©The Author(s) 2025.
World J Clin Oncol. Nov 24, 2025; 16(11): 112514
Published online Nov 24, 2025. doi: 10.5306/wjco.v16.i11.112514
Published online Nov 24, 2025. doi: 10.5306/wjco.v16.i11.112514
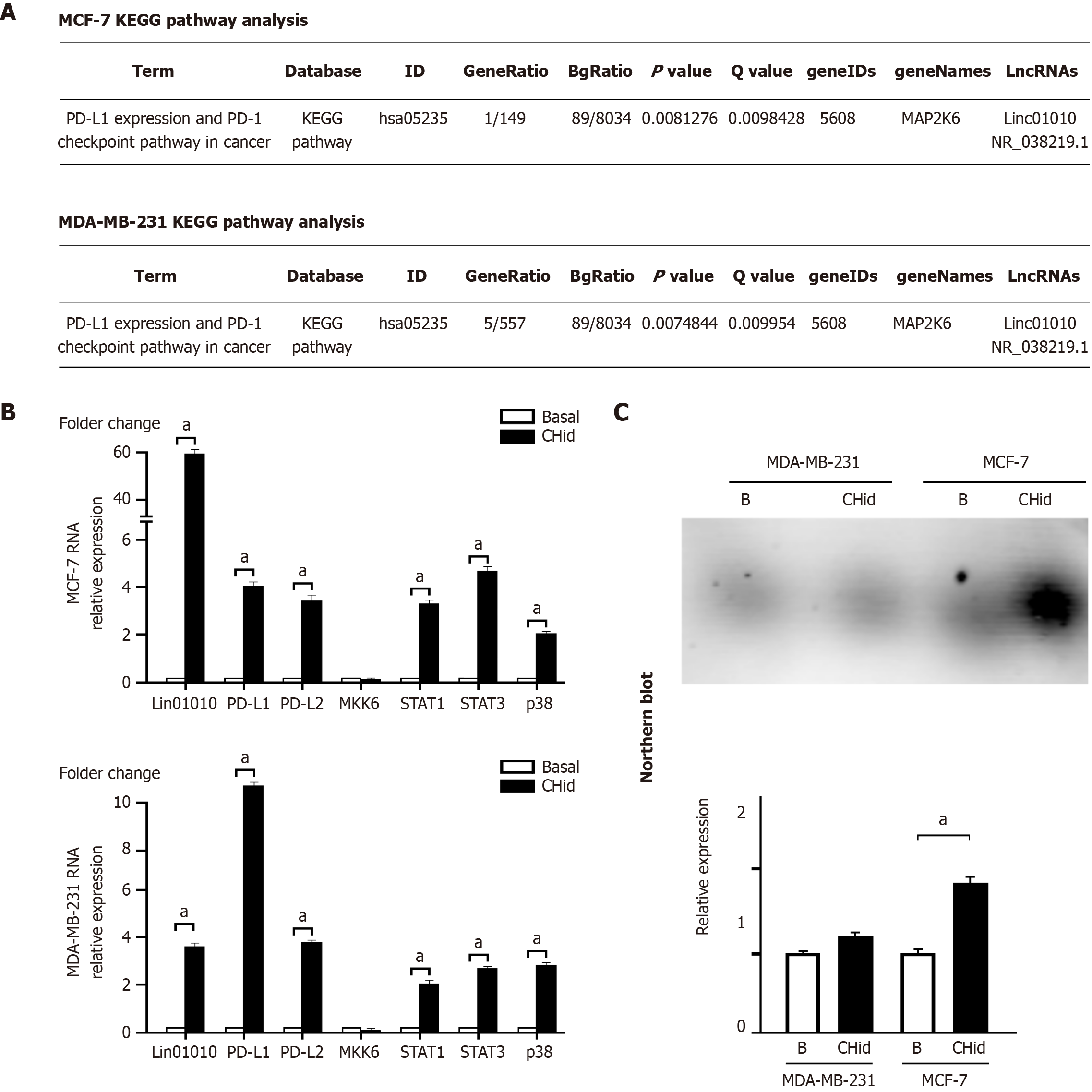
Figure 1 The accumulation of Linc01010 induced by Chidamide was associated with the activation of the mitogen-activated protein kinases pathway and the initiation of the programmed death-1/programmed death-ligand 1 checkpoint.
A: According to our previous RNA high-throughput sequencing data, we applied Kyoto Encyclopedia of Genes and Genomes analysis to try to reveal the relevant signaling pathways; B: The quantitative polymerase chain reaction was used to investigate the mRNA changes of related factors in breast cancer cells; C: Northern blot was utilized to investigate the actual RNA amount of Linc01010 in Chidamide-treated cells. The mean ± SD for three independent experiments and analyzed using Statistical Package for Social Science software, 20 μM Chidamide vs Basal at aP < 0.05. CHid: Chidamide; KEGG: Kyoto Encyclopedia of Genes and Genomes; LncRNAs: Long chain non-coding RNAs; MKK6: Mitogen-activated protein kinase kinase 6; PD-L: Programmed death-ligand; PD-1: Programmed death-1.
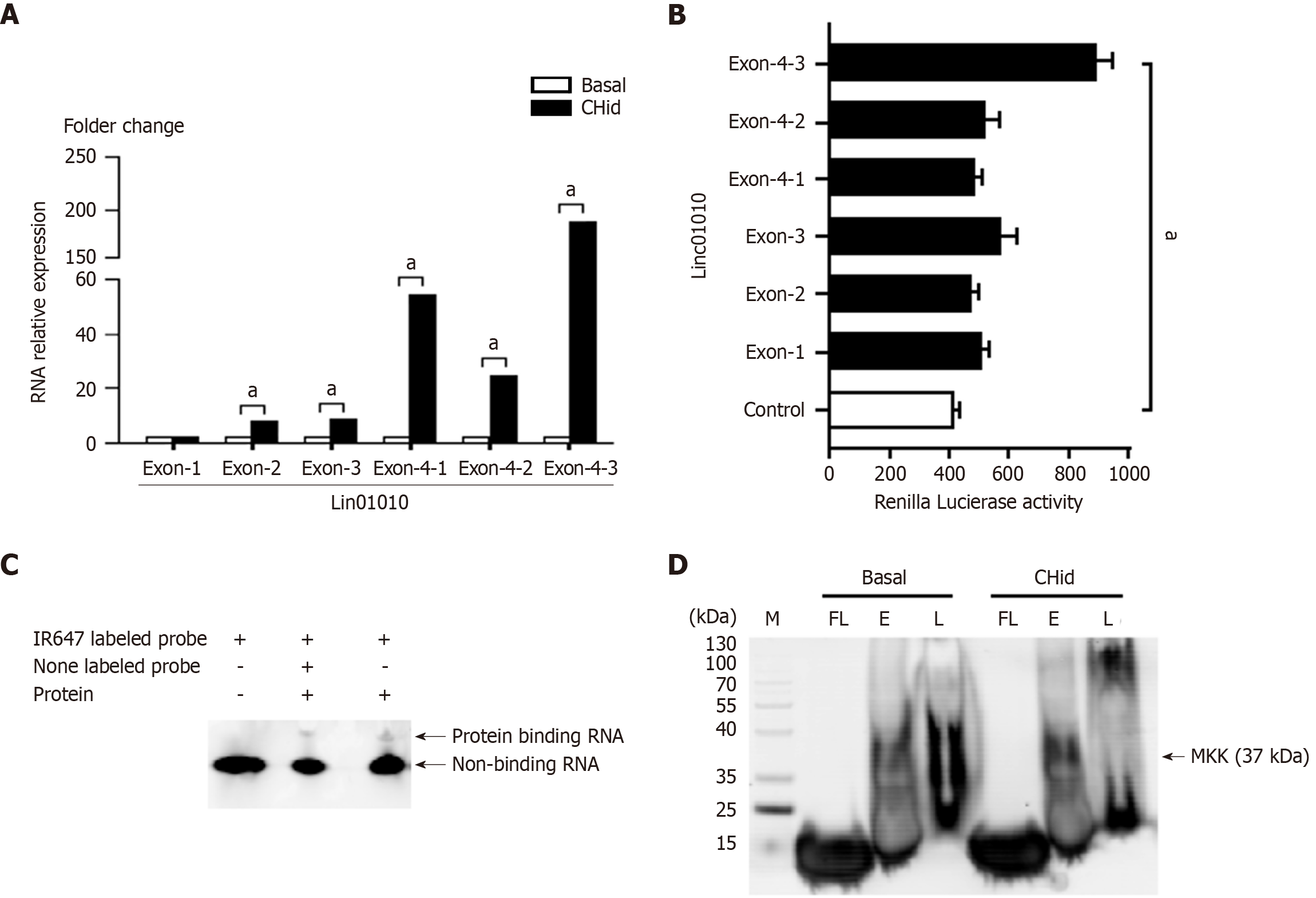
Figure 2 Linc01010 exercised its regulatory goal by targeting binding to mitogen-activated protein kinase kinase 6 protein through specific exon domains.
A and B: Quantitative polymerase chain reaction and luciferase assay for MCF-7 cells were conducted to reveal the functional localization of Linc01010 exon components in breast cancer cells; C and D: Electrophoretic mobility shift assays and RNA-Protein Pull-Down Assay were utilized to investigate evidence of Linc01010 binding to target protein in breast cancer cells. The mean ± SD for three independent experiments and analyzed using Statistical Package for Social Science software, 20 μM Chidamide vs Basal at aP < 0.05. CHid: Chidamide; MKK: Mitogen-activated protein kinase kinase.
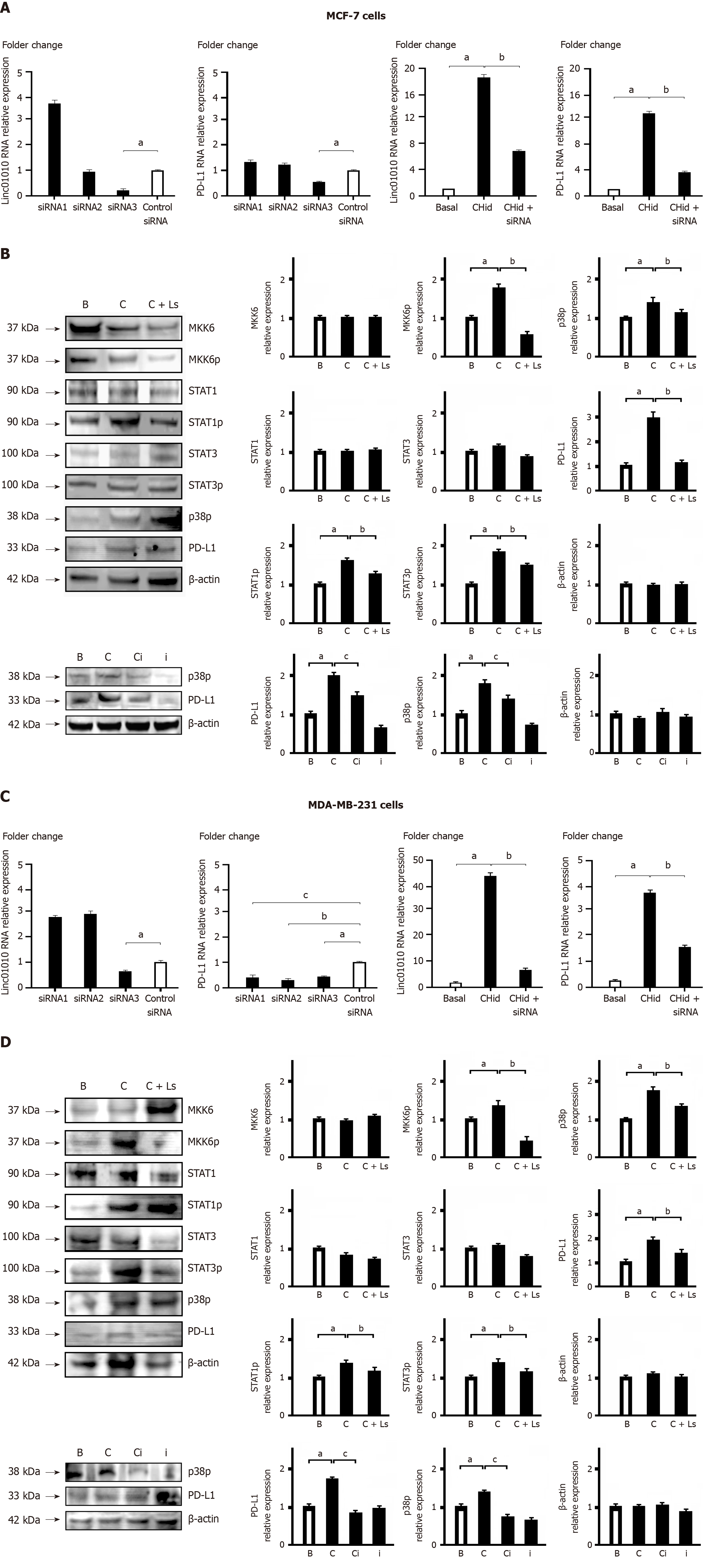
Figure 3 The phosphorylation alteration of key factors in the mitogen-activated protein kinases signaling involved in the Linc01010-mitogen-activated protein kinase kinase 6 interaction.
A: Silence Linc01010 performance in MCF-7 cells; B: Linc01010-mitogen-activated protein kinase kinase 6 (MKK6) in MCF-7 cells; C: Silence Linc01010 performance in MDA-MB-231 cells; D: Linc01010-MKK6 in MDA-MB-231 cells. After synthesizing and transfecting three oligonucleotides based on the sequence of Linc01010, we screened the small interfering RNA (siRNA) molecules to effectively silence Linc01010 performance in MCF-7 and MDA-MB-231 cells. Western blot was examined the alterations in key factors of the signal transduction pathway during the interaction of Linc01010-MKK6 in MCF-7 and MDA-MB-231 cells. The mean ± SD for three independent experiments and analyzed using Statistical Package for Social Science software. In A and C: Specific siRNA vs control siRNA at aP < 0.05, 20 μM Chidamide and specific siRNA vs 20 μM Chidamide at bP < 0.05, 20 μM Chidamide and p38 inhibitor vs 20 μM Chidamide at cP < 0.05. In B and D: 20 μM Chidamide vs Basal at aP < 0.05, 20 μM Chidamide and Linc01010 siRNA vs 20 μM Chidamide at bP < 0.05, 20 μM Chidamide and p38 inhibitor vs 20 μM Chidamide at cP < 0.05. CHid: Chidamide; MKK6: Mitogen-activated protein kinase kinase 6; PD-L: Programmed death-ligand; siRNA: Small interfering RNA.
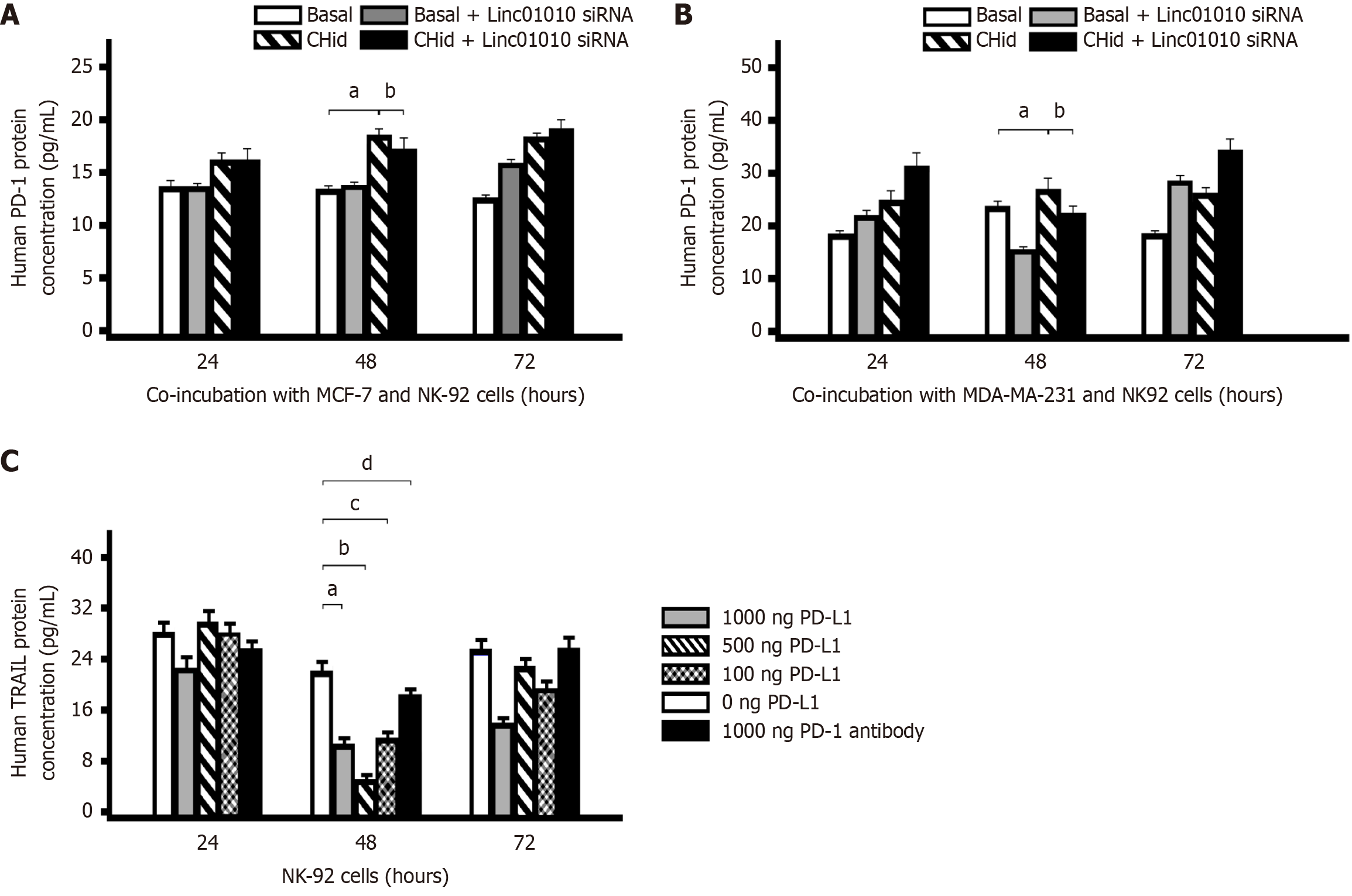
Figure 4 Chidamide induced programmed death-1 expression regulated tumor necrosis factor-related apoptosis-inducing ligand secretion in natural killer cells.
A and B: Content of programmed death-1 (PD-1) in the cell lysate; C: An impact on the secretion of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) in natural killer (NK) cells. First, a co-culture model was adopted that the breast cancer MCF-7 or MDA-MB-231 cells treated with 20 μM Chidamide were cultured with NK-92 cells for 24 hours, 48 hours and 72 hours respectively, after transfecting with Linc01010 specific small interfering RNA (siRNA). We measured the content of PD-1 in the cell lysate with enzyme-linked immunosorbent assay (ELISA) to screen the best time for Chidamide to induce PD-1 expression. Next, ELISA assay was determined whether programmed death-ligand 1 proteins had an impact on the secretion of TRAIL in NK cells. The mean ± SD for three independent experiments and analyzed using Statistical Package for Social Science software. In A and B: 20 μM Chidamide vs Basal at aP < 0.05, 20 μM Chidamide and Linc01010 siRNA vs 20 μM Chidamide at bP < 0.05. In C: 1000 ng PD-L1 vs 0 ng PD-L1 at aP < 0.05, 500 ng PD-L1 vs 0 ng PD-L1 at bP <0.05, 100 ng PD-L1 vs 0 ng PD-L1 at cP <0.05, 1000 ng PD-1 antibody vs 0 ng PD-L1 at dP <0.05. CHid: Chidamide; NK: Natural killer; PD-L1: Programmed death-ligand 1; PD-1: Programmed death-1; TRAIL: Tumor necrosis factor-related apoptosis-inducing ligand.
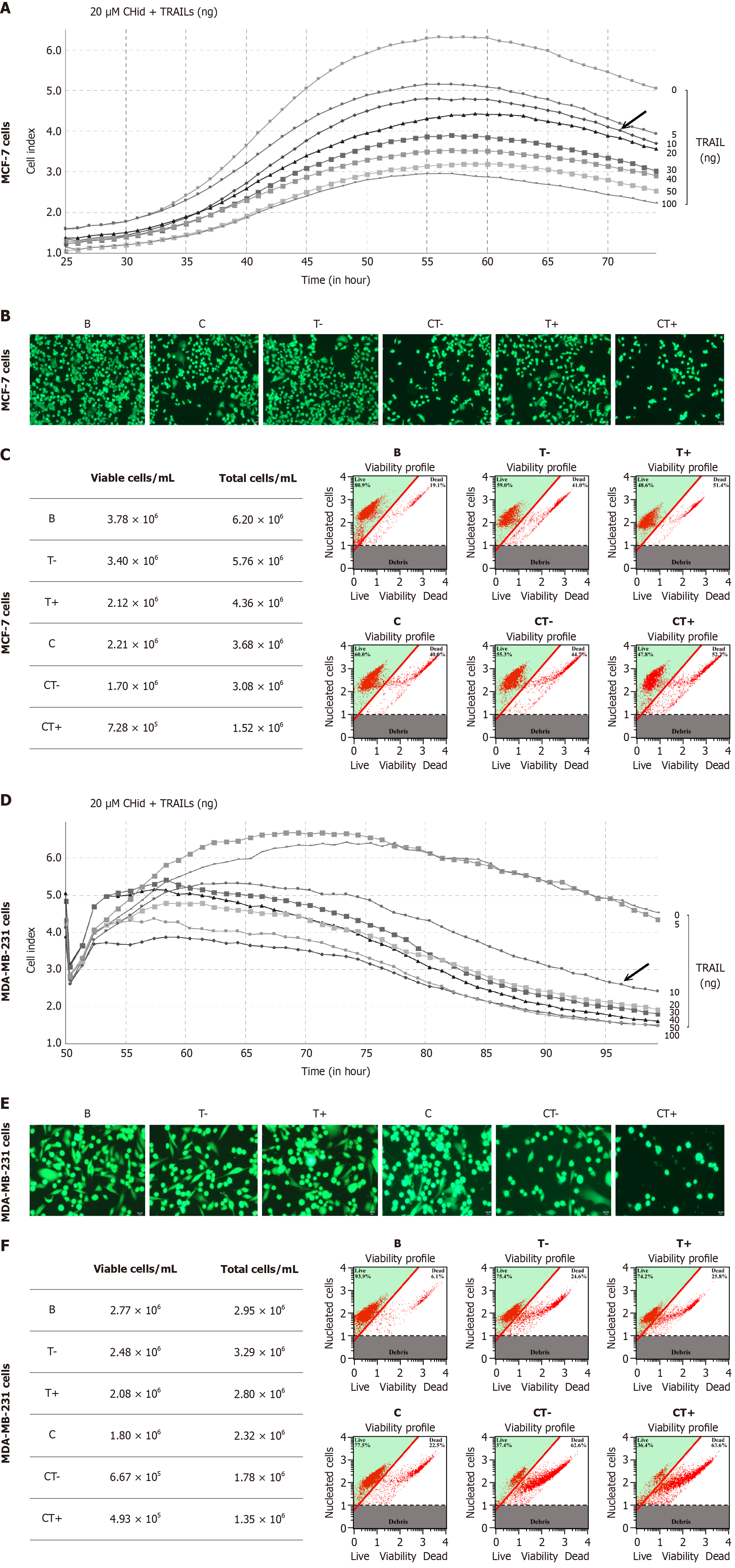
Figure 5 Exogenous secreted tumor necrosis factor-related apoptosis-inducing ligand could enhance the pharmacological effect of Chidamide on the growth of breast cancer cells.
A: Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) (0-100 ng/mL) and 20 μM Chidamide were treated with breast cancer MCF-7; B: Percentage of viable cells for MCF-7 cells; C: The pharmacological effects of different doses of TRAIL and Chidamide for MCF-7 cells; D: TRAIL (0-100 ng/mL) and 20 μM Chidamide were treated with breast cancer MDA-MB-231 cells; E: Percentage of viable cells for MDA-MB-231 cells; F: The pharmacological effects of different doses of TRAIL and Chidamide for MDA-MB-231 cells. Different doses of TRAIL (0-100 ng/mL) and 20 μM Chidamide were treated with breast cancer MCF-7 or MDA-MB-231 cells, and then detected the growth of breast cancer cells in the drug treatment environment with a cell dynamic analysis device. Cell viability assay was performed by the Muse Count and Viability reagent (Millipore) following the manufacturer’s protocols. The results were obtained with Muse Count and Viability software module and the statistics were shown the percentage of viable cells. Fluorescence microscopy was to confirm the pharmacological effects of different doses of TRAIL and Chidamide. The mean ± SD for three independent experiments and analyzed using Statistical Package for Social Science software. CHid: Chidamide; TRAIL: Tumor necrosis factor-related apoptosis-inducing ligand.
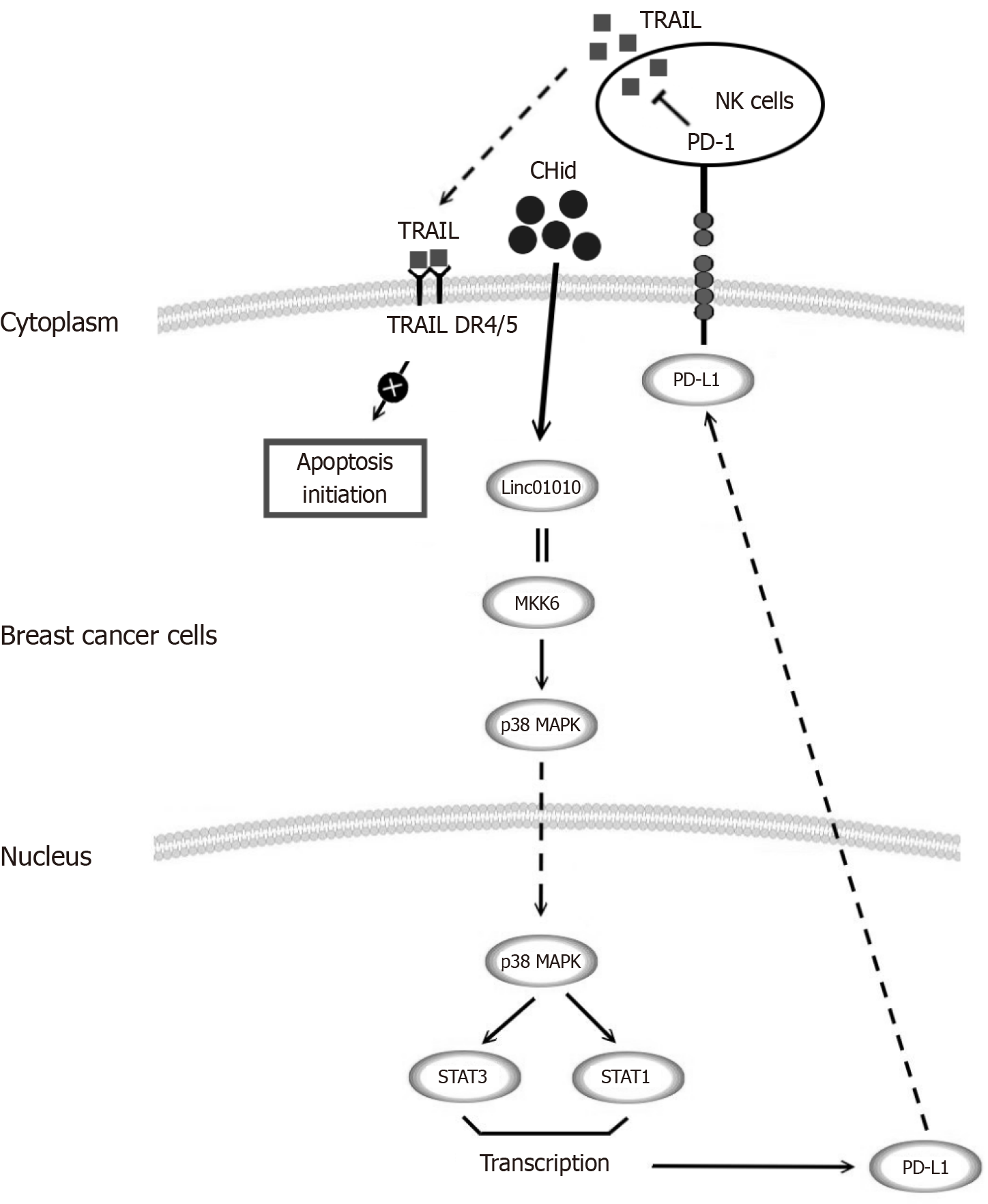
Figure 6 Schematic representation of tumor microenvironment model with function of Linc01010-mitogen-activated protein kinase kinase 6-programmed death-ligand 1 with breast cancer cells and natural killer cells.
We investigated the mechanism of long chain non-coding RNA Linc01010 when provoked by Chidamide with mitogen-activated protein kinase kinase 6 interaction, and its ability to induce the p38-mitogen-activated protein kinases pathway, resulting in an increase in downstream programmed death-ligand 1 (PD-L1). The increase of PD-L1 reduced the secretion of tumor necrosis factor-related apoptosis-inducing ligand by natural killer cells in the tumor microenvironment through programmed death-1, and ultimately moderated the pharmacological effect of Chidamide on breast cancer cells. The results obtained from this study can provide a strong basis for enhancing the effectiveness of current anti-breast cancer medications and identifying new targets for drug resistance. CHid: Chidamide; MAPK: Mitogen-activated protein kinases; MKK6: Mitogen-activated protein kinase kinase 6; NK: Natural killer; PD-L1: Programmed death-ligand 1; PD-1: Programmed death-1; TRAIL: Tumor necrosis factor-related apoptosis-inducing ligand.
- Citation: Han H, Guo XY, Wen JX, Zhao XM, Zhou WQ. Immune regulation of Chidamide-induced Linc01010 accumulation in breast cancer cell death. World J Clin Oncol 2025; 16(11): 112514
- URL: https://www.wjgnet.com/2218-4333/full/v16/i11/112514.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i11.112514