Copyright
©The Author(s) 2025.
World J Cardiol. Dec 26, 2025; 17(12): 112062
Published online Dec 26, 2025. doi: 10.4330/wjc.v17.i12.112062
Published online Dec 26, 2025. doi: 10.4330/wjc.v17.i12.112062
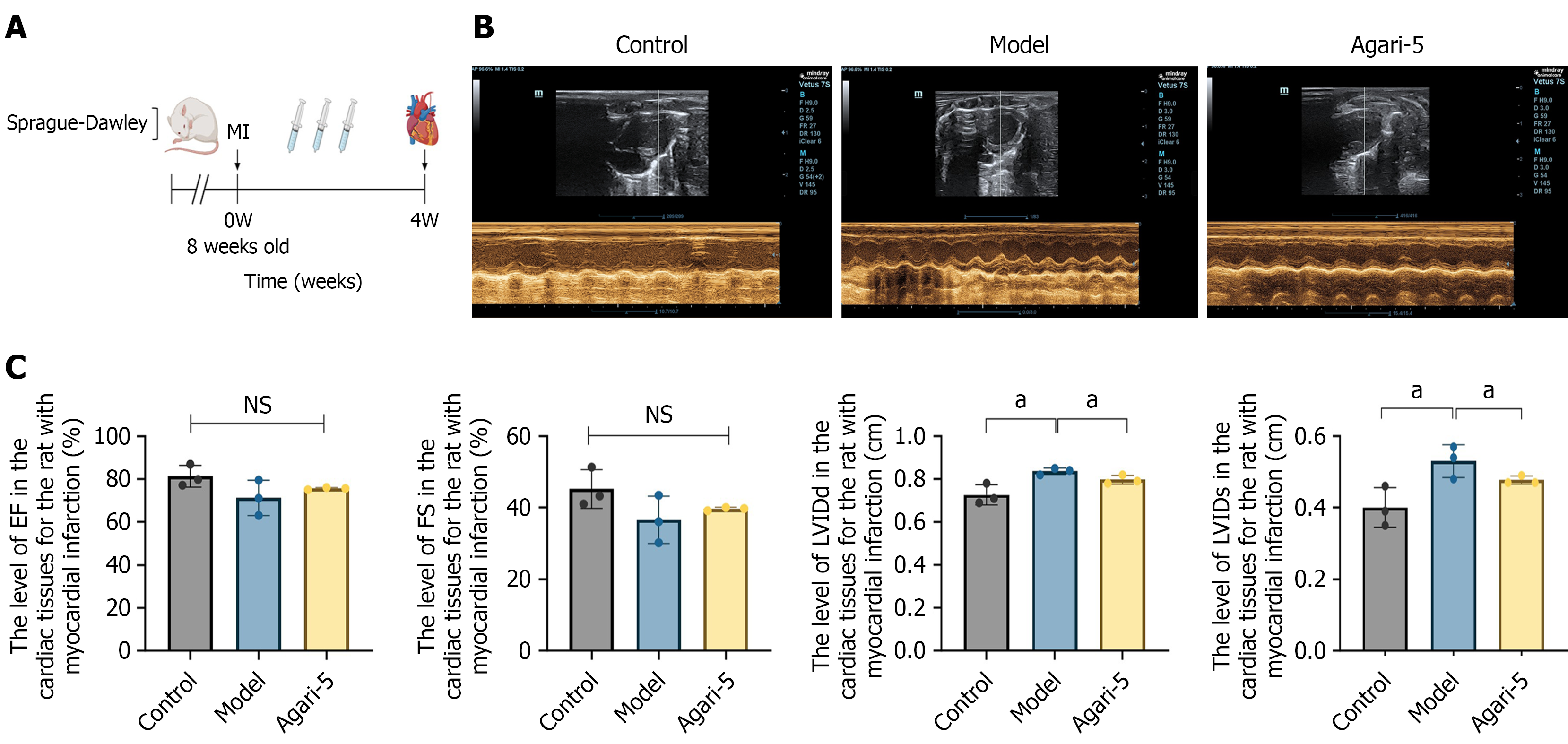
Figure 1 Agari-5 restored cardiac function after myocardial infarction in rats.
A: Overall experimental flowchart; B: Cardiac ultrasound of rats in each group (n = 3 hearts/group); C: Statistical histograms of ejection fraction, fractional shortening, left ventricular end-diastolic dimension, and left ventricular end-systolic dimension data of cardiac ultrasound in rats of each group. All data are expressed as mean ± SD. All statistics are shown in bar graphs. aP < 0.05. EF: Ejection fraction; FS: Fractional shortening; LVIDs: Left ventricular end-systolic dimension; LVIDd: Left ventricular end-diastolic dimension; MI: Myocardial infarction; NS: Not significant.
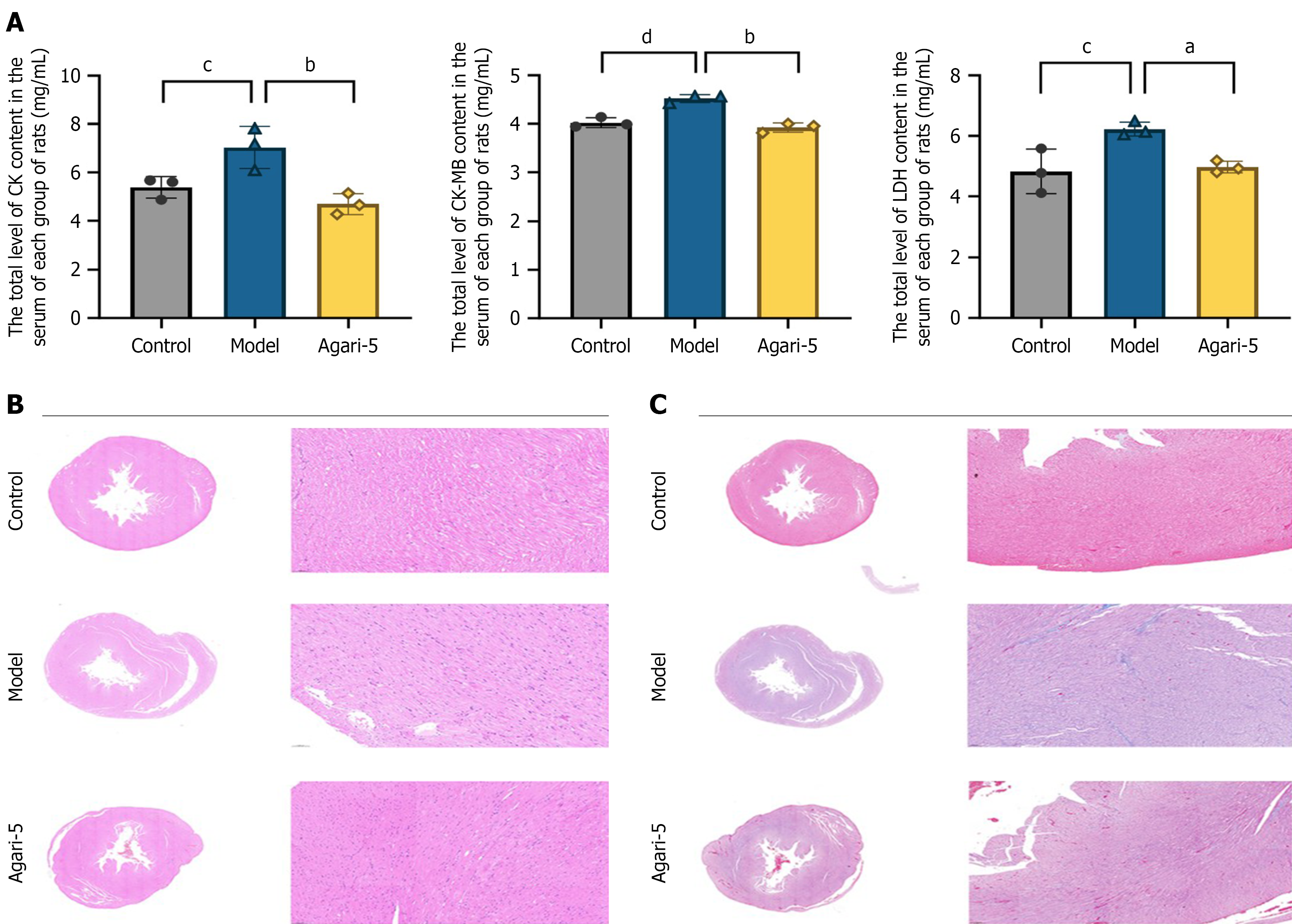
Figure 2 Common indicators for detecting myocardial infarction in rats.
A: Agari-5 restored indicators of cardiac function after myocardial infarction in rats from left to right: (1) Creatine kinase; (2) Creatine kinase isoenzyme; and (3) Lactate dehydrogenase. All data are expressed as mean ± SD. aP < 0.05 vs model, bP < 0.01 vs model, cP < 0.05 vs control, dP < 0.01 vs control; B and C: Histopathological pictures of heart tissue sections were stained with hematoxylin and eosin and Masson’s staining. These parameters were recorded after 4 weeks of gavage administration of Agari-5 following myocardial infarction in rats (n = 3). CK: Creatine kinase; CK-MB: Creatine kinase isoenzyme; LDH: Lactate dehydrogenase.
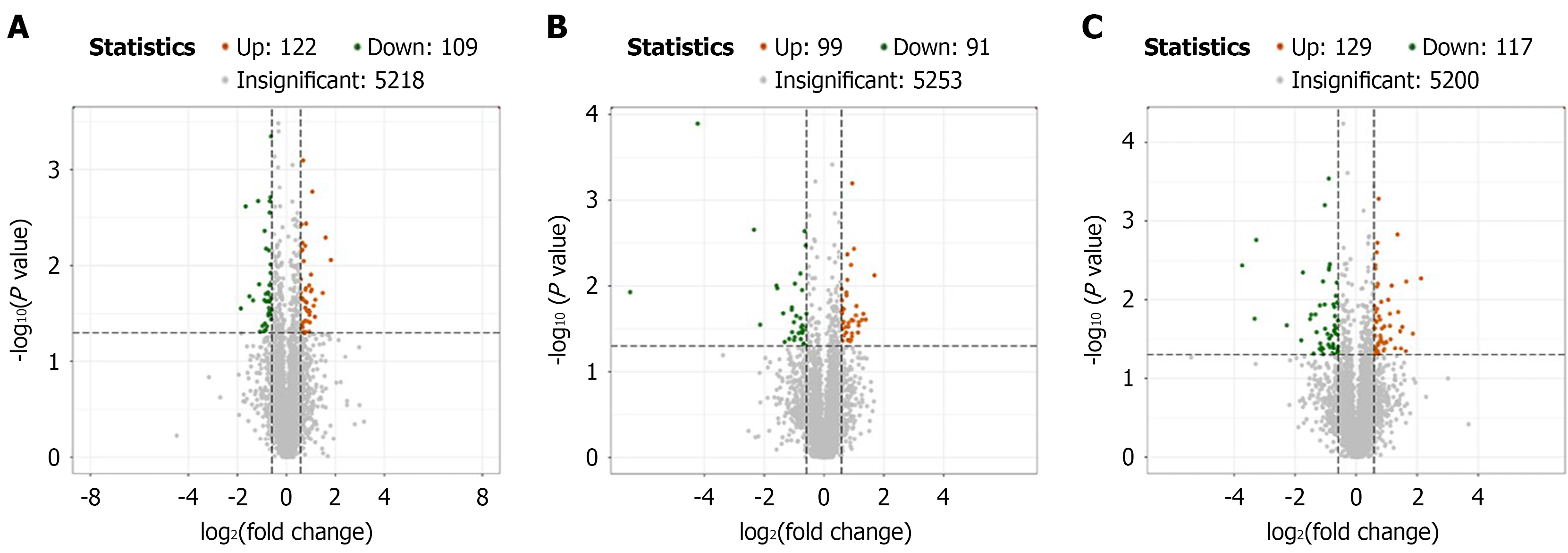
Figure 3 Volcano plot of differential proteins between groups of rats.
A: The control group compared with the model group with 122 proteins upre
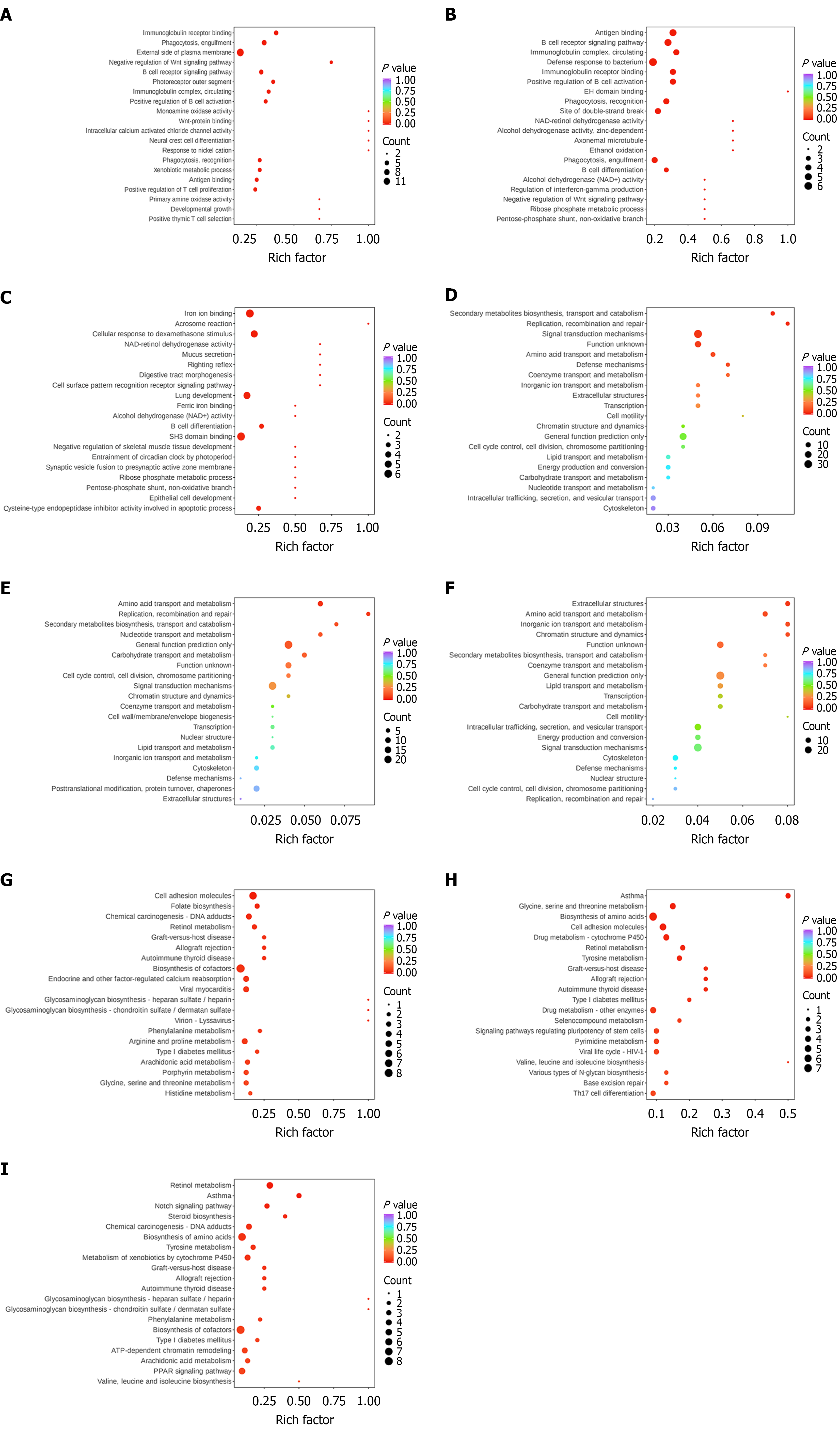
Figure 4 Bubble diagram of enrichment analysis of differentially expressed proteins for Gene Ontology, EuKaryotic Orthologous Groups, and Kyoto Encyclopedia of Genes and Genomes.
A: Gene Ontology (GO) enrichment analysis (control vs model); B: GO enrichment analysis (Agari-5 vs model); C: GO enrichment analysis (Agari-5 vs control); D: EuKaryotic Orthologous Groups (KOG) enrichment analysis (control vs model); E: KOG enrichment analysis (Agari-5 vs model); F: KOG enrichment analysis (Agari-5 vs control); G: KEGG enrichment analysis (control vs model); H: Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Agari-5 vs Model); I: KEGG enrichment analysis (Agari-5 vs Control). The horizontal coordinate represents the enrichment multiplicity, and the vertical coordinate represents the name of the entries of GO, KOG, and KEGG.
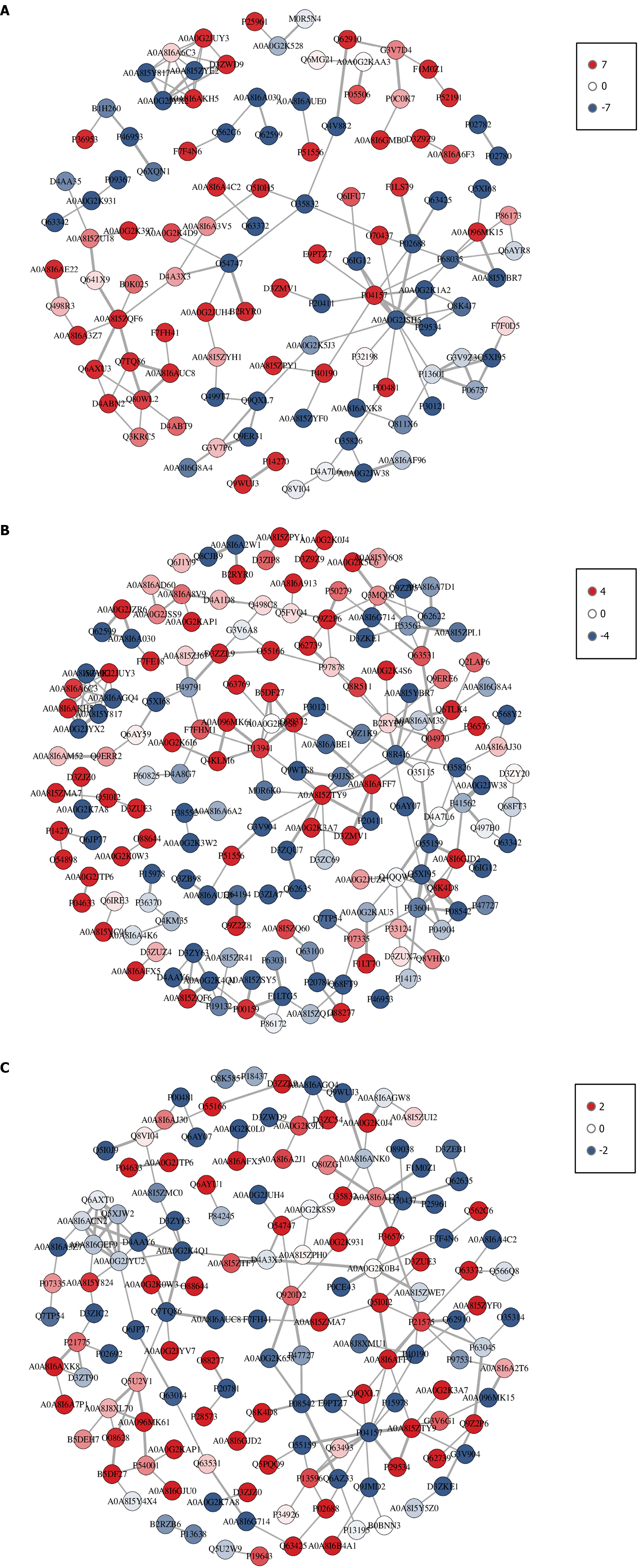
Figure 5 Interaction network diagram of differentially expressed proteins among groups.
A: Control group; B: Model group; C: Agari-5 group. Each node in the interaction network represents a differentially expressed protein; node red (up) indicates that the expression level of differentially expressed proteins is upregulated, and node blue (down) indicates that the expression level of differentially expressed proteins is downregulated.
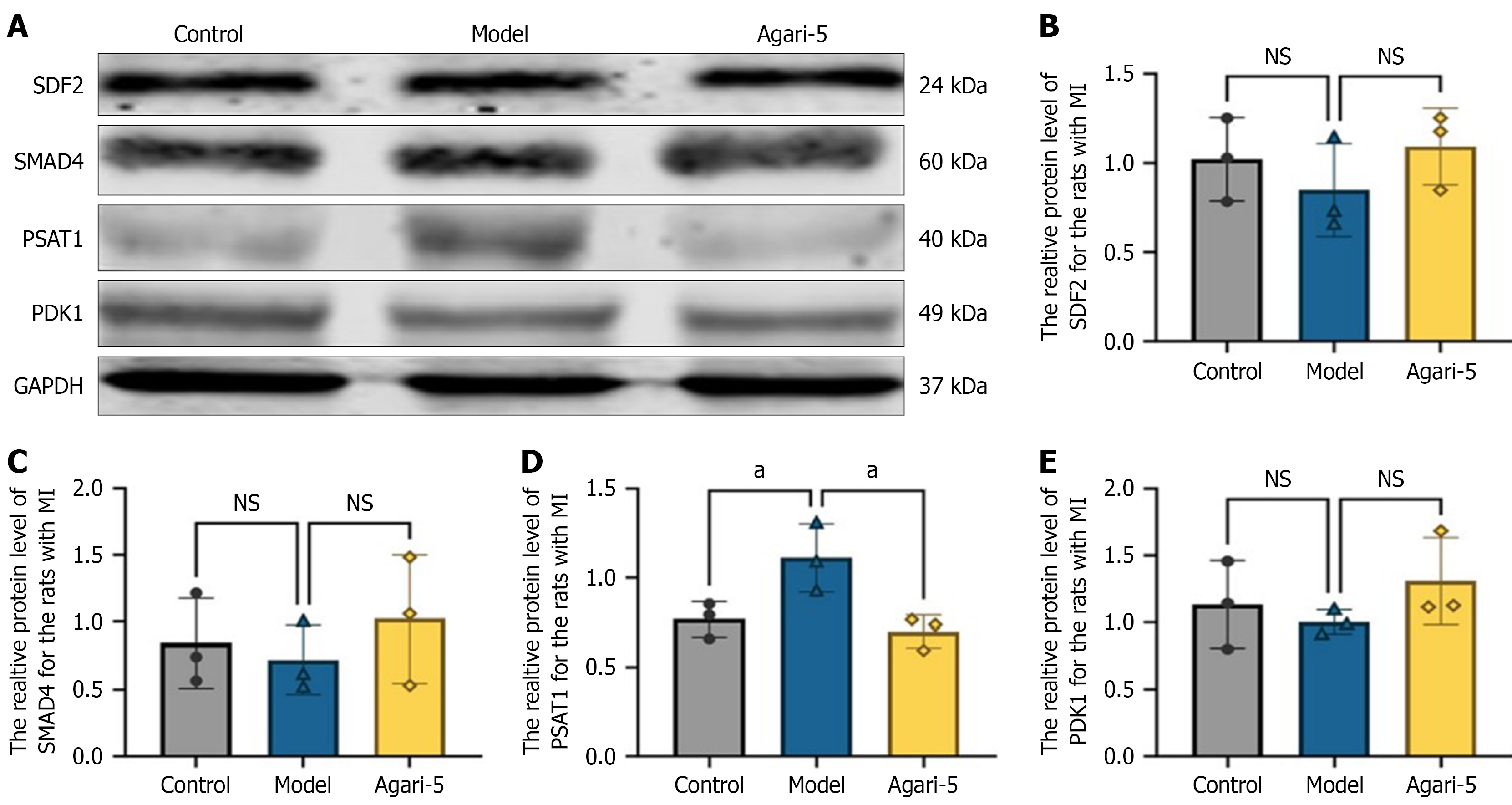
Figure 6 Protein bands.
A: Protein bar graph; B-E: The statistical histograms of each protein (n = 3, mean ± SD). aP < 0.05; NS: Not significant.
- Citation: Zhao YB, Bao ZH, Tu Y, Qiu X, Bao YL, Su M, Qi HJ, Wan Q. Proteomics-based investigation of the protective effect and mechanism of Agari-5 in rats with myocardial infarction. World J Cardiol 2025; 17(12): 112062
- URL: https://www.wjgnet.com/1949-8462/full/v17/i12/112062.htm
- DOI: https://dx.doi.org/10.4330/wjc.v17.i12.112062