic map.
A total of 9426 publications on this topic were found worldwide. According to the keywords analysis, we found that the studies of miRNAs focused on their expression level, effects, and mechanisms on the biological behaviour of HCC. Keywords bursting analysis showed that in the early years (2013–2017), “microRNA expression”, “gene expression”, “expression profile”, “functional polymorphism”, “circulating microRNA”, “susceptibility” and “mir 21” started to attract attention. In the latest phase (2018–2022), the hot topics turned to “sorafenib resistance”, “tumor microenvironment” and so on.
This study provides a comprehensive overview of the role of miRNAs in HCC development based on bibliometric analysis. The hotspots in this field focus on miRNAs expression level, effects, and mechanisms on the biological behavior of HCC. The frontiers turned to sorafenib resistance, tumor microenvironment and so on.
Future research should focus on the application value of miRNAs as biomarkers in predicting clinical treatment outcomes and prognosis in HCC patients.
We would like to thank “Guangxi Zhuang Autonomous Region Clinical Medicine Research Center for Molecular Pathology and Intelligent Pathology Precision Diagnosis” for providing technical support.
| 1. | Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, Laversanne M, McGlynn KA, Soerjomataram I. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77:1598-1606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 1425] [Article Influence: 356.3] [Reference Citation Analysis (1)] |
| 2. | Li Q, Cao M, Lei L, Yang F, Li H, Yan X, He S, Zhang S, Teng Y, Xia C, Chen W. Burden of liver cancer: From epidemiology to prevention. Chin J Cancer Res. 2022;34:554-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 3. | Wei Q, Zhou H, Hou X, Liu X, Chen S, Huang X, Chen Y, Liu M, Duan Z. Current status of and barriers to the treatment of advanced-stage liver cancer in China: a questionnaire-based study from the perspective of doctors. BMC Gastroenterol. 2022;22:351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345-1362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1284] [Cited by in RCA: 1591] [Article Influence: 397.8] [Reference Citation Analysis (42)] |
| 5. | Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1487] [Cited by in RCA: 1464] [Article Influence: 183.0] [Reference Citation Analysis (0)] |
| 6. | Shi YH, Wen TF, Xiao DS, Dai LB, Song J. Predicting miRNA targets for hepatocellular carcinoma with an integrated method. Transl Cancer Res. 2020;9:1752-1760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 7. | Ho PTB, Clark IM, Le LTT. MicroRNA-Based Diagnosis and Therapy. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 427] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 8. | Chen S, Wang Y, Li D, Wang H, Zhao X, Yang J, Chen L, Guo M, Zhao J, Chen C, Zhou Y, Liang G, Xu L. Mechanisms Controlling MicroRNA Expression in Tumor. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 9. | Zeng Z, Cai J, Liao Y, Sun S, Xie L. Progress in the effect of microRNA-21 on diseases via autophagy. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2022;47:936-941. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 10. | Hamidi AA, Taghehchian N, Basirat Z, Zangouei AS, Moghbeli M. MicroRNAs as the critical regulators of cell migration and invasion in thyroid cancer. Biomark Res. 2022;10:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 11. | Luo Y, Peng L, Shan W, Sun M, Luo L, Liang W. Machine learning in the development of targeting microRNAs in human disease. Front Genet. 2022;13:1088189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 12. | Kousar K, Ahmad T, Abduh MS, Kanwal B, Shah SS, Naseer F, Anjum S. miRNAs in Regulation of Tumor Microenvironment, Chemotherapy Resistance, Immunotherapy Modulation and miRNA Therapeutics in Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 42] [Reference Citation Analysis (0)] |
| 13. | Entezari M, Taheriazam A, Orouei S, Fallah S, Sanaei A, Hejazi ES, Kakavand A, Rezaei S, Heidari H, Behroozaghdam M, Daneshi S, Salimimoghadam S, Mirzaei S, Hashemi M, Samarghandian S. LncRNA-miRNA axis in tumor progression and therapy response: An emphasis on molecular interactions and therapeutic interventions. Biomed Pharmacother. 2022;154:113609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 14. | Yao D, Nong L, Qin M, Wu S, Yao S. Identifying circRNA-miRNA interaction based on multi-biological interaction fusion. Front Microbiol. 2022;13:987930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 15. | Huang T, Wu Z, Zhu S. The roles and mechanisms of the lncRNA-miRNA axis in the progression of esophageal cancer: a narrative review. J Thorac Dis. 2022;14:4545-4559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Nishita-Hiresha V, Varsha R, Jayasuriya R, Ramkumar KM. The role of circRNA-miRNA-mRNA interaction network in endothelial dysfunction. Gene. 2023;851:146950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 17. | Ishaq Y, Ikram A, Alzahrani B, Khurshid S. The Role of miRNAs, circRNAs and Their Interactions in Development and Progression of Hepatocellular Carcinoma: An Insilico Approach. Genes (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 18. | Wang C, Su K, Lin H, Cen B, Zheng S, Xu X. Identification and Verification of a Novel MAGI2-AS3/miRNA-374-5p/FOXO1 Network Associated with HBV-Related HCC. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Hwang H, Chang HR, Baek D. Determinants of Functional MicroRNA Targeting. Mol Cells. 2023;46:21-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 32] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 20. | Wang H, Meng Q, Qian J, Li M, Gu C, Yang Y. Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer. Pharmacol Ther. 2022;234:108123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 21. | Ninkov A, Frank JR, Maggio LA. Bibliometrics: Methods for studying academic publishing. Perspect Med Educ. 2022;11:173-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 484] [Article Influence: 121.0] [Reference Citation Analysis (0)] |
| 22. | Hong Y, Ye M, Wang F, Fang J, Wang C, Luo J, Liu J, Liu L, Zhao Q, Chang Y. MiR-21-3p Promotes Hepatocellular Carcinoma Progression via SMAD7/YAP1 Regulation. Front Oncol. 2021;11:642030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 23. | Zhang YQ, Chen RL, Shang LQ, Yang SM. Nicotine-induced miR-21-3p promotes chemoresistance in lung cancer by negatively regulating FOXO3a. Oncol Lett. 2022;24:260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 24. | Jiao W, Leng X, Zhou Q, Wu Y, Sun L, Tan Y, Ni H, Dong X, Shen T, Liu Y, Li J. Different miR-21-3p isoforms and their different features in colorectal cancer. Int J Cancer. 2017;141:2103-2111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Amirfallah A, Knutsdottir H, Arason A, Hilmarsdottir B, Johannsson OT, Agnarsson BA, Barkardottir RB, Reynisdottir I. Hsa-miR-21-3p associates with breast cancer patient survival and targets genes in tumor suppressive pathways. PLoS One. 2021;16:e0260327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 26. | Fang W, Xia Y. LncRNA HLA-F-AS1 attenuates the ovarian cancer development by targeting miR-21-3p/PEG3 axis. Anticancer Drugs. 2022;33:671-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Lokhande HA. Bioinformatics Analysis of miRNA Sequencing Data. Methods Mol Biol. 2023;2595:225-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 28. | Murakami Y, Tanahashi T, Okada R, Toyoda H, Kumada T, Enomoto M, Tamori A, Kawada N, Taguchi YH, Azuma T. Comparison of hepatocellular carcinoma miRNA expression profiling as evaluated by next generation sequencing and microarray. PLoS One. 2014;9:e106314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (1)] |
| 29. | Rao K, Zheng K, Zhao Q, He J, Zhou B, Hou G, Sha N, Wang W, Yan M, Zhou Y, Jin Y, Jiang Y, Xia Q. The negative effect of G1958A polymorphism on MTHFD1 protein stability and HCC growth. Cell Oncol (Dordr). 2023;46:735-744. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 30. | Ambrozkiewicz F, Trailin A, Červenková L, Vaclavikova R, Hanicinec V, Allah MAO, Palek R, Třeška V, Daum O, Tonar Z, Liška V, Hemminki K. CTNNB1 mutations, TERT polymorphism and CD8+ cell densities in resected hepatocellular carcinoma are associated with longer time to recurrence. BMC Cancer. 2022;22:884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 31. | Mohamed AA, Esmail OE, Ibrahim AMA, Makled S, Al-Hussain E, Elsaid A, Alboraie M, El-Awady RR. The role of PRDM1 gene polymorphism in the progression of hepatocellular carcinoma in Egyptian patients. J Med Virol. 2023;95:e28343. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 32. | Youssef SS, Youness RA, Abbas EAE, Osman NM, ELFiky A, El-Kassas M. miR-516a-3P, a potential circulating biomarker in hepatocellular carcinoma, correlated with rs738409 polymorphism in PNPLA3. Per Med. 2022;19:483-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 33. | Seo W, Gao Y, He Y, Sun J, Xu H, Feng D, Park SH, Cho YE, Guillot A, Ren T, Wu R, Wang J, Kim SJ, Hwang S, Liangpunsakul S, Yang Y, Niu J, Gao B. ALDH2 deficiency promotes alcohol-associated liver cancer by activating oncogenic pathways via oxidized DNA-enriched extracellular vesicles. J Hepatol. 2019;71:1000-1011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 34. | Yu B, Zhou S, Liang H, Ye Q, Wang Y. Development and Validation of a Novel Circulating miRNA-Based Diagnostic Score for Early Detection of Hepatocellular Carcinoma. Dig Dis Sci. 2022;67:2283-2292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 35. | Yousuf T, Dar SB, Bangri SA, Choh NA, Rasool Z, Shah A, Rather RA, Rah B, Bhat GR, Ali S, Afroze D. Diagnostic implication of a circulating serum-based three-microRNA signature in hepatocellular carcinoma. Front Genet. 2022;13:929787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Ding LH, Fallgren CM, Yu Y, McCarthy M, Edmondson EF, Ullrich RL, Weil MM, Story MD. Orthologs of human circulating miRNAs associated with hepatocellular carcinoma are elevated in mouse plasma months before tumour detection. Sci Rep. 2022;12:10927. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 37. | Tork ASE, Kamel AAF, Zaki MA, Abo El-Wafa RAH, El-Assar OS, Ibrahim Abdelkarem OA. Circulating MiRNA-373 as a Predictor of Response to Super-selective Transarterial Chemoembolization Bridging Therapy in Hepatocellular Carcinoma Patients Awaiting Liver Transplantation. Asian Pac J Cancer Prev. 2023;24:291-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 38. | Zhang Q, Xu X, Wu M, Qin T, Wu S, Liu H. MiRNA Polymorphisms and Hepatocellular Carcinoma Susceptibility: A Systematic Review and Network Meta-Analysis. Front Oncol. 2020;10:562019. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 39. | Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810-1820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 583] [Cited by in RCA: 557] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 40. | Ding J, Li M, Wan X, Jin X, Chen S, Yu C, Li Y. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci Rep. 2015;5:13729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 217] [Cited by in RCA: 216] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 41. | Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871-2883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 638] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 42. | Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol. 2009;23:1876-1884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 186] [Article Influence: 10.9] [Reference Citation Analysis (1)] |
| 43. | Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56:1946-1957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 532] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 44. | Wei J, Feng L, Li Z, Xu G, Fan X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed Pharmacother. 2013;67:387-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 101] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 45. | Lin Y, Deng W, Pang J, Kemper T, Hu J, Yin J, Zhang J, Lu M. The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy. Cell Microbiol. 2017;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 46. | Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 184] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 47. | Abdel-Al A, El-Ahwany E, Zoheiry M, Hassan M, Ouf A, Abu-Taleb H, Abdel Rahim A, El-Talkawy MD, Zada S. miRNA-221 and miRNA-222 are promising biomarkers for progression of liver fibrosis in HCV Egyptian patients. Virus Res. 2018;253:135-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 48. | Blaya D, Coll M, Rodrigo-Torres D, Vila-Casadesús M, Altamirano J, Llopis M, Graupera I, Perea L, Aguilar-Bravo B, Díaz A, Banales JM, Clària J, Lozano JJ, Bataller R, Caballería J, Ginès P, Sancho-Bru P. Integrative microRNA profiling in alcoholic hepatitis reveals a role for microRNA-182 in liver injury and inflammation. Gut. 2016;65:1535-1545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 49. | Liu RY, Diao CF, Zhang Y, Wu N, Wan HY, Nong XY, Liu M, Tang H. miR-371-5p down-regulates pre mRNA processing factor 4 homolog B (PRPF4B) and facilitates the G1/S transition in human hepatocellular carcinoma cells. Cancer Lett. 2013;335:351-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 50. | Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, Farci P. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133:816-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 51. | Hishida M, Nomoto S, Inokawa Y, Hayashi M, Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S, Nakayama G, Fujii T, Sugimoto H, Koike M, Fujiwara M, Takeda S, Kodera Y. Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis. Int J Oncol. 2013;43:88-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 52. | Wagenaar TR, Zabludoff S, Ahn SM, Allerson C, Arlt H, Baffa R, Cao H, Davis S, Garcia-Echeverria C, Gaur R, Huang SM, Jiang L, Kim D, Metz-Weidmann C, Pavlicek A, Pollard J, Reeves J, Rocnik JL, Scheidler S, Shi C, Sun F, Tolstykh T, Weber W, Winter C, Yu E, Yu Q, Zheng G, Wiederschain D. Anti-miR-21 Suppresses Hepatocellular Carcinoma Growth via Broad Transcriptional Network Deregulation. Mol Cancer Res. 2015;13:1009-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 53. | Gnoni A, Licchetta A, Memeo R, Argentiero A, Solimando AG, Longo V, Delcuratolo S, Brunetti O. Role of BRAF in Hepatocellular Carcinoma: A Rationale for Future Targeted Cancer Therapies. Medicina (Kaunas). 2019;55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 54. | Wang YL, Liu JY, Yang JE, Yu XM, Chen ZL, Chen YJ, Kuang M, Zhu Y, Zhuang SM. Lnc-UCID Promotes G1/S Transition and Hepatoma Growth by Preventing DHX9-Mediated CDK6 Down-regulation. Hepatology. 2019;70:259-275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 55. | Zhang Z, He CZ, Qin YQ, Liao JJ, Huang ST, Mo S, Li HM, Lin JY. Exploring the mechanism of resistance to sorafenib in two hepatocellular carcinoma cell lines. Aging (Albany NY). 2020;12:24255-24269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 56. | Lin Z, Xia S, Liang Y, Ji L, Pan Y, Jiang S, Wan Z, Tao L, Chen J, Lin C, Liang X, Xu J, Cai X. LXR activation potentiates sorafenib sensitivity in HCC by activating microRNA-378a transcription. Theranostics. 2020;10:8834-8850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 57. | Xu Y, Huang J, Ma L, Shan J, Shen J, Yang Z, Liu L, Luo Y, Yao C, Qian C. MicroRNA-122 confers sorafenib resistance to hepatocellular carcinoma cells by targeting IGF-1R to regulate RAS/RAF/ERK signaling pathways. Cancer Lett. 2016;371:171-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 140] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 58. | Ji L, Lin Z, Wan Z, Xia S, Jiang S, Cen D, Cai L, Xu J, Cai X. miR-486-3p mediates hepatocellular carcinoma sorafenib resistance by targeting FGFR4 and EGFR. Cell Death Dis. 2020;11:250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 59. | Chen MY, Yadav VK, Chu YC, Ong JR, Huang TY, Lee KF, Lee KH, Yeh CT, Lee WH. Correction: Chen et al Hydroxychloroquine (HCQ) Modulates Autophagy and Oxidative DNA Damage Stress in Hepatocellular Carcinoma to Overcome Sorafenib Resistance via TLR9/SOD1/hsa-miR-30a-5p/Beclin-1 Axis. Cancers 2021, 13, 3227. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 60. | Pratama MY, Pascut D, Massi MN, Tiribelli C. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann Transl Med. 2019;7:577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 61. | Tang W, Chen Z, Zhang W, Cheng Y, Zhang B, Wu F, Wang Q, Wang S, Rong D, Reiter FP, De Toni EN, Wang X. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduct Target Ther. 2020;5:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 797] [Article Influence: 132.8] [Reference Citation Analysis (1)] |
| 62. | Lu Y, Chan YT, Tan HY, Zhang C, Guo W, Xu Y, Sharma R, Chen ZS, Zheng YC, Wang N, Feng Y. Epigenetic regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates sorafenib resistance in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2022;41:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 156] [Article Influence: 39.0] [Reference Citation Analysis (1)] |
| 63. | Shao YY, Chen PS, Lin LI, Lee BS, Ling A, Cheng AL, Hsu C, Ou DL. Low miR-10b-3p associated with sorafenib resistance in hepatocellular carcinoma. Br J Cancer. 2022;126:1806-1814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 64. | Gao Y, Yin Z, Qi Y, Peng H, Ma W, Wang R, Li W. Golgi phosphoprotein 3 promotes angiogenesis and sorafenib resistance in hepatocellular carcinoma via upregulating exosomal miR-494-3p. Cancer Cell Int. 2022;22:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 65. | Wang L, Wang L, Xiao B, Cui M, Zhang B. Differences Between Sorafenib and Lenvatinib Treatment from Genetic and Clinical Perspectives for Patients with Hepatocellular Carcinoma. Med Sci Monit. 2022;28:e934936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 66. | Li J, Bao H, Huang Z, Liang Z, Wang M, Lin N, Ni C, Xu Y. Little things with significant impact: miRNAs in hepatocellular carcinoma. Front Oncol. 2023;13:1191070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 67. | Zhou XH, Xu H, Xu C, Yan YC, Zhang LS, Sun Q, Wang WL, Shi YJ. Hepatocellular carcinoma-derived exosomal miRNA-761 regulates the tumor microenvironment by targeting the SOCS2/JAK2/STAT3 pathway. World J Emerg Med. 2022;13:379-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 68. | Yugawa K, Yoshizumi T, Mano Y, Itoh S, Harada N, Ikegami T, Kohashi K, Oda Y, Mori M. Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur J Surg Oncol. 2021;47:384-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Beenet L, United States S-Editor: Liu JH L-Editor: A P-Editor: Zhang XD