©The Author(s) 2025.
World J Radiol. Dec 28, 2025; 17(12): 112911
Published online Dec 28, 2025. doi: 10.4329/wjr.v17.i12.112911
Published online Dec 28, 2025. doi: 10.4329/wjr.v17.i12.112911
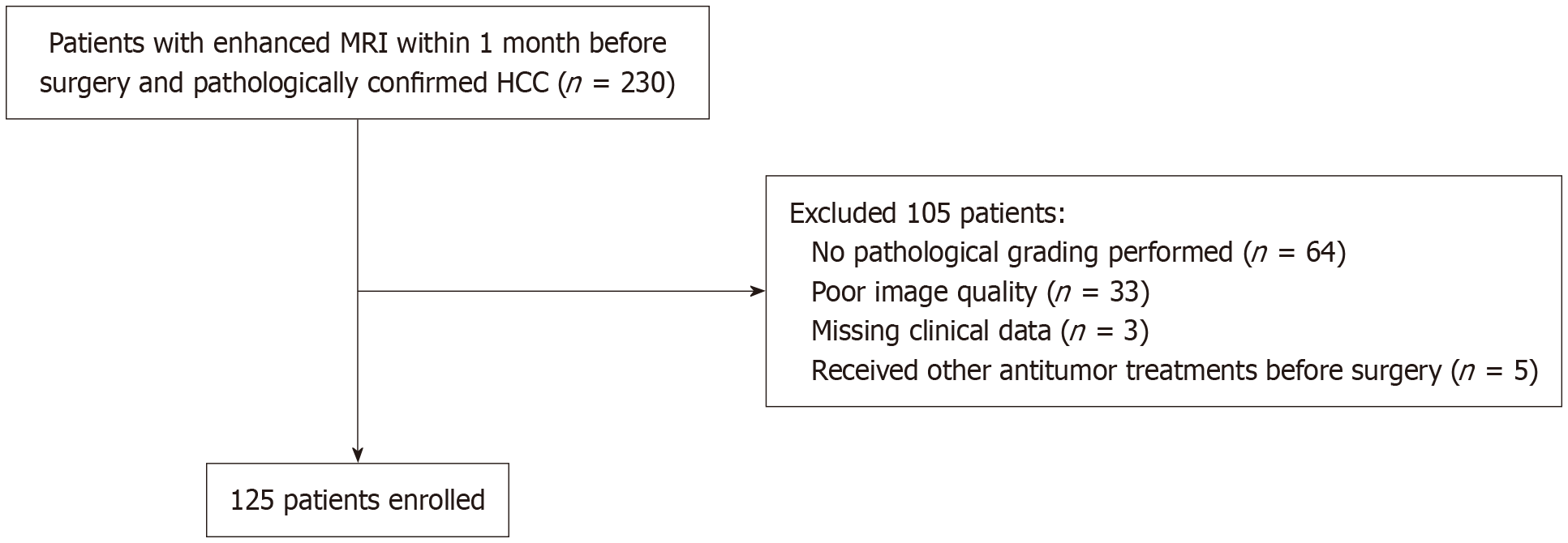
Figure 1 Flowchart of patient enrollment.
MRI: Magnetic resonance imaging; HCC: Hepatocellular carcinoma.
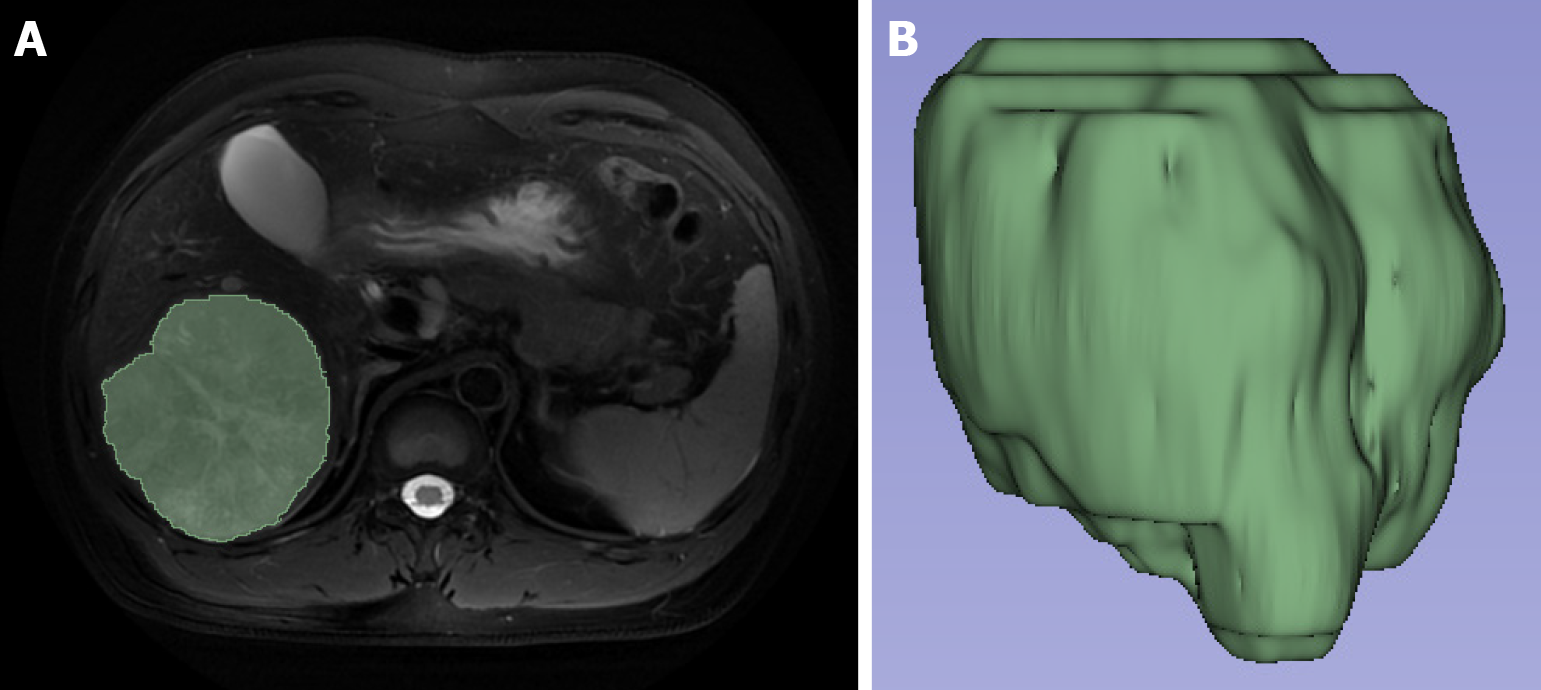
Figure 2 Target volume sketch and 3D volumetric regions of interest view.
A: The volumetric regions of interest were obtained by manually sketching the region of interest layer by layer on the fat-suppressed T2-weighted imaging image; B: The 3D view was automatically generated from the volumetric regions of interest.
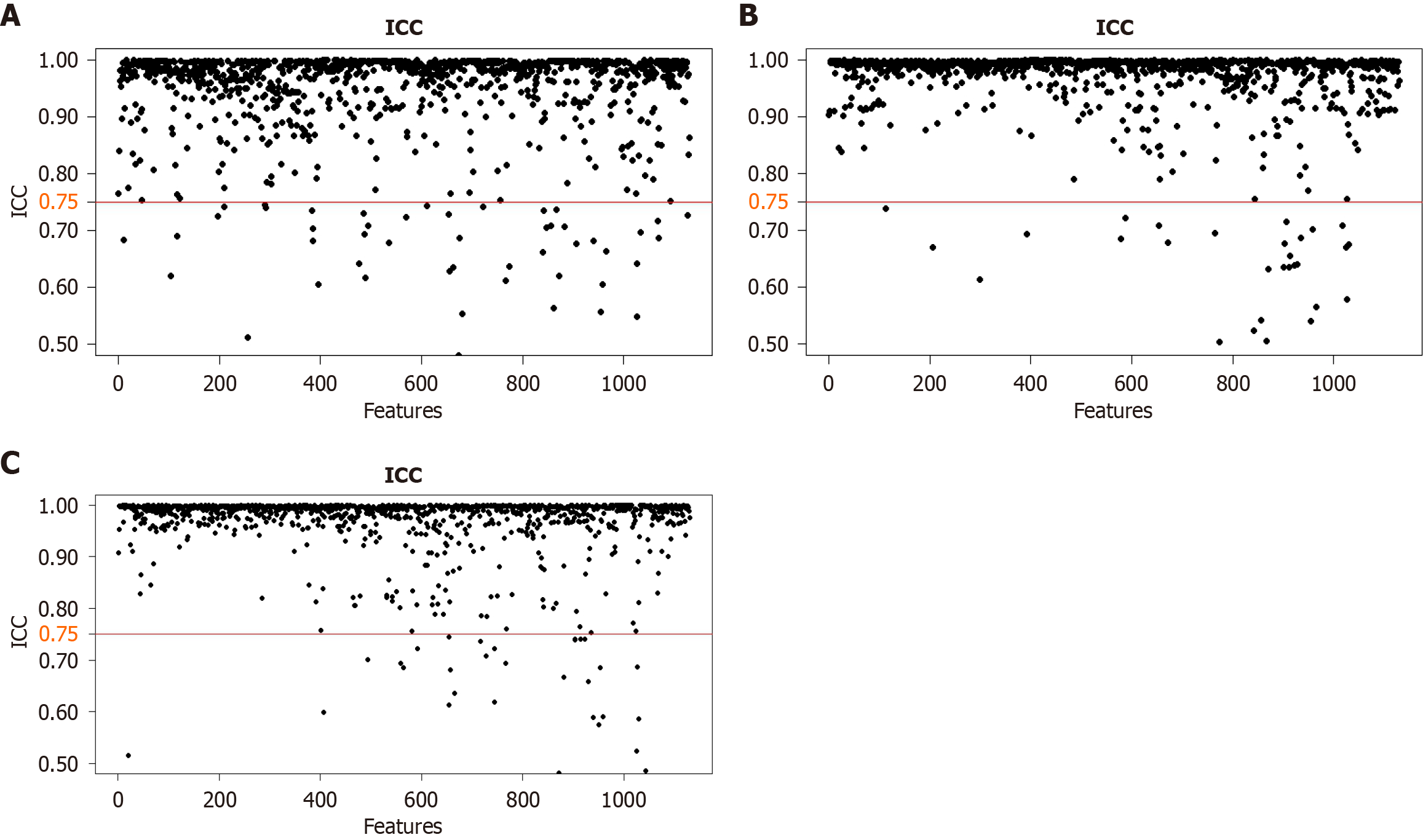
Figure 3 Consistency assessment results for each series.
A: Fat-suppressed T2-weighted image; B: Arterial phase; C: Portal venous phase. ICC: Interclass correlation coefficient.
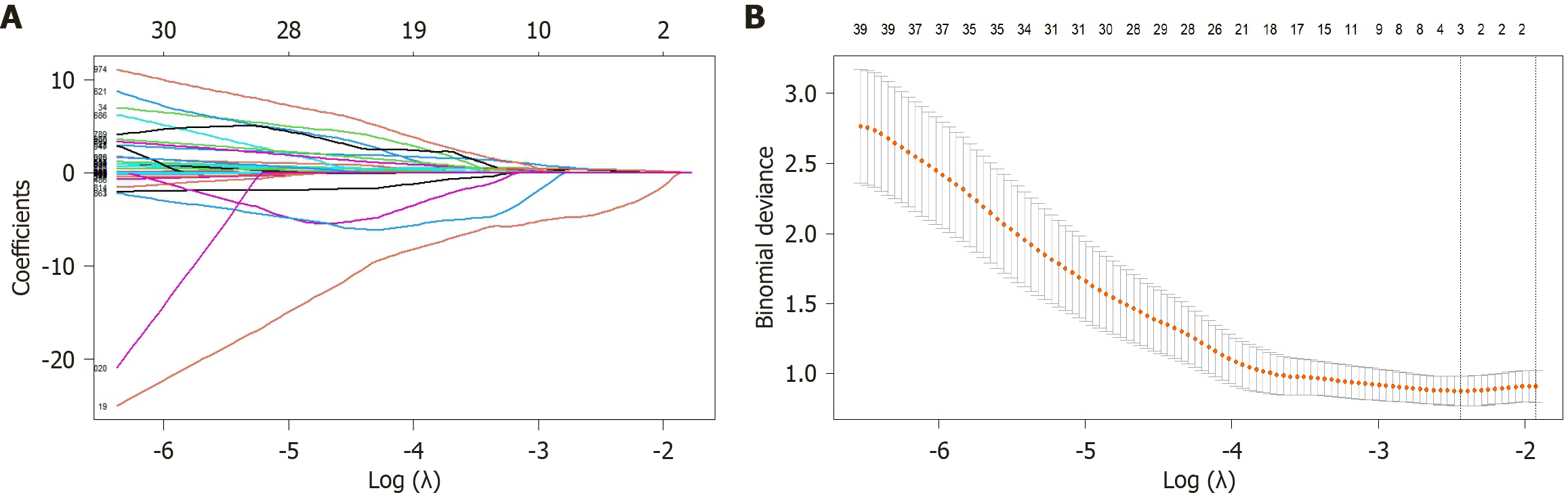
Figure 4 Radiomics features screened via least absolute selection and shrinkage operator regression.
The vertical axis shows the model misclassification rate. The horizontal axis shows log(λ). The two vertical lines were drawn at the selected values using cross-validation, and the optimal values were obtained by applying the minimum criteria and 1 of the minimum criteria (1-standard error criteria). A: Coefficient change spectra of the radiomics features; B: The process of feature selection for determining the parameter (λ) in the least absolute selection and shrinkage operator model by 10-fold cross-validation.
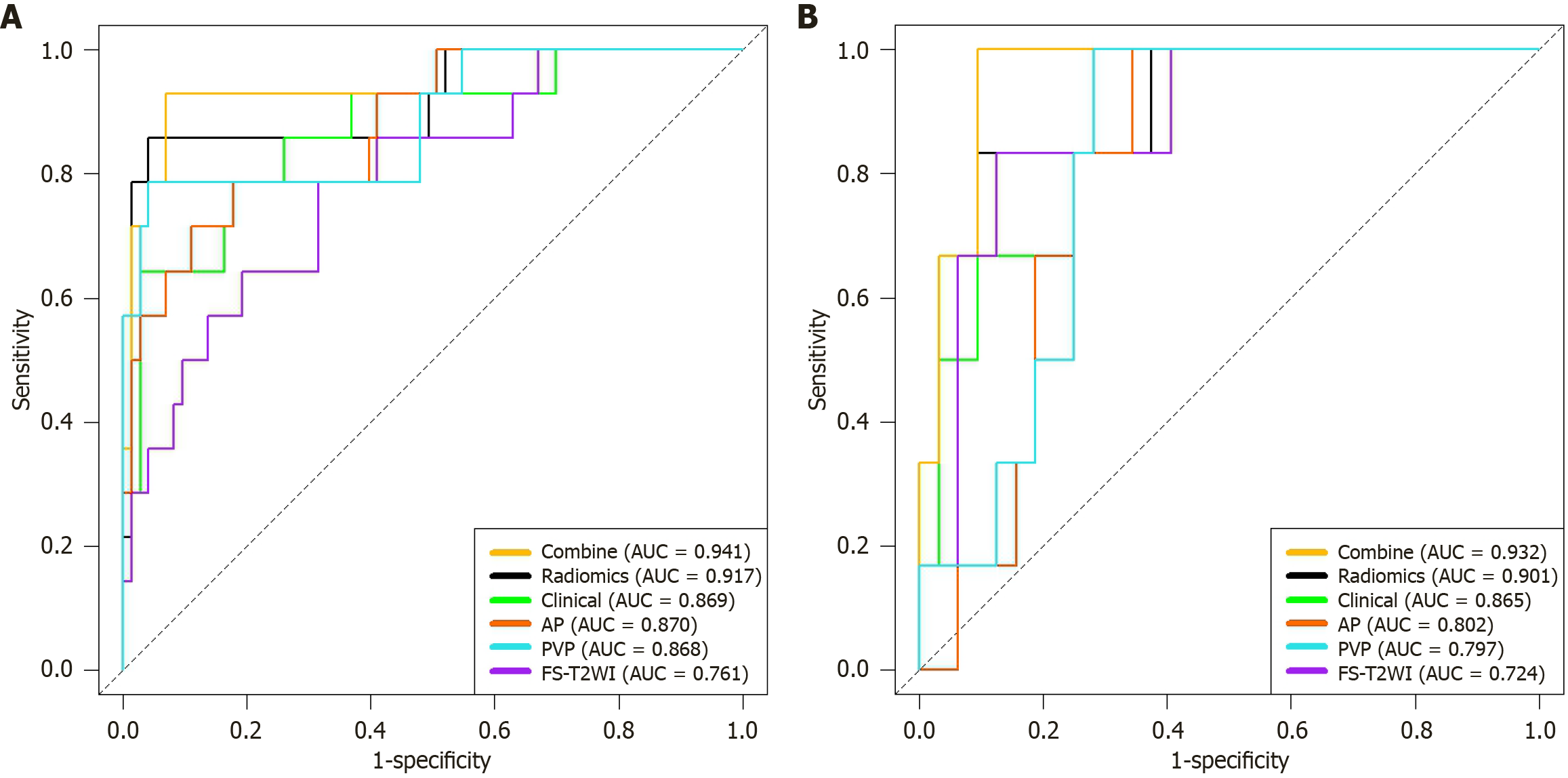
Figure 5 Receiver operating characteristic curves for each model.
A: Training group; B: Validation group. AUC: Area under the receiver operating characteristic curve; AP: Arterial phase; PVP: Portal venous phase; FS-T2WI: Fat-suppressed T2-weighted imaging.
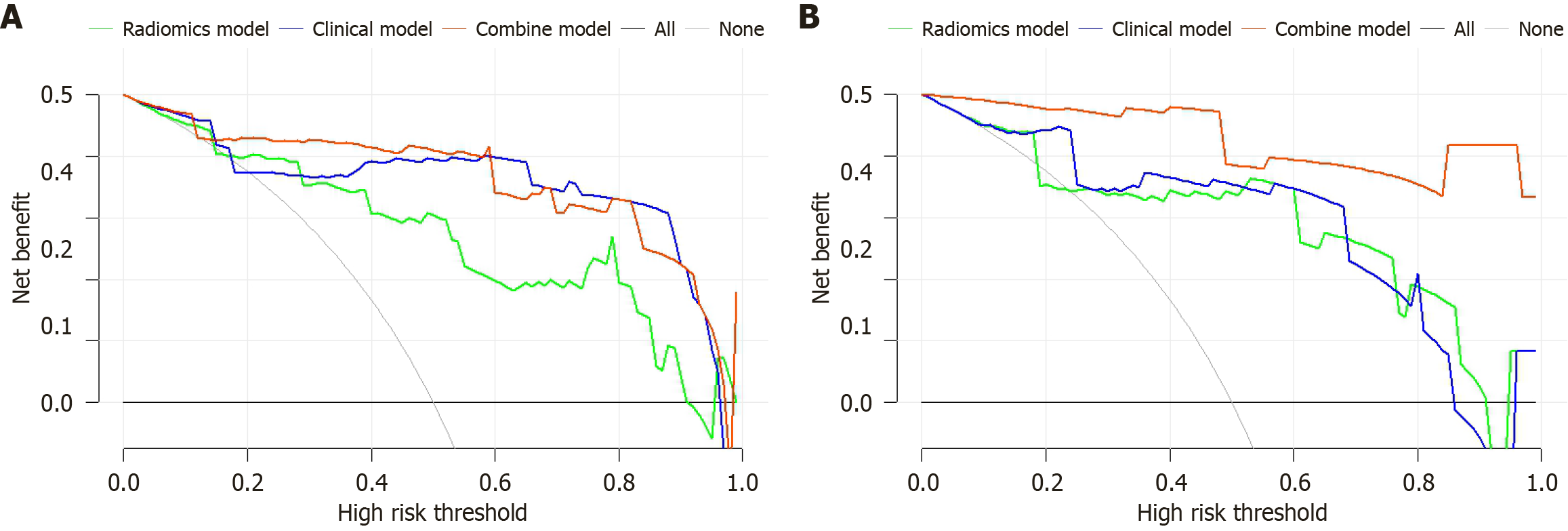
Figure 6 Decision curve analysis curves.
A: Training group; B: Validation group.
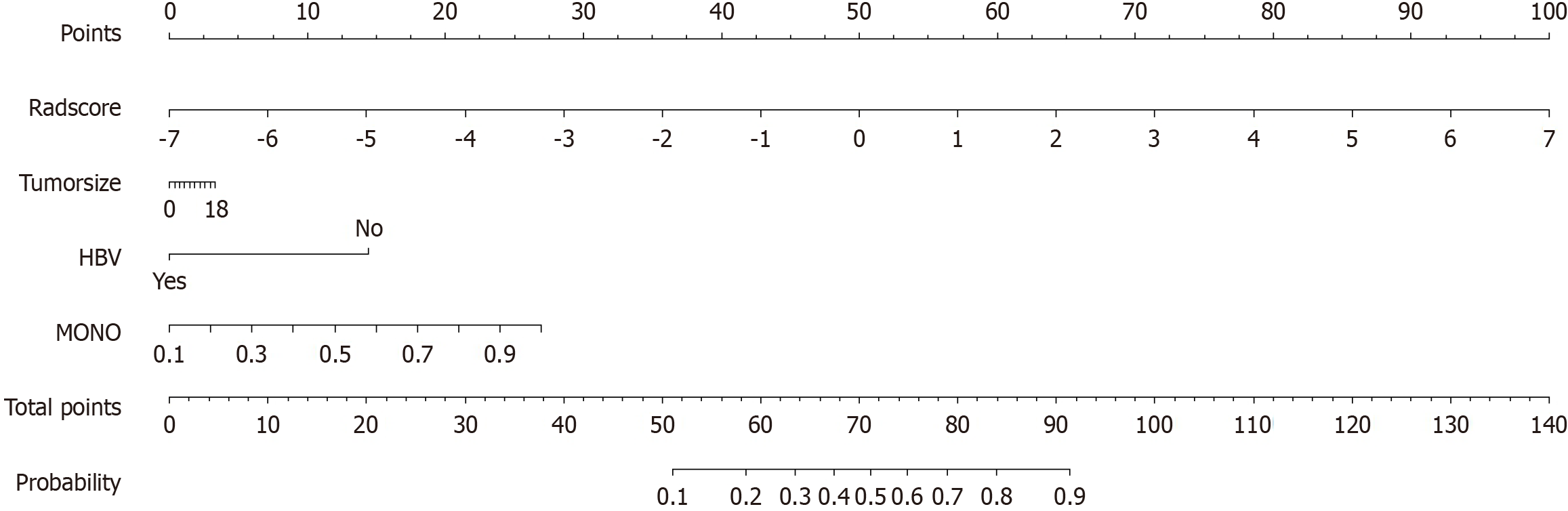
Figure 7 Traditional nomogram based on the combined radiomics-clinical model.
HBV: Hepatitis B virus; MONO: Monocyte count.
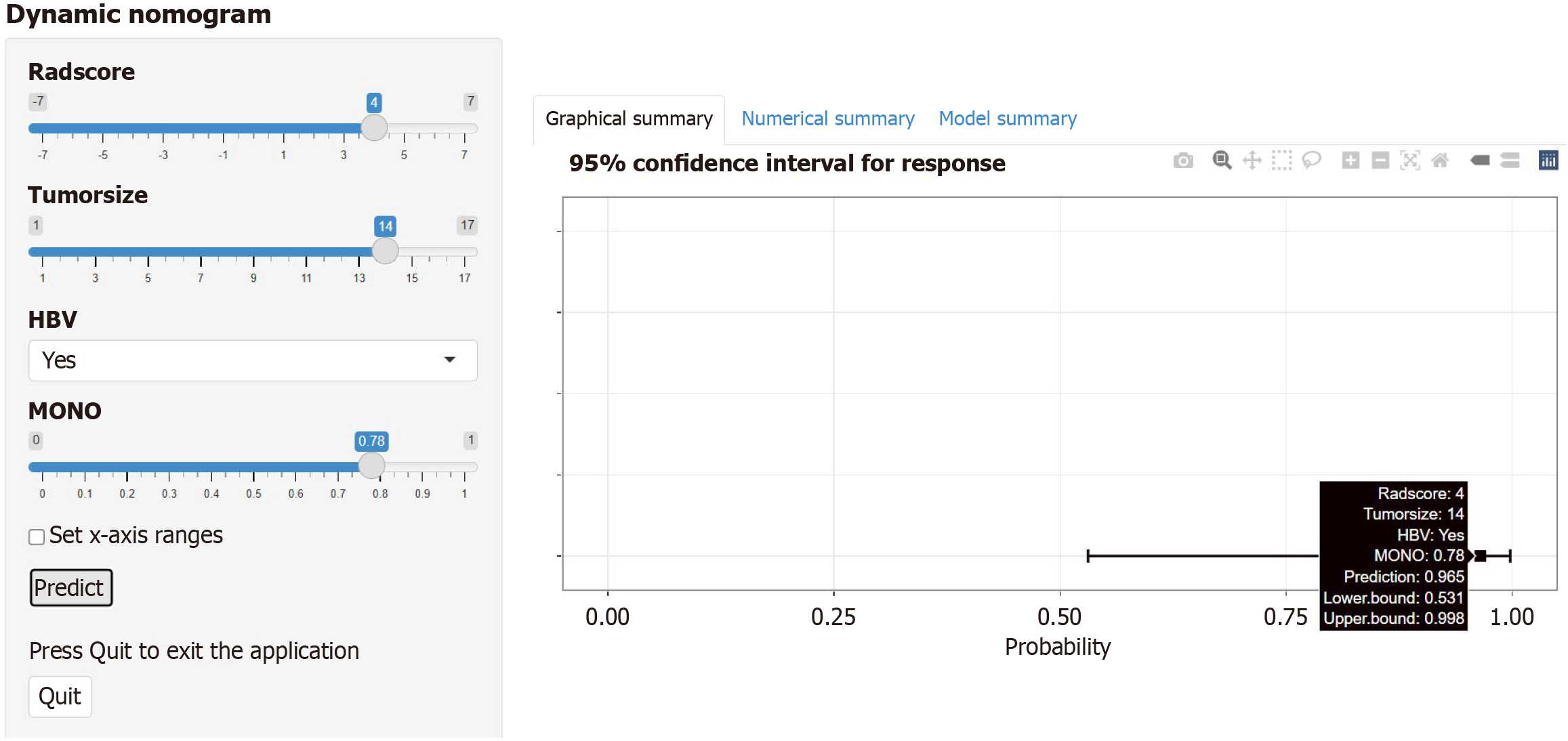
Figure 8 Dynamic nomogram based on the combined radiomics-clinical model via the web interface.
A dynamic nomogram was developed for the training group by combining the following four parameters: Rad score, tumor size, hepatitis B virus status, and monocyte count; and an example of predicting the probability of poor differentiation in hepatocellular carcinoma patients is shown. HBV: Hepatitis B virus; MONO: Monocyte count.
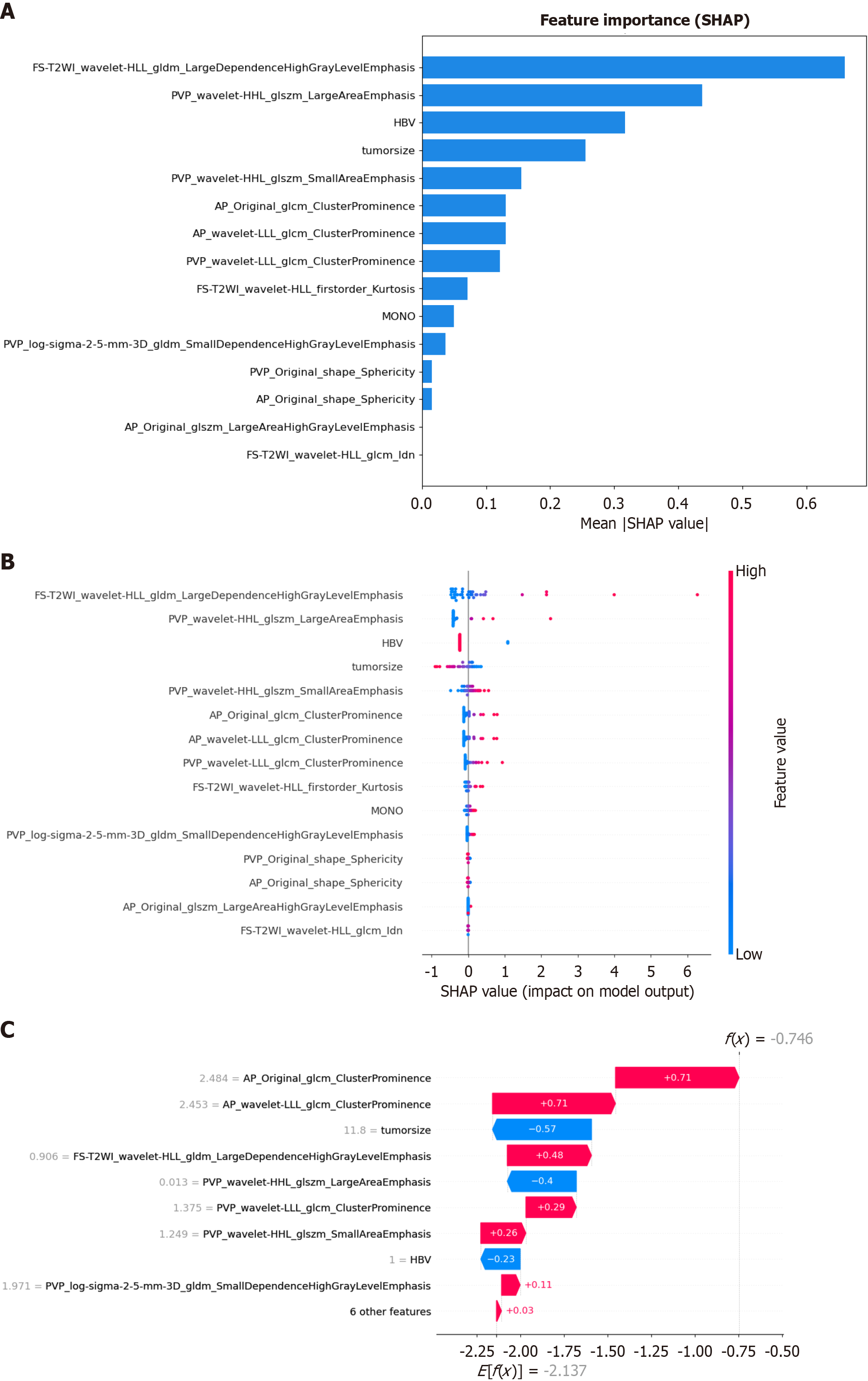
Figure 9 SHapley Additive exPlanations analysis diagram of each feature in the optimal model.
A: SHapley Additive exPlanations (SHAP) value bar graph, showing the mean absolute SHAP value of all the features, arranged in descending order of importance; B: SHAP value scatter graph, showing the average degree of influence of each feature in the model. The color of the dot represents the original value of the feature, with blue indicating a low value and red indicating a high value, and the position of the dot represents the SHAP value. A positive SHAP value indicates an increased risk of predicting poorly differentiated hepatocellular carcinoma, and vice versa. A higher value corresponds to a higher risk. Each point corresponds to a prediction for each patient; C: Waterfall diagram of a random sample. The calculation process of the degree of contribution of each feature in a single random sample to the prediction result and the final predicted value, which is represented by bars of different colors, with blue representing a negative value and red representing a positive value. SHAP: SHapley Additive exPlanations; FS-T2WI: Fat-suppressed T2-weighted imaging; PVP: Portal venous phase; AP: Arterial phase; LLL: Low-low-low frequency band; HBV: Hepatitis B virus; MONO: Monocyte count.
- Citation: Shi Y, Zhang P, Li L, Yang HM, Li ZM, Zheng J, Yang L. Interpretable model based on multisequence magnetic resonance imaging radiomics for predicting the pathological grades of hepatocellular carcinomas. World J Radiol 2025; 17(12): 112911
- URL: https://www.wjgnet.com/1949-8470/full/v17/i12/112911.htm
- DOI: https://dx.doi.org/10.4329/wjr.v17.i12.112911