Published online Dec 26, 2025. doi: 10.4330/wjc.v17.i12.112062
Revised: August 17, 2025
Accepted: October 28, 2025
Published online: December 26, 2025
Processing time: 160 Days and 4.7 Hours
Myocardial infarction (MI) occupies a very high mortality and morbidity rate, and the search for effective pharmacological treatments has far-reaching implications for clinical research.
To explore the protective effects of Mongolian medicine Agari-5 on rats with MI.
Sprague-Dawley rats were used, and both the Agari-5 and model groups had their coronary arteries clamped to induce MI. Proteomics was used to research the potential mechanism of action while ELISA, hematoxylin and eosin, and Masson’s staining were used to preliminarily investigate the protective impact of Agari-5 on rats with MI.
The current study has shown that Agari-5 might enhance cardiac function indicators, including echocardiography results of rats and creatine kinase, creatine kinase isoenzyme, and lactate dehydrogenase, in rats that had MI. According to the results of pathological staining, Agari-5 may lessen inflammatory cell infiltration and cardiomyocyte fibrosis, among other things. The proteome analysis revealed that there were 60 distinct proteins in total, four of which were asso
Potential therapeutic effects of Agari-5 for MI and its mechanism of action may be related to PSAT1, PDK1, SMAD4, and SDF2.
Core Tip: We demonstrated that myocardial infarction (MI) rats treated with Agari-5 expressed similar levels of SDF2, PDK1, SMAD4, and PSAT1 as the control group, suggesting that these proteins be important targets influenced by Agari-5. Through proteomic analysis the research delved deeper into the protective mechanisms of Mongolian traditional medicine formulas, finding that these traditional formulas may protect MI rats through the regulation of multiple protein targets and molecular mechanisms, such as multicomponent interactions and synergistic effects, providing a new direction for research on national medicines and offering a reference for clinical treatments of cardiovascular diseases.
- Citation: Zhao YB, Bao ZH, Tu Y, Qiu X, Bao YL, Su M, Qi HJ, Wan Q. Proteomics-based investigation of the protective effect and mechanism of Agari-5 in rats with myocardial infarction. World J Cardiol 2025; 17(12): 112062
- URL: https://www.wjgnet.com/1949-8462/full/v17/i12/112062.htm
- DOI: https://dx.doi.org/10.4330/wjc.v17.i12.112062
Myocardial infarction (MI) is a leading cause of death globally, particularly among middle-aged and elderly populations, and is one of the four major non-communicable diseases with high mortality and morbidity rates worldwide along with cardiovascular disease, cancer, diabetes mellitus, and chronic respiratory disease[1,2]. MI occurs when atherosclerotic lesions in the coronary arteries cause narrowing or occlusion, leading to hypoxia and ischemia, which result in the necrosis of myocardial tissue[3,4]. During the hypoxia-reoxygenation process of cardiomyocytes, a series of damaging changes occur in myocardial ultrastructure, energy metabolism, cardiac function, and electrophysiology, which continue to affect cardiac function and structural remodeling (key pathological processes in MI)[5]. Current treatments for MI include percutaneous coronary intervention and pharmacological therapies[6], and there is growing recognition of the potential of ethnomedicine in treating this condition.
Mongolian medicine Agari-5 is a widely used traditional remedy for cardiovascular diseases consisting of five herbs: (1) Descending incense; (2) Sumac; (3) Umeboshi; (4) Borax; and (5) Danshen. It primarily works by promoting blood circulation and resolving blood stasis and is used to treat conditions such as vascular fever, heart deficiency heat, heart stabbing pain, hypertension, and arteriosclerosis. It is particularly effective for treating heart diseases like Haye disease. The compound formula reflects the holistic approach and evidence-based principles of Mongolian medicine, offering a multicomponent and multitarget therapeutic effect.
Proteomics is the study of the entire set of proteins expressed in a cell or organism, utilizing techniques such as protein purification and mass spectrometry analysis to identify and quantify proteins[7,8]. Unlike genomics, proteomics is more complex as it seeks to understand not only the total protein expression but also various aspects like protein modifications, structures, and protein-protein interactions (PPIs). This comprehensive analysis makes proteomics a critical tool for understanding disease processes and cellular metabolism.
In this study a rat MI model was constructed, and proteomics technology was applied to investigate the therapeutic effects of the Mongolian medicine Agari-5. By analyzing protein expression differences and utilizing bioinformatics methods, the study aimed to elucidate the molecular basis and advantages of Agari-5 in treating MI[9]. The research focused on uncovering the protein-level mechanisms behind the multitarget, multipathway, and multifunctional therapeutic effects of Agari-5, providing insights into its mechanism of action. These findings will offer a theoretical basis for the use of Agari-5 in treating cardiovascular diseases and guiding future research.
Agari-5 (No. M20201334000) was obtained from DL-Dithiothreitol (Solarbio), Iodoacetamide (Aladdin), and Tetraethylammonium bromide (Sigma). Creatine kinase (CK), CK isoenzyme (CK-MB), and lactate dehydrogenase (LDH) were supplied by Mlbio Biotechnology Ltd. Other reagents, such as EDTA, xylene brilliant cyanine G, and sodium dodecyl sulfate, were sourced from Sinopharm Chemical Reagent Co.
Animal experiments were conducted in compliance with the guidelines set by the Experimental Animal Ethics Committee of the Affiliated Hospital of Inner Mongolia Minzu University (No. NM-LL-2024-05-16-08). The study followed the National Institute of Health Guidelines for the Ethical Use of Animals. Sprague-Dawley rats were obtained from the Changsheng Experimental Animal Centre of Liaoning Province, China [SCXK (Liao) 2020-0001] and housed in a specific pathogen-free experimental facility at Inner Mongolia Minzu University. The rats were kept under controlled conditions with a temperature of 23 ± 1 °C, humidity of 55% ± 5%, and a 12-h light-dark cycle. They were given ad libitum access to food, water, and physical activity throughout the study period.
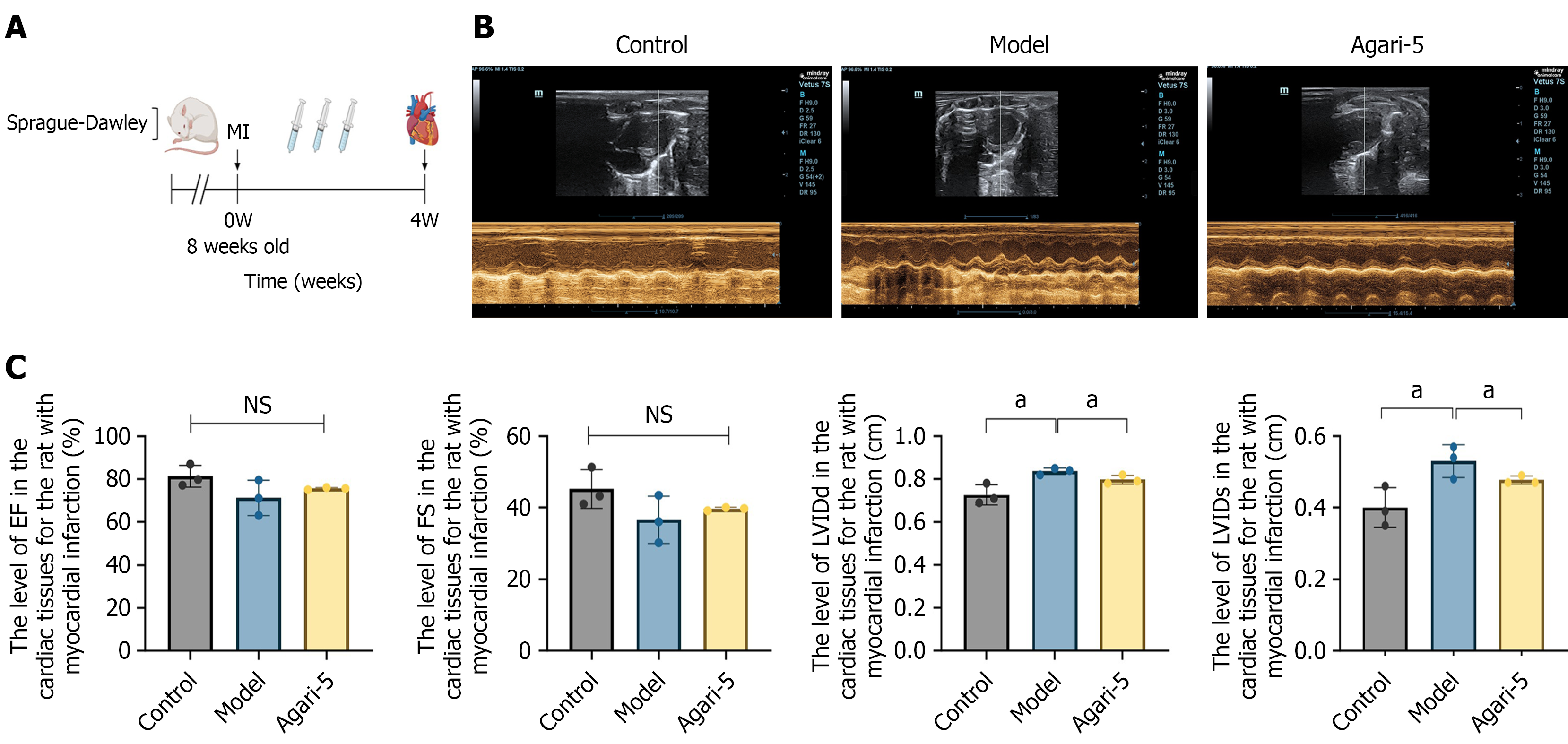
After 1 week of adaptive feeding, the rats were randomly assigned to a control group, model group, and Agari-5 group (n = 10). Rats in the model and Agari-5 groups underwent MI induction. Under isoflurane anesthesia the animals were disinfected with alcohol, and a thoracotomy was performed to expose the heart. The pericardium was carefully torn to fully expose the heart, and the left anterior descending coronary artery was ligated with a No. 4-0 black silk thread to induce MI[9-11]. The specific experimental flow is shown in Figure 1A.
In the control group and model group, an equivalent volume of saline was administered by gavage once daily. In the Agari-5 group a dose of 0.8127 g/kg was given daily for 4 weeks at a fixed time. The gavage doses for all rats were adjusted based on the equivalent doses for animals and humans.
Using a small animal ultrasound imaging system, echocardiograms were monitored for each group of rats, and several key cardiac function indicators were measured. These indicators included left ventricular ejection fraction, left ventricular short-axis shortening fraction, left ventricular end-systolic diameter, and left ventricular end-diastolic dimension.
Rats were anesthetized with isoflurane, and the abdominal cavity was opened to collect blood from the abdominal aorta. The blood was left at room temperature for 30 min and then centrifuged at 3500 rpm for 10 min. The supernatant was further centrifuged at 12000 rpm for 10 min at 4 °C and stored at -80 °C. Heart tissue from the rats was also rapidly collected and stored at -80 °C for further analysis.
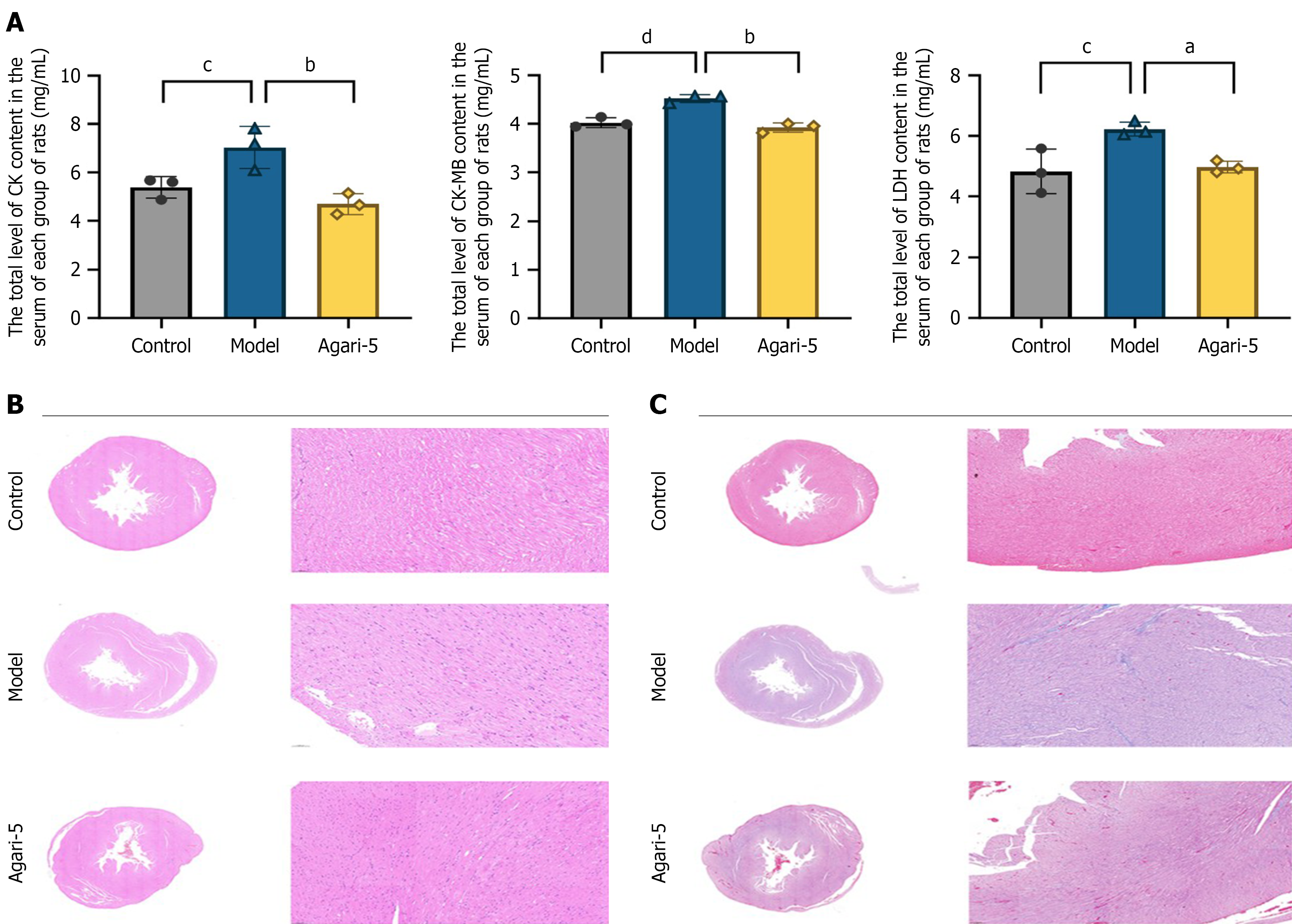
After the final gastric lavage, the rats were fasted for 12 h and anesthetized. After routine disinfection the abdominal cavity was incised along the midline of the abdomen using surgical scissors. Postoperatively, the needle tip of the puncture needle was inserted at an angle of approximately 25° to 30° toward the heart end with a depth of approximately 5 mm, and blood was aspirated. Samples were centrifuged at room temperature (15000 × g, 20 min), and supernatant was removed. Then a second centrifugation step (15000 × g, 20 min) was performed at 4 °C. Supernatant was removed, and samples were then analyzed. Serum CK (No. ml107007, mlbio Co., Ltd., China), CK-MB isoenzyme (No. ml107008, mlbio Co., Ltd., China), LDH (No. ml106660, mlbio Co., Ltd., China) levels were assayed by commercially ELISA kits., following the manufacturer’s instructions. A plate reader was used to measure the absorbance of each well at a wavelength of 450 nm.
After euthanizing the rats under anesthesia, tissue samples were collected and fixed in 10% neutral formaldehyde for 24 h. The samples underwent gradient ethanol dehydration, xylene clearing, and paraffin embedding. Sections were then cut, stained with hematoxylin and eosin stain and Masson’s stain, and sealed with neutral gum. The lesions were subsequently observed under a light microscope.
Cardiac samples were collected and ground to a powder in liquid nitrogen. An appropriate amount of the powder was transferred to a 1.5 mL centrifuge tube and incubated with lysis buffer (8 M urea) containing 1 mmol/L phenylmethylsulfonyl fluoride and 2 mmol/L EDTA (final concentrations) for 5 min. The mixture was then sonicated on ice for 5 min (2 seconds on, 3 seconds off). The lysate was centrifuged at 4 °C at 15000 g for 20 min, and the supernatant was collected. The total protein concentration was determined using the bicinchoninic acid protein quantification assay. An aliquot of the protein solution was taken based on the protein concentration and adjusted to a final volume of 200 µL with 8 M urea. The solution was reduced with 10 mmol/L dithiothreitol for 45 min at 37 °C and alkylated with 50 mmol/L iodoacetamide for 15 min in the dark at room temperature. Precooled acetone, four times the volume of the protein solution, was added to precipitate the proteins for 2 h at -20 °C. After centrifugation the protein precipitate was air-dried and resuspended in 200 µL of 25 mmol/L ammonium bicarbonate solution, followed by the addition of 3 µL of trypsin (Promega) for overnight digestion at 37 °C. After digestion the peptides from each sample were desalted using a C18 column, concentrated by vacuum centrifugation, and redissolved in 0.1% (v/v) formic acid[12].
Nanoliter liquid chromatography detection: Samples were separated using a Vanquish Neo ultra-high-performance liquid chromatography (UHPLC) system[13]. The mobile phase consisted of phase A, which was a 0.1% formic acid aqueous solution, and phase B, which was a 0.1% formic acid acetonitrile solution (100% acetonitrile). The injection mode utilized a trap-analytical dual-column method in which the trap column was a PepMap Neo Trap Cartridge (300 μm × 5 mm, 5 μm) and the analytical column was an Easy-Spray™ PepMap™ Neo UHPLC column (150 µm × 15 cm, 2 µm). The temperature of the analytical column was maintained at 55 °C using an integrated column oven. The sample volume was set at 200 ng with a flow rate of 2.5 µL/minute and an effective gradient duration of 6.9 min, resulting in a total run time of 8 min.
Data-independent acquisition (DIA) analysis was carried out using a nanoscale Vanquish Neo system (Thermo Fisher Scientific) for chromatographic separation. Samples, following nanoscale UHPLC separation, were analyzed by DIA mass spectrometry using an Orbitrap Astral high-resolution mass spectrometer (Thermo Scientific). The detection mode was set to positive ion with a parent ion scanning range of 380-980 m/z and a primary mass spectral resolution of 240000 at 200 m/z. The normalized automatic gain control target was set at 500% with a maximum ion injection time of 5 ms. For MS2 analysis the DIA data acquisition mode was employed, utilizing 299 scanning windows. The isolation window was set to 2 Th with a higher energy collision dissociation collision energy of 25%. The normalized automatic gain control target was also 500%, and the maximum injection time was set at 3 ms.
The library search software used for DIA mass spectrometry data in this study was DIA-NN (v1.8.1). The library was searched using the library-free method with the following search parameters: (1) The database used was uniprot-proteome_UP000002494_Rat_20220719.fasta database, which contained a total of 46069 sequences; and (2) The iRT2.fasta database, which included 1 sequence. The deep learning-based parameter was enabled to predict a spectral library. Additionally, the multiple bypass retrieval option was selected to generate a spectral library using DIA data, which was then utilized for reanalyzing the DIA data for protein quantification. Both precursor ions and protein-level false discovery rates were filtered at 1%, ensuring that the filtered data could be used for subsequent raw data analyses[14,15].
To gain a comprehensive understanding of the functional properties of the identified and differentially expressed proteins, a thorough enrichment analysis was conducted. This analysis included the following components: (1) Gene Ontology (GO) annotation; (2) EuKaryotic Orthologous Groups (KOG) functional classification; (3) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis; (4) Protein domain characterization; (5) Subcellular localization assessment; and (6) Signal peptide prediction (using SignalP). The differentially expressed proteins in each comparative group were enriched and analyzed at three levels: (1) GO classification; (2) KOG functional classification; and (3) KEGG pathway analysis.
PPI analysis was performed using the StringDB protein interaction database (http://string-db.org/). If a corresponding species was available in the database, the protein sequences of that species were directly extracted. If no corresponding species was found, protein sequences from a closely related species were utilized. The differentially expressed protein sequences were compared with the extracted sequences using BLAST, and interactions among the differentially expressed proteins were identified based on a confidence score greater than 400 (medium confidence). Static and dynamic network diagrams were constructed using R packages, specifically qgraph for static visualizations and networkD3 for dynamic visualizations.
Protein extraction was performed from the hearts of rats in each experimental group, and protein concentrations were quantified using a bicinchoninic acid kit. For protein analysis, 20 µg of each sample was loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel in which the proteins were separated according to their molecular weight during electrophoresis. After separation the proteins were transferred from the gel to polyvinylidene fluoride membranes (Millipore, IPVH00010). To prevent nonspecific binding, the polyvinylidene fluoride membranes were subjected to a 2-h blocking process. Subsequently, the membranes were incubated overnight at 4 °C with primary antibodies diluted 1:1000 to ensure optimal specificity and sensitivity. After the primary antibody incubation, the membranes were washed three times with PBS to remove any unbound antibodies. The membranes were then incubated with secondary antibodies diluted 1:5000 for 1 h at room temperature, facilitating the detection of protein bands of interest. Finally, protein expression levels in the different experimental groups were compared.
Statistical analysis was conducted using GraphPad Prism 8.0 (GraphPad Software). The data are presented as mean ± SD. Student’s t-tests were used to evaluate differences between two groups while one-way analysis of variance was applied for multiple comparisons. Spearman’s correlation analysis and the χ2 test were performed to assess correlations between different gene pairs. Survival differences were evaluated using the log-rank test. All statistical analyses were two-sided, with a significance level set at P < 0.05, indicating statistical significance.
General condition of rats: As shown in Figure 1A, this is the overall animal experiment flowchart. The rats in the control group exhibited bright, lustrous fur, were highly responsive, and demonstrated normal eating and drinking behaviors with no mortality occurring during the experimental period. In contrast, the rats in the model group displayed poor vitality, dull fur, cold skin, and a tendency to sleep excessively along with considerably reduced food and water intake; some of these rats also had scanty stools. The rats in the Agari-5 group appeared to have slightly better vitality though their fur lacked luster, and their complexion was somewhat warmer; however, they also exhibited slightly lower food and water intake compared with the control group. Body weight measurements revealed a steady increase in the control group each week while the model group experienced a significant decrease in body weight. In contrast, the Agari-5 group showed an upward trend in body weight compared with the model group, indicating weight gain as detailed in Supplementary Figure 1A.
The effect of Agari-5 on cardiac function in rats with MI: As shown in Figure 1B and C, rat cardiac ultrasound images revealed a decreasing trend in left ventricular ejection fraction and left ventricular short-axis shortening fraction levels in the model group compared with the control group (P > 0.05), and an increasing trend in the Agari-5 group compared with the model group (P > 0.05). At the left ventricular diameter in diastole and left ventricular internal diameters levels, the model group showed an increasing trend compared with the control group (P < 0.05). Compared with the model group, the Agari-5 group showed significant differences (P < 0.05).
Cardiac function indicators: MI resulted in the destruction of cardiomyocytes, leading to increased activity levels of cardiac function enzymes, which are common indicators for detecting MI in rats. Compared with the control group, levels of CK, CK-MB, and LDH were significantly elevated in the model group (P < 0.05), confirming the successful establishment of the model. In contrast, treatment with Agari-5 significantly reduced the cardiac function markers in rats with MI compared with the model group (P < 0.05). For further details, refer to Figure 2A.
Hematoxylin and eosin staining and Masson’s staining: As shown in Figure 2B, hematoxylin and eosin staining results indicated that the myocardial fibers in the control group were organized in bundles with intact myocardial tissue structure and no evidence of inflammatory cell infiltration. Meanwhile, the model group exhibited disordered myocardial tissue with blurred transverse striations and a significant enlargement of the myocardial interstitial space, accompanied by extensive inflammatory cell infiltration. Compared with the model group, the myocardial fibers in the Agari-5 group were more organized, showing improved cytoplasmic edema and more intact cytoplasmic nuclei. As illustrated in Figure 2C, Masson’s staining revealed no significant collagen fiber deposition in the myocardial tissue of the control group. In the model group normal myocardial fibers were damaged with extensive collagen fiber deposits observed, indicating pronounced myocardial fibrosis and atrophic degeneration along with inflammatory cell infiltration in the infarcted areas. In the Agari-5 group myocardial fibrosis was significantly reduced with fewer collagen fibers deposited and diminished inflammatory cell infiltration.
Quality assessment of quantitative proteomics results: In constructing the mathematical model, principal component analysis was initially employed to process the overall data, allowing for the observation of the natural distribution of experimental samples and group relationships. The distribution of each group, as shown in Supplementary Figure 1B and C, was classified satisfactorily, indicating clear differences in protein expression among the groups. Correlation analysis between samples within each group is presented in Supplementary Figure 1D. This analysis enabled the observation of biological replicates among samples within the same group. The results demonstrated that the correlation coefficients of samples within each group were higher than those of samples between groups, suggesting that the differential proteins identified in this experiment are more reliable.
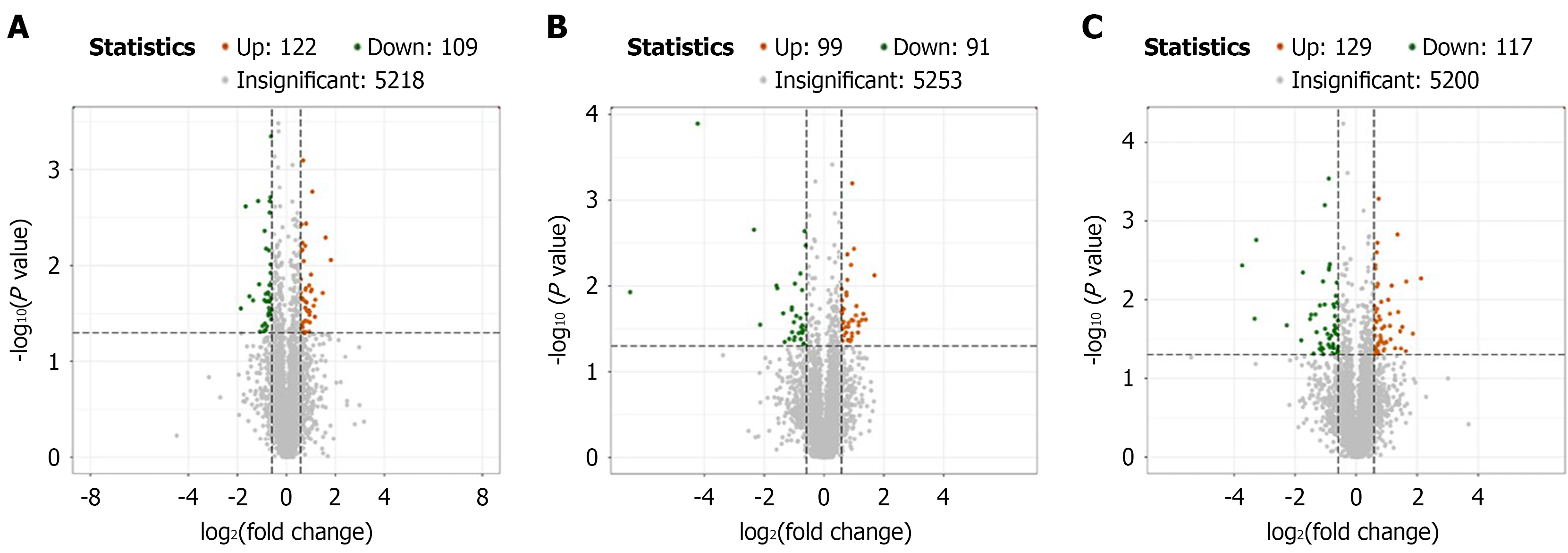
Differential protein expression: In the analysis of differential protein expression, a total of 231 proteins were identified in the control group compared with the model group with 122 proteins being upregulated and 109 downregulated as shown in Figure 3A. In the comparison between the Agari-5 group and the model group, 190 proteins were identified of which 99 were upregulated and 91 were downregulated as displayed in Figure 3B. Finally, Figure 3C shows that in the Agari-5 group compared with the control group, 246 proteins were identified with 129 proteins upregulated and 117 downregulated.
Functional annotation of differentially expressed proteins: GO is an international standard classification system for gene functions, comprising three main components: (1) Biological process; (2) Molecular function; and (3) Cellular component. The differentially expressed proteins annotated to secondary GO entries were counted, and each classification displayed the top 20 GO terms with the highest-ranked differentially expressed proteins (in descending order). The results are presented in Supplementary Figure 2A-C. In the biological process category, the groups include cellular process, metabolic process, biological regulation, and cellular component. Molecular function is primarily characterized by cellular anatomical entities and protein-containing complexes. Cellular component is mainly represented by binding activities.
This study was annotated using the KOG database, and the number of differentially expressed proteins within each KOG category was counted and displayed in a bar chart (Supplementary Figure 2D-F). The categories included posttranslational modification, protein turnover, chaperones, general function prediction only, signal transduction mechanisms, and others, highlighting the functional groups of proteins identified in each experimental comparison.
KEGG pathways mainly encompass areas such as metabolism, genetic information processing, environmental information processing, cellular processes, human diseases, and drug development. Pathway analysis allows for the identification of critical biochemical, metabolic, and signal transduction pathways in which the differentially expressed proteins are involved. Supplementary Figure 2G-I presents the top KEGG pathways identified, including biosynthesis of amino acids, carbon metabolism, purine metabolism, thermogenesis, and T helper type 17 cell differentiation.
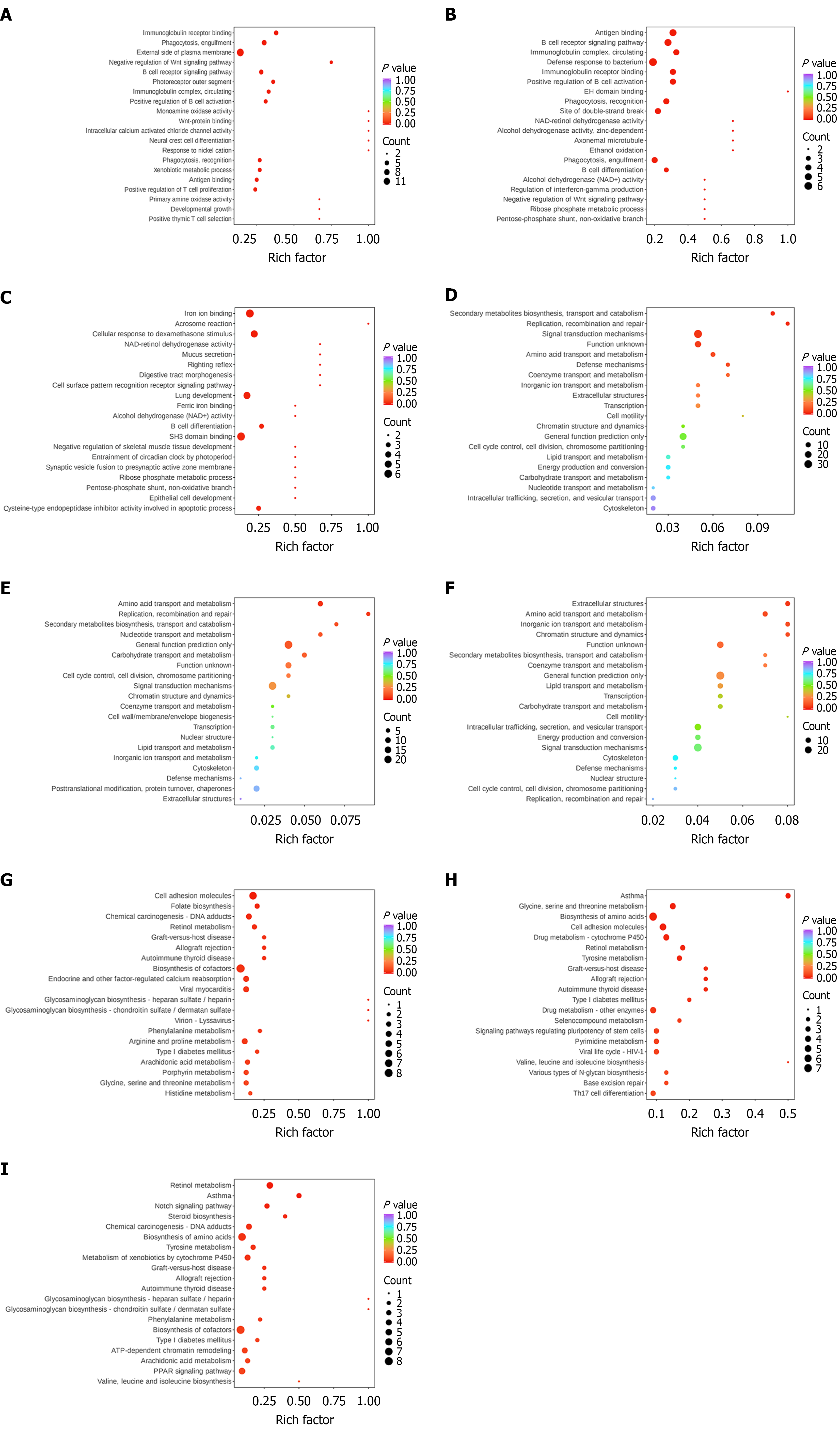
Functional enrichment of differentially expressed proteins: In the enrichment analysis the 50 GO terms with the highest P values (in descending order) were selected and visualized in bar charts as shown in Figure 4A-C. For biological processes the dominant terms included signal transduction, negative regulation of transcription by RNA polymerase II, and innate immune response. In molecular function the prominent categories were metal ion binding, ATP binding, and protein homodimerization activity. In cellular component key categories included extracellular space, integral component of the membrane, and Golgi apparatus. From the KOG enrichment analysis, 20 functional categories with the top P value rankings (sorted from smallest to largest) were selected and visualized using bubble diagrams (Figure 4D-F). The dominant pathways included general function prediction only, signal transduction mechanisms, amino acid transport and metabolism, and carbohydrate transport and metabolism. For the KEGG enrichment analysis results (Figure 4G-I), bubble diagrams were used to represent the degree of enrichment based on fold enrichment, P value, and the number of differentially expressed proteins involved. Key enriched pathways included biosynthesis of amino acids, cell adhesion molecules, drug metabolism, T helper type 17 cell differentiation, and biosynthesis of cofactors.
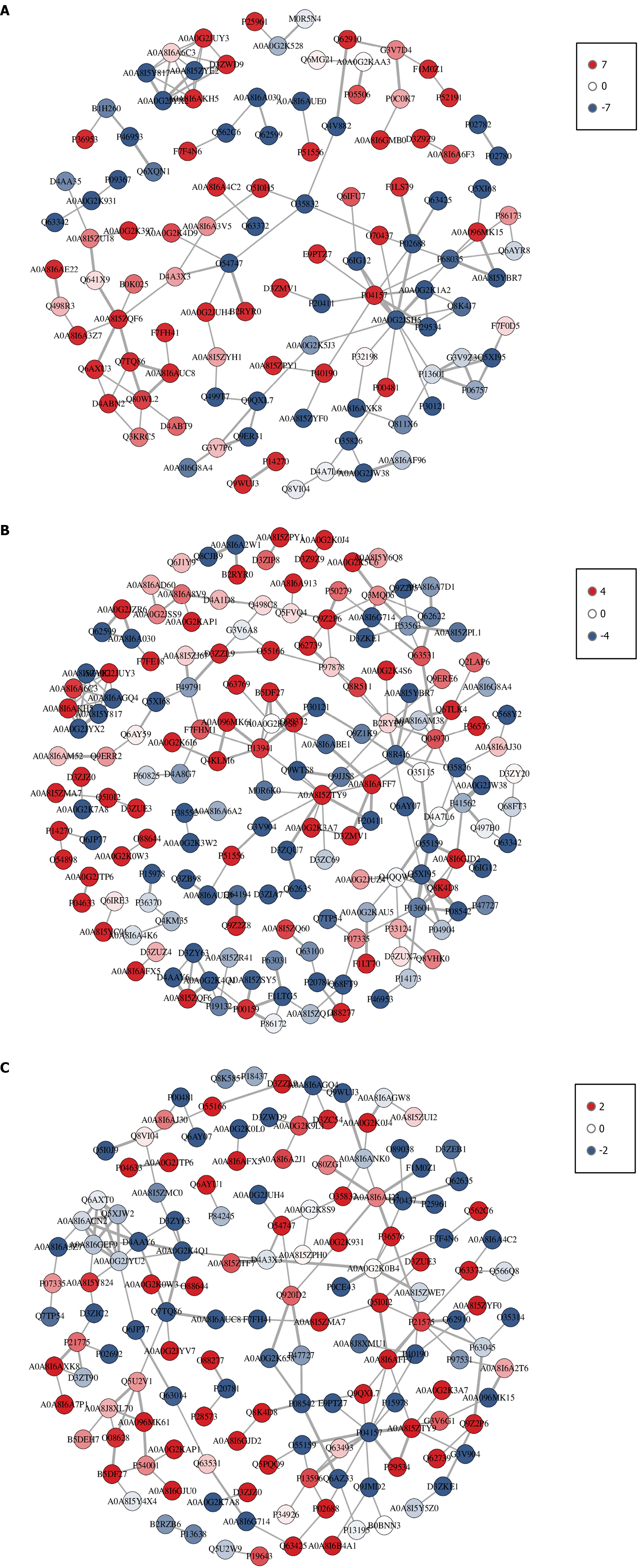
Interaction network analysis of differentially expressed proteins: The results of the PPI interaction network analysis are displayed in Figure 5. These networks can be imported into Cytoscape software for further visual editing and exploration. In the comparison between the control group and model group, 144 nodes were identified. In the Agari-5 group vs the model group, 112 nodes were found. Between the Agari-5 group and the control group, 168 nodes were identified. In total, the analysis revealed 60 differential proteins, detailed in Table 1.
| Gene | Agari5 vs model regulation | Model vs control regulation | Agari5 vs control regulation |
| Neb | Up | Down | Insig |
| Pnkp | Up | Down | Insig |
| Papss1 | Up | Down | Insig |
| Lonrf1 | Up | Down | Insig |
| Psat1 | Down | Up | Insig |
| Ccser2 | Up | Down | Insig |
| Man1c1 | Down | Up | Insig |
| Smoc1 | Down | Up | Insig |
| AC095947.1 | Up | Down | Insig |
| Nxn | Up | Down | Insig |
| Tanc1 | Up | Down | Insig |
| Sdf2 | Up | Down | Insig |
| Syap1 | Up | Down | Insig |
| Dmc1 | Up | Down | Insig |
| Nom1 | Up | Down | Insig |
| Decr1 | Down | Up | Insig |
| Rnpep | Down | Up | Insig |
| Pdk1 | Up | Down | Insig |
| Isg15 | Up | Down | Insig |
| Trio | Up | Down | Insig |
| Trim32 | Up | Down | Insig |
| Csnk2a2 | Up | Down | Insig |
| Chgb | Up | Down | Insig |
| Cdk18 | Down | Up | Insig |
| Pold1 | Down | Up | Insig |
| Smad4 | Up | Down | Insig |
| Otc | Up | Down | Insig |
| Mbp | Down | Up | Insig |
| Psbpc1 | Down | Up | Insig |
| Ptprc | Up | Down | Insig |
| Grb10 | Up | Down | Insig |
| Pth1r | Up | Down | Insig |
| Slc6a7 | Down | Up | Insig |
| Vcam1 | Down | Up | Insig |
| Map1a | Down | Up | Insig |
| Afm | Up | Down | Insig |
| Il6st | Up | Down | Insig |
| Padi3 | Up | Down | Insig |
| Ermard | Down | Up | Insig |
| Gramd1a | Up | Down | Insig |
| Znf830 | Down | Up | Insig |
| Lztfl1 | Down | Up | Insig |
| Cep70 | Down | Up | Insig |
| Armcx3 | Up | Down | Insig |
| Synj1 | Up | Down | Insig |
| Nrxn1 | Down | Up | Insig |
| Prx | Down | Up | Insig |
| Ces1f | Down | Up | Insig |
| Isg20 L2 | Up | Down | Insig |
| Dync2 Li1 | Down | Up | Insig |
| Blvra | Up | Down | Insig |
| Krt42 | Up | Down | Insig |
| Tepp | Down | Up | Insig |
| Fmo2 | Up | Down | Insig |
| Wls | Up | Down | Insig |
| Rsl1d1 L1 | Up | Down | Insig |
| Asrgl1 | Down | Up | Insig |
| Ttl | Up | Down | Insig |
| Nme7 | Down | Up | Insig |
| Pde4dip | Up | Down | Insig |
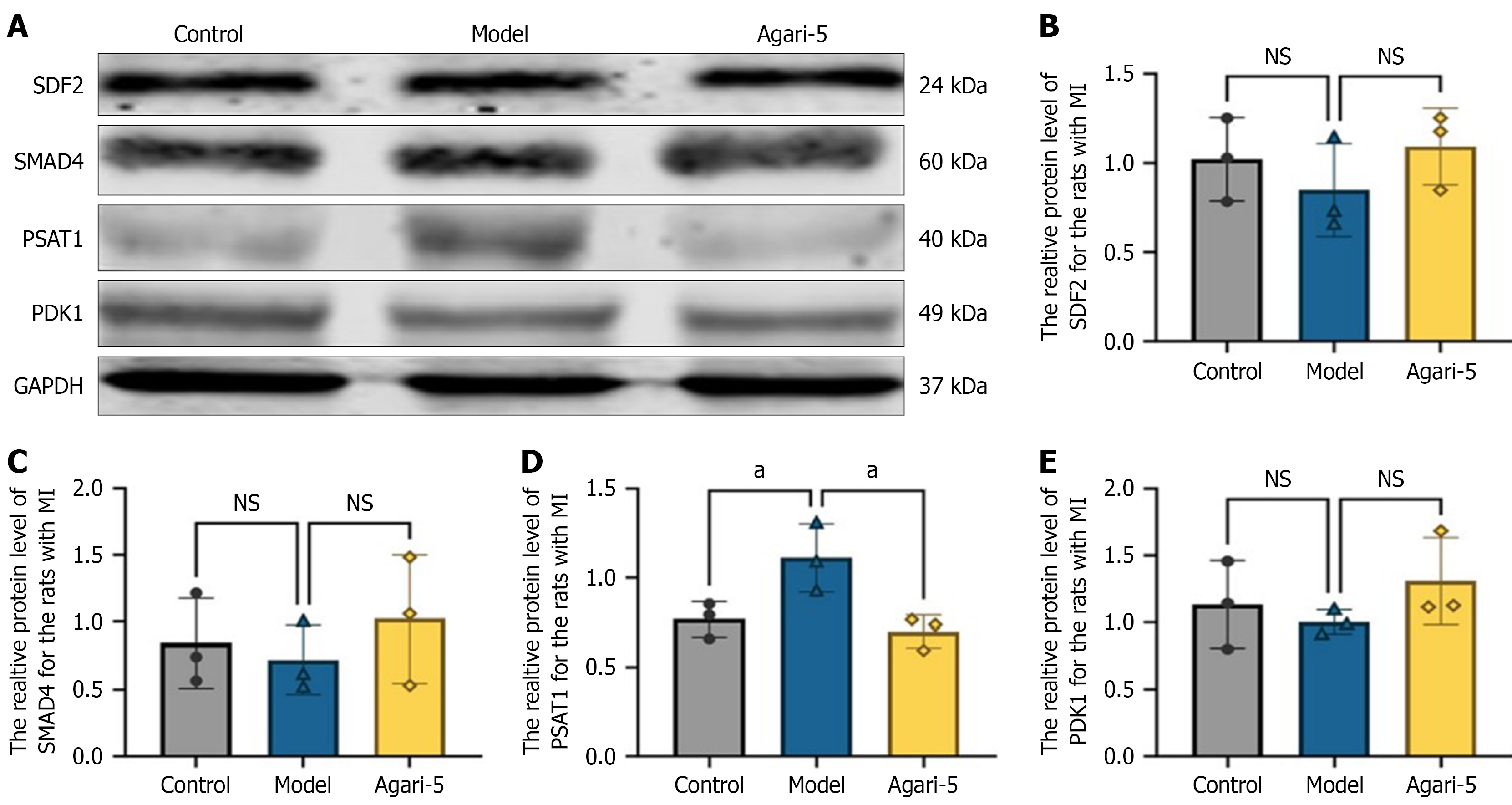
Immunoblot analysis of differentially expressed proteins in rat heart tissue: The levels of differentially expressed proteins in rat heart tissues were evaluated through western blot analysis. Based on the proteomics data, four key proteins were identified: (1) SDF2; (2) PDK1; (3) SMAD4; and (4) PSAT1. For SDF2, PDK1, and SMAD4, protein expression levels in the model group were decreased compared with the control group though the differences were not statistically significant (P > 0.05). In contrast, the Agari-5 group showed an upregulation trend in the expression levels of these proteins compared with the model group again without statistical significance (P > 0.05). Regarding PSAT1, its expression in the model group was upregulated relative to the control group (P > 0.05). However, the Agari-5 group exhibited a trend of downregulated PSAT1 expression compared with the model group although this change was not statistically significant (P > 0.05). The protein bands are shown in Figure 6A, and the corresponding statistical bar graphs are displayed in Figure 6B-E.
In recent decades natural medicines have gained significant use in clinical treatment for various diseases[16-18]. Cardiovascular disease remains a leading cause of mortality and morbidity worldwide. Studies have demonstrated that traditional Mongolian medicine preparations, such as Sanwei Tanxiang Decoction, can alleviate acute ischemia-reperfusion injury by modulating the electrophysiological properties of rat cardiomyocytes[19]. Similarly, the extract from Sugmul-3, another Mongolian medicine, has shown protective effects against isoproterenol-induced heart failure in Wistar rats by regulating mitochondrial dynamics[20].
Mongolian medicine emphasizes a holistic view of the human body, treating diseases through the interaction of multiple drugs. This approach offers advantages such as fewer adverse reactions and side effects, particularly in preventing and treating cardiovascular diseases. Agari-5 is a widely used traditional compound in Mongolian medicine for managing cardiovascular diseases. Its multicomponent and multitarget characteristics enhance its effectiveness in treating cardiovascular diseases[21].
Among the key components of Agari-5 is Salvia miltiorrhiza, which plays a critical role in reducing myocardial fibrosis, delaying cardiac remodeling[22], reducing myocardial inflammation and injury[23,24], and decreasing oxidative stress and injury[25,26]. Therefore, understanding the protective mechanisms of Agari-5 is essential for guiding clinical use, improving patient outcomes, and enhancing the prognosis, treatment, and overall quality of life for individuals with cardiovascular diseases.
In this study an MI rat model was established to assess cardiac function indices and the pathological changes in cardiac tissues. The results demonstrated that compared with the control group the cardiac function indices in the model group were significantly elevated while pathological findings showed disorganized myocardial tissue structure with blurred transverse striations, markedly enlarged myocardial gaps, and extensive infiltration by inflammatory cells. However, following treatment with Agari-5, both cardiac function and myocardial tissue pathology improved. Notably, the degree of myocardial fibrosis was markedly reduced with less collagen fiber deposition and decreased inflammatory cell infiltration. These findings suggest that the therapeutic effect of the Agari-5 compound may be attributed to the interactions of its components.
In the proteomics study the results showed a clear distribution and high dispersion between groups, indicating reliable findings for differential protein expression. Comparisons between the groups revealed a range of 190 to 246 differentially expressed proteins while enrichment analyses suggested that the therapeutic mechanism of Agari-5 in rats following MI may involve alterations in processes such as fatty acid degradation, metabolism, elongation, carbon metabolism, and protein digestion and absorption, all of which are vital for energy metabolism. Furthermore, differential protein function annotation analysis identified cardiovascular-related proteins in biological processes. These findings also indicated that Agari-5 may act on pathways related to myocardial fibrosis and mitochondrial energy metabolism in cardiomyocytes after MI. Together, the differential protein annotations and enrichment analyses suggest that Agari-5 may exert a therapeutic effect on MI by modulating these pathways.
In the PPI analysis a total of 60 proteins were identified with four specific proteins (SDF2, PDK1, SMAD4, and PSAT1) receiving attention due to their potential role as biomarkers for MI diagnosis. The SMAD4 signaling pathway has been reported to have a close association with cardiovascular disease in which its activation can lead to myocardial injury[27] and is significantly involved in the progression of myocardial fibrosis. PDK1 plays a key role in cellular phosphorylation processes related to mitochondrial energy metabolism, such as maintaining mitochondrial membrane potential, ATP generation, and calcium uptake[28].
In this study we used western blotting to investigate the expression levels of these differential proteins. These results showed that in MI rats treated with Agari-5 the expression levels of these proteins were comparable with those in the control group, suggesting that SDF2, PDK1, SMAD4, and PSAT1 may be important targets influenced by Agari-5. This study applied a logical approach of “disease-drug-target” to preliminarily verify the efficacy of Agari-5 in treating MI in rats.
Moreover, through proteomic analysis the research delved deeper into the protective mechanisms of Mongolian traditional medicine formulas, finding that these traditional formulas may protect MI rats through the regulation of multiple protein targets and molecular mechanisms, such as multicomponent interactions and synergistic effects, providing a new direction for research on national medicines and offering a reference for clinical treatments of cardiovascular diseases.
This study successfully identified the potential therapeutic benefits and mechanisms by which Agari-5 exerts its effects against MI. The findings provide initial evidence supporting the use of Agari-5 as a treatment for cardiovascular diseases and cardiac injuries, highlighting its possible role as an effective strategy in clinical settings.
| 1. | Nowbar AN, Gitto M, Howard JP, Francis DP, Al-Lamee R. Mortality From Ischemic Heart Disease. Circ Cardiovasc Qual Outcomes. 2019;12:e005375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 533] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 2. | GBD 2015 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1603-1658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1562] [Cited by in RCA: 1490] [Article Influence: 149.0] [Reference Citation Analysis (0)] |
| 3. | Davis BH, Morimoto Y, Sample C, Olbrich K, Leddy HA, Guilak F, Taylor DA. Effects of myocardial infarction on the distribution and transport of nutrients and oxygen in porcine myocardium. J Biomech Eng. 2012;134:101005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 4. | Fernandez-Jimenez R, Vivas D, García-Rubira JC, Fernandez-Ortiz A, Balbacid E, Kallmeyer A, Macaya C. Profound myocardial ischemia associated to occlusion of the right coronary artery. Int J Cardiol. 2011;149:e123-e124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 5. | Frangogiannis NG. Pathophysiology of Myocardial Infarction. Compr Physiol. 2015;5:1841-1875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 464] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 6. | Wang J, Li L, Ma N, Zhang X, Qiao Y, Fang G, Li G, Zhong T. Clinical investigation of acute myocardial infarction according to age subsets. Exp Ther Med. 2020;20:120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 7. | Hernandez AF, Udell JA, Jones WS, Anker SD, Petrie MC, Harrington J, Mattheus M, Seide S, Zwiener I, Amir O, Bahit MC, Bauersachs J, Bayes-Genis A, Chen Y, Chopra VK, A Figtree G, Ge J, G Goodman S, Gotcheva N, Goto S, Gasior T, Jamal W, Januzzi JL, Jeong MH, Lopatin Y, Lopes RD, Merkely B, Parikh PB, Parkhomenko A, Ponikowski P, Rossello X, Schou M, Simic D, Steg PG, Szachniewicz J, van der Meer P, Vinereanu D, Zieroth S, Brueckmann M, Sumin M, Bhatt DL, Butler J. Effect of Empagliflozin on Heart Failure Outcomes After Acute Myocardial Infarction: Insights From the EMPACT-MI Trial. Circulation. 2024;149:1627-1638. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 68] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 8. | Wasinger VC, Cordwell SJ, Cerpa-Poljak A, Yan JX, Gooley AA, Wilkins MR, Duncan MW, Harris R, Williams KL, Humphery-Smith I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis. 1995;16:1090-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 650] [Cited by in RCA: 542] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 9. | Hou Y, Huang C, Cai X, Zhao J, Guo W. Improvements in the establishment of a rat myocardial infarction model. J Int Med Res. 2011;39:1284-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Zhang HY, Chen X, Hu P, Liang QL, Liang XP, Wang YM, Luo GA. Metabolomic profiling of rat serum associated with isoproterenol-induced myocardial infarction using ultra-performance liquid chromatography/time-of-flight mass spectrometry and multivariate analysis. Talanta. 2009;79:254-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Wang J, Liu X, Ren B, Rupp H, Takeda N, Dhalla NS. Modification of myosin gene expression by imidapril in failing heart due to myocardial infarction. J Mol Cell Cardiol. 2002;34:847-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154-1169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3400] [Cited by in RCA: 3324] [Article Influence: 151.1] [Reference Citation Analysis (0)] |
| 13. | Wu J, Xie X, Liu Y, He J, Benitez R, Buckanovich RJ, Lubman DM. Identification and confirmation of differentially expressed fucosylated glycoproteins in the serum of ovarian cancer patients using a lectin array and LC-MS/MS. J Proteome Res. 2012;11:4541-4552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 14. | Prianichnikov N, Koch H, Koch S, Lubeck M, Heilig R, Brehmer S, Fischer R, Cox J. MaxQuant Software for Ion Mobility Enhanced Shotgun Proteomics. Mol Cell Proteomics. 2020;19:1058-1069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 137] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 15. | Yu F, Haynes SE, Teo GC, Avtonomov DM, Polasky DA, Nesvizhskii AI. Fast Quantitative Analysis of timsTOF PASEF Data with MSFragger and IonQuant. Mol Cell Proteomics. 2020;19:1575-1585. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 232] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 16. | Li TT, Wang ZB, Li Y, Cao F, Yang BY, Kuang HX. The mechanisms of traditional Chinese medicine underlying the prevention and treatment of atherosclerosis. Chin J Nat Med. 2019;17:401-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 17. | Hao PP, Jiang F, Chen YG, Yang J, Zhang K, Zhang MX, Zhang C, Zhao YX, Zhang Y. Traditional Chinese medication for cardiovascular disease. Nat Rev Cardiol. 2015;12:318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Lee W, Ku SK, Min BW, Lee S, Jee JG, Kim JA, Bae JS. Vascular barrier protective effects of pellitorine in LPS-induced inflammation in vitro and in vivo. Fitoterapia. 2014;92:177-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Wan Q, Wang Y, Yong M, Hu P, Le Liang CG, Yang XJ, Zhao X, San D, Bai TT, Tong G, Zhai J, Zhao M, Zhang Q. Sanwei sandalwood decoction ameliorates acute ischemiareperfusion injury in rats by modulating myocyte electrophysiological characteristics. Biomed Pharmacother. 2023;158:114103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 20. | Zhen D, Na RS, Wang Y, Bai X, Fu DN, Wei CX, Liu MJ, Yu LJ. Cardioprotective effect of ethanol extracts of Sugemule-3 decoction on isoproterenol-induced heart failure in Wistar rats through regulation of mitochondrial dynamics. J Ethnopharmacol. 2022;292:114669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Zhang S, Shan L, Li Q, Wang X, Li S, Zhang Y, Fu J, Liu X, Li H, Zhang W. Systematic Analysis of the Multiple Bioactivities of Green Tea through a Network Pharmacology Approach. Evid Based Complement Alternat Med. 2014;2014:512081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 22. | Liu T, Song D, Dong J, Zhu P, Liu J, Liu W, Ma X, Zhao L, Ling S. Current Understanding of the Pathophysiology of Myocardial Fibrosis and Its Quantitative Assessment in Heart Failure. Front Physiol. 2017;8:238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 157] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 23. | Dutka M, Bobiński R, Ulman-Włodarz I, Hajduga M, Bujok J, Pająk C, Ćwiertnia M. Various aspects of inflammation in heart failure. Heart Fail Rev. 2020;25:537-548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 24. | Dawuti A, Sun S, Wang R, Gong D, Liu R, Kong D, Yuan T, Zhou J, Lu Y, Wang S, Du G, Fang L. Salvianolic acid A alleviates heart failure with preserved ejection fraction via regulating TLR/Myd88/TRAF/NF-κB and p38MAPK/CREB signaling pathways. Biomed Pharmacother. 2023;168:115837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 25. | van der Pol A, van Gilst WH, Voors AA, van der Meer P. Treating oxidative stress in heart failure: past, present and future. Eur J Heart Fail. 2019;21:425-435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 613] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 26. | Chen R, Chen W, Huang X, Rui Q. Tanshinone IIA attenuates heart failure via inhibiting oxidative stress in myocardial infarction rats. Mol Med Rep. 2021;23:404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 27. | Xue D, Sun JL, Yang J. Early L-T4 intervention improves fetal heart development in pregnant rats with subclinical hypothyroidism rats by activating BMP4/Smad4 signaling pathway. BMC Cardiovasc Disord. 2020;20:369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 28. | Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci U S A. 2013;110:12526-12534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 451] [Article Influence: 34.7] [Reference Citation Analysis (0)] |