Published online Jan 15, 2026. doi: 10.4251/wjgo.v18.i1.113553
Revised: September 30, 2025
Accepted: November 18, 2025
Published online: January 15, 2026
Processing time: 132 Days and 14 Hours
Colorectal cancer remains one of the leading causes of morbidity and mortality worldwide. Despite notable advances in early detection and therapeutic stra
Core Tip: MicroRNAs are crucial regulators of colorectal cancer progression, influencing tumor development, therapeutic response, and serving as potential biomarkers or therapeutic targets. Their expression is strongly shaped by the dynamic interaction between tumor cells and the tumor microenvironment. This comprehensive review summarizes the key microRNAs implicated in tumor progression and discusses the main therapeutic targets, highlighting current knowledge and strategies designed to modulate their expression and enhance treatment response.
- Citation: Quiroz-Reyes AG, Delgado-Gonzalez P, Islas JF, Loaiza-Gutierrez VL, Santoyo-Suarez MG, Garcia-Loredo JA, Gonzalez-Villarreal CA, Ramirez-Fernandez F, Garza-Treviño EN. Tumor microenvironment-driven microRNA dysregulation: Key interactions in colorectal cancer progression. World J Gastrointest Oncol 2026; 18(1): 113553
- URL: https://www.wjgnet.com/1948-5204/full/v18/i1/113553.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v18.i1.113553
Colorectal cancer (CRC) is one of the most common and aggressive types of cancer, ranking third in incidence and second in mortality worldwide[1]. Fortunately, CRC is characterized by hallmarks related to cellular and molecular mechanisms throughout its development, progression, and metastasis. Although its complexity is still being unraveled, factors such as specific mutations, the diverse pool of cells contributing to tumor parenchyma formation, immune suppression, and paracrine protein communication indicate that our understanding remains limited. As with other major diseases, various therapeutic strategies have been developed, including surgery, radiotherapy, chemotherapy, and immunotherapy. However, due to the nature and progression of the disease, nearly 50% of cases remain incurable[2]. Therefore, novel alternatives, such as microRNAs (miRNAs or miRs) regulation, represent promising options to explore.
MiRNAs are short non-coding RNAs ranging from 18 to 25 nucleotides in length. Their structure enables them to interact with messenger RNAs at the 3’ untranslated region (UTR) region, blocking translation and thereby regulating gene expression[3]. Over the past decade, it has become evident that altered expression of several miR’s is strongly associated with the etiology and clinical outcomes of many human cancers, including CRC, highlighting their potential roles in carcinogenesis[4]. While many miR’s work at a local nuclear level, recent developments have determined that miR’s are carried outside the cell via exosomes, hence establishing important communication amongst effector-affected cells, i.e., tumor to adjacent cells, developing crosstalk, which is vital for establishing the tumor microenvironment (TME). Thus, understanding the roles of miR’s is essential for clarifying the mechanisms underlying TME regulation and development. In particular, this review focuses on literature from 2015 to 2025, examining miRNAs and their validated molecular targets, incorporating, where applicable, studies participating in CRC progression, using in vitro and/or in vivo models, as well as patient samples. Special attention is given to alterations in the TME and to molecular pathways driving CRC progression such as proliferation, angiogenesis, migration, and therapeutic response.
MiRNAs act as chemical messengers that mediate intercellular communication in cancer regulation by functioning either as tumor suppressors, blocking malignant transformation, or as oncogenes, promoting cell proliferation and invasion[5]. MiRNAs often exhibit altered expression patterns that contribute to the acquisition of cancer hallmarks. Over time, studies have shown that epigenetic modifications, such as hypermethylation or hypomethylation of promoter CpG islands and dysregulated histone acetylation, can suppress tumor-suppressive miRNAs or enhance oncogenic miRNAs[6]. These alterations promote cell proliferation, resistance to apoptosis, invasiveness, and angiogenesis[7]. Therefore, changes in miR expression profiles may provide valuable insights into disease stage, therapeutic response, and patient prognosis, representing promising therapeutic targets across diverse cancer types[8].
During miR biogenesis, alterations in processing steps can lead to dysregulation. Mutations in key components of the miRNA machinery, such as DGCR8, DROSHA, and DICER1, have been linked to cellular transformation and tumor progression. In addition, many miRs are located in genomic regions prone to deletion, amplification, or translocation in cancer[8]. Alterations in the 3’ UTR and other structural changes can affect RNA structure and binding sites, impairing pre-miR transcription and miR expression, which in turn dysregulate target genes. Some miRs, such as miR-29b, miR-148a, and miR-152, directly target DNA methyltransferases and indirectly modulate gene expression through epigenetic mechanisms[7]. Furthermore, both tumor suppressors and oncogenic factors influence miRNA expression and contribute to cancer pathogenesis[7,8].
Given that miR expression is tightly regulated, extraneous factors such as cellular stress, reduced oxygen availability, and hypoxia within the TME can alter the production and activity of mature miRs. Moreover, miRs can be encapsulated within extracellular vesicles (EVs) to facilitate intercellular communication in the TME[7]. EV-miRs can transform normal fibroblasts into cancer-associated fibroblasts (CAFs), which secrete cytokines and growth factors such as transforming growth factor (TGF)-β, interleukin (IL)-1, IL-6, and tumor necrosis factor (TNF)-α, all of which strongly contribute to tumor progression[5].
The CRC TME is a heterogeneous microenvironment that includes cancer cells, fibroblasts, endothelial cells, and immune cells. The TME develops under hypoxic conditions, which trigger metabolic reprogramming associated with inflammation and promote tumor progression, immune evasion, and therapeutic resistance[9,10]. Under these conditions, hypoxia-inducible factors (HIFs) are expressed, leading to acidification of the microenvironment, which further drives stromal remodeling while suppressing the immune response[11]. HIF activity is regulated by signaling pathways including phosphatidylinositol 3-kinase (PI3K)-mammalian target of rapamycin (mTOR), Janus kinase (JAK)-signal transducer and activator of transcription (STAT) 3, nuclear factor kappa-B, mitogen-activated protein kinase (MAPK), Wnt/β-catenin, and Notch, all of which have been implicated in CRC development[12]. Table 1 includes miR’s presented in key alterations of CRC TME.
| miR | Cell type | Conditions | Context (hipoxy/inflammation/acidosis) | Key effect on TME | Ref. |
| miR-210 | Epitelial tumor CRC | Progression tumor and metastasis | Hypoxia-upregulated | Classic “hypoxamiR”: Induced by HIF-1α; promotes adaptation to hypoxia, invasion and resistance | Coronel-Hernández et al[132] |
| miR-21 | Tumor/epithelial cells and exosome-mediated transfer to stromal, endothelial, and immune cells | Primary tumor and progression tumor | Inflammation (IL-6/STAT3) and angiogenesis upregulated | Role as an oncomiR; suppresses PTEN and PDCD4; potentiates IL-6/STAT3 signaling, thereby promoting invasion and metastasis; contributes to the establishment of a pro-angiogenic TME | Lai et al[133] |
| miR-25-3p | Exosomes released from tumor epithelial cells to target endothelial cells | Progression tumor | Hypoxia/angiogenesis (TME) upregulated | Enhances vascular permeability and angiogenesis through the KLF2/KLF4 axis regulating VEGFR2, ZO-1, occludin, and claudin-5; contributing to the establishment of pre-metastatic niche | Xiong et al[134] |
| miR-1229 | Exosomes released from tumor epithelial cells to target endothelial cells | Progression tumor | Hypoxia/angiogenesis (TME) upregulated | Promotes tube formation by inhibiting HIPK2 and enhancing VEGF | Soheilifar et al[135] |
| miR-320 | Epithelial/estromal (colon) IL-6R/STAT3 | Primary tumor and metastasis | Inflammation (CAC) downregulated | Inhibits IL-6R STAT3 signaling and reduces tumorigenesis in colitis-associated CRC | Wu et al[136]; Mjelle et al[137] |
| miR-590-3p (CAF-exosomal) | CAFs (exosomes) tumoral cells | Progression tumor | Damage response/TME stress upregulated | Confers radioresistance and activates PI3K/AKT; an example of TME remodeling by CAFs | Gou et al[138] |
| miR-34a | Epithelial cells to tumoral cells | Primary tumor supress metastasis | Hipoxia-inflammation/TME downregulated | p53mt-miR-34a suppresses EMT; IL-6/STAT3 downregulates miR-34a, establishing a pro-inflammatory and pro-EMT feedback loop | Włodarczyk et al[139]; Zhang et al[140] |
| miR-338-5p | Epithelial | Primary tumor, progression and drug resistance | Hypoxia/inflammation downregulated | Deficiency of miR-338-5p enhances IL-6/STAT3 signaling and confers resistance to oxaliplatin, fostering a pro-inflammatory TME | Valencia-Cervantes and Sierra-Vargas[141] |
| miR-19a | Epithelial | Progression tumor | Inflammation/survival upregulated (hypoxia conditions) | Suppression of PTEN-PI3K/AKT signaling promotes proliferation and invasion, further sustained by IL-6/STAT3 activation | Rahbar Farzam et al[142] |
| miR-135b-5p (CAF-exosomal) | CAFs (exosomes) epithelial and endothelial cells | Progression tumor | Hypoxia/inflammation upregulated | Exosomes derived from CAFs upregulate miR-135b-5p, leading to TXNIP suppression and enhanced tumor growth and angiogenesis | Umezu et al[143]; Shao et al[144] |
| miR-425-5p (exosomal) | Tumor (exosomes) macrophages/T | Progression tumor | Inmunosupression upregulated | Induction of M2-like polarization along with suppression of the pro-inflammatory T-cell response contributes to tumor progression and increased vascular permeability | Feng et al[145] |
| miR-934 (exosomal) | Tumor (exosomes) macrophages (liver) | Upregulated metastasis | Inflammation/metastasis | Induces M2 polarization and facilitates hepatic metastasis | Zhao et al[105] |
| miR-128-3p | Tumor (exosomes) epithelial | Primary tumor and progression | Inflammation (STAT3) upregulated | Activation of JAK/STAT3 and TGF-β/SMAD signaling promotes EMT and metastatic progression | Rahbar Farzam et al[142] |
| miR-9-5p | Epithelial tumoral to SLC9A1/NHE1 (antiport Na+/H+) | Progression tumor and metastasis | Acidosis upregulated | Modulation of NHE1 contributes to extracellular acidification, which in turn facilitates tumor invasion and metastasis | Wang et al[146] |
| miR-224-5p | Epithelial tumoral (HT29) SLC4A4/NBCe1 (Na+/HCO3-) | Progression | Acidosis upregulated | Repression of HCO3- transport diminishes pH buffering capacity, thereby exacerbating tumor acidosis | Yi and Yu[147] |
| miR-34a | Epithelial tumoral LDHA (lactate dehydrogenase A) | Primary tumor and progression | Acidosis downregulated | Acidosis suppress p53wt downregulation of miR-34a increases LDHA expression, leading to elevated lactate levels and acidosis; it also promotes EMT and therapy resistance | Li et al[14]; Xiong et al[134] |
| miR-143 | Epithelial tumoral hexokinase 2 | Primary tumor overexpresssion metastasis | Acidosis downregulated | Loss of this factor promotes glycolytic flux and lactate accumulation, exacerbating tumor acidosis | Gregersen et al[148]; Guo et al[149] |
The progression of the TME in CRC, driven by alterations in miR expression, is highly complex. Evidence suggests that dysregulated expression of specific miRs promotes the formation of aberrant crypts and rectal polyps while inducing microenvironmental changes in mouse models. These findings underscore the importance of detecting early miRs expression alterations within normal intestinal mucosa, as such changes may facilitate TME initiation and progression[13].
Among the diverse features of the TME, hypoxia is particularly prominent and fosters cellular proliferation, angio
Several studies have identified oxygen deprivation as a driver for subsets of miRs collectively termed hypoxiamirs. In cancer, these include miRs that reinforce TME remodeling, such as miR-210, miR-2, and miR-30d[15]. Yang et al[16] further identified miR-197 and miR-26a as potential remodelers, whereas miR-375 appeared to play a protective role. These findings were correlated with poorer patient prognosis. Low oxygen conditions also correlate with activation of inflammatory signals such as TGF-β, platelet-derived growth factor, and other cytokines, as well as the induction of CAFs.
CAFs are important because they produce extracellular matrix components that facilitate tumor invasion, angiogenesis, and therapeutic resistance[10]. Savardashtaki et al[11] reported that CAF-derived exosomal miRs, miR-21, miR-199a, miR-181a, and miR-329, which are modulators of tumor-stromal crosstalk, contribute to aggressive cancer phenotypes. Inflammation and associated markers, i.e., TNF-α and nuclear factor kappa-B, are also subject to regulation by miRs[17].
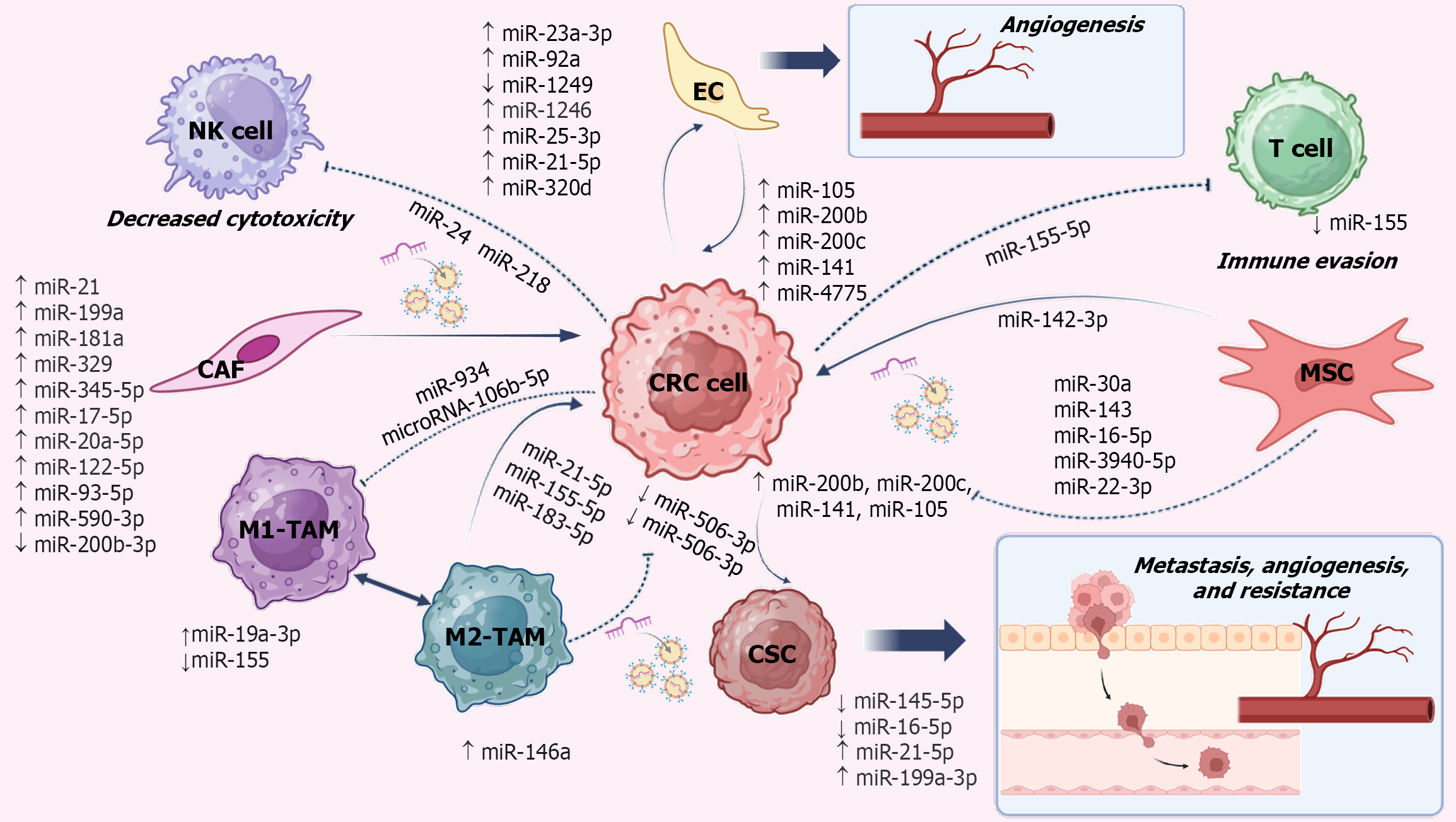
Also, such effects have been observed in tumor-associated macrophages (TAMs), where elevated miR-155 activates Toll-like receptor signaling through nuclear factor kappa-B, hence mediating remodeling[18]. TAMs predominantly display an M2-like phenotype, accumulate in hypoxic tumor niches, and secrete pro-angiogenic and immunosuppressive factors such as vascular endothelial growth factor (VEGF), TNF-α, and matrix metalloproteinase-9, thereby supporting tumor growth and invasion[17]. Moreover, miR-24 and miR-218 reduce the cytotoxic effects of natural killer (NK) cells in CRC and lung adenocarcinoma, respectively, contributing to immune tolerance in the TME[19]. Remodeling of the TME is further promoted by the angiogenic activity of miR-155[20]. Another effect occurs when miR-21 is induced by chronic inflammation, enhancing chemoresistance and immune evasion through the PTEN and Toll-like receptor pathways[21].
IL-6 secreted by TAMs activates the JAK/STAT pathway and suppresses miR-506-3p expression in CRC cells. miR-506-3p functions as a tumor suppressor by targeting FoxQ1, thereby inhibiting CCL2 expression and macrophage recruitment. IL-6 also increases vimentin expression while reducing E-cadherin expression[22]. Furthermore, IL-6 activates the IL-6R/STAT3 pathway, transcriptionally activates STAT3, and suppresses the tumor suppressor miR-204-5p, which increases chemoresistance to 5-fluorouracil (5-FU) and oxaliplatin[23]. M2 macrophages with high expression of miR-21-5p and miR-155-5p bind to BRG1 in CRC, promoting metastasis[24].
Epithelial-mesenchymal transition (EMT) is a key step in metastasis, driven by cancer stem cells (CSCs), which main
Feedback communication between CRC and endothelial cells allows transfer of miR’s to promote angiogenesis. CRC-derived miR-25-3p enhances vascular permeability and angiogenesis by targeting KLF2 and KLF4 in endothelial cells[30]. In addition, miR-21-5p suppresses KRIT1 in human umbilical vein endothelial cells (HUVECs), activating the β-catenin pathway and increasing VEGFA and CCND1 expression[31]. CRC-derived micro-vesicles containing miR-1246 promote angiogenesis by activating SMAD signaling in HUVECs through HIPK2 targeting. Suppression of HIPK2 allows MEF2C-mediated VEGF activation[32]. Furthermore, miR-320 loaded into micro-vesicles reduces GNAI1 levels in endothelial cells, enhancing JAK2/STAT3 signaling, VEGF production, proliferation, invasion, and angiogenesis[33].
MiRNAs regulate gene expression by coordinating complex molecular networks involved in CRC initiation, progression, metastasis, and response to therapy. Table 2 summarizes the main miRNAs associated with each consensus molecular subtype (CMS)(CMS1-CMS4) of CRC and highlights their clinical relevance. Notable miRNAs in CRC research include miR-145-5p[34,35], miR-16-5p[36,37], miR-199a-3p[38], miR-21-3p[39,40], and miR-21-5p[41], which collectively modulate overlapping pathways, such as PI3K/AKT, Ras/MAPK, Wnt/β-catenin, apoptosis regulators, DNA repair, angiogenesis, and EMT[42], as shown in Table 3. Dysregulation of these miRNAs disrupts cellular homeostasis, leading to uncontrolled proliferation, enhanced survival, EMT activation, stemness, metastatic potential, and resistance to chemotherapy, radiotherapy, and targeted therapies[31-84].
| CMS class | Molecular features | Frequency (%) | Immune phenotype | Prognosis | miR | Ref. |
| CMS1: Immune MSI | CIMP (increase); BRAFV600E m; hypermutated; KRASwt; TP53wt | 14 | Immune activation and infiltration LTC and NK | Intermediate prognosis; good early disease control but poor survival after relapse | miR-625 (increase), miR-31 (increase), miR-155 (increase) | Adam et al[150] |
| CMS2: Canonical (epithelial differentiation) | CIMP negative; BRAFwt; KRASwt; TP53m | 37 | WNT and MYC activation. Immune dessert | Best overall | miR-592 (increase), miR-552 (increase) | Adam et al[150] |
| CMS3: Metabolic | CIMP negative; BRAFwt; KRASm; TP53wt | 13 | Metabolic deregulation | Poor immunogenicity | miR-625 (increase) | Adam et al[150] |
| CMS4: Mesenchymal | CIMP negative; BRAFwt; KRASwt | 23 | Stromal infiltration (macrophages) TGF-β activator-CSC EMT and angiogenesis | Worse and poor survival. Resistant standard treatment | miR-625 (decrease), miR-143 (increase) (CMS4 vs CMS2); miR-200 (decrease), miR-218 (increase) | Adam et al[150]; Gherman et al[151] |
| miR | Expression | Study model | Target genes | Modulated pathways | Ref. |
| miR-145-5p | Downregulated | In vitro | N-RAS and IRS1 | Cell proliferation by AKT inactivation | Yin et al[152] |
| miR-145-5p | Downregulated | In vitro | CDCA3 | Cell proliferation, migration, invasion, EMT | Chen et al[84] |
| miR-145-5p | Downregulated | In vitro | TWIST1 | Migration and invasion | Shen et al[153] |
| miR-145-5p | Downregulated | In vitro | MAPK1 | Cell proliferation, migration, and invasion | Yang et al[154] |
| miR-145-5p | Downregulated | In vitro | SIP1 | Cell proliferation, migration, and invasion | Sathyanarayanan et al[155] |
| miR-145-5p | Downregulated | In vitro | PAK4 | Migration and invasion | Sheng et al[59] |
| miR-145-5p | Downregulated | In vitro and in vivo | p70S6K1 | Tumor growth and angiogenesis by HIF-1 and VEGF | Xu et al[156] |
| miR-145-5p | Downregulated | In vitro and in vivo | LASP1 | Invasion and metastasis | Wang et al[58] |
| miR-145-5p | Downregulated | In vitro | CXCL1 and ITGA2 | Cell proliferation and migration | Zhuang et al[60] |
| miR-16-5p | Downregulated | In vitro and in vivo | PVT1 | Cell proliferation, migration, and invasion by VEGFA and p-AKT | Rahmati et al[98] |
| miR-16-5p | Downregulated | In vitro and in vivo | ITGA2 | Apoptosis and tumor growth | Xu et al[36] |
| miR-16-5p | Downregulated | In vitro | BIRC5 | Apoptosis, cell proliferation, and angiogenesis | Aslan et al[47] |
| miR-16-5p | Downregulated | In vitro | FOXK1 | Cell proliferation and angiogenesis by PI3K/AKT/mTOR signaling | Huang et al[37] |
| miR-16-5p | Downregulated | In vitro and in vivo | HMGA2 | Migration, invasion, and EMT by β-catenin pathway | Cai et al[63] |
| miR-199a-3p | Downregulated | In vitro | PAK4 and BCAR3 | Cell proliferation, migration and invasion | Hou et al[70] |
| miR-199a-3p | Downregulated | In vitro | FN1 | EMT by N-cadherin and vimentin | Lin et al[71] |
| miR-199a-3p | Downregulated | In vitro | NLK | Metastasis | Han et al[72] |
| miR-199a-3p | Downregulated | In vitro | TGFBR1 and PDGFRB | Cell proliferation by MAPK-signaling | Slattery et al[157] |
| miR-21-3p | Upregulated | In vitro and in vivo | SMAD7 | EMT through the increase of N-cadherin | Jiao et al[39] |
| miR-21-3p | Upregulated | In vitro | RBPMS | Migration, invasion, and apoptosis by Smad4/ERK signaling | Hou et al[53] |
| miR-21-5p | Upregulated | In vitro and in vivo | KRIT1 | Angiogenesis through β-catenin signaling pathway, VEGFA and CCND1 | He et al[31] |
| miR-21-5p | Upregulated | In vitro and in vivo | PDCD4 and TGFBR2 | Stemness promotion by upregulation of β-catenin, c-MYC and cyclin-D1 | Yu et al[158] |
| miR-21-5p | Upregulated | In vitro and in vivo | PTEN | Apoptosis, cell proliferation and invasion | Wu et al[76]; Lin et al[77] |
| miR-21-5p | Upregulated | In vitro and in vivo | CHL1 | Cell proliferation, invasion and tumor growth | Yu et al[79] |
| miR-21-5p | Downregulated | In vitro | TGFBI | Pyroptosis | Jiang et al[82] |
| Downregulated | In vitro | SATB1 | Cells sensitive to chemoradiation | Lopes-Ramos et al[83] | |
| miR-4461 | Downregulated | In vitro | COPB2 | Cell proliferation, migration, and invasion | Chen et al[159] |
| miR-449a | Downregulated | In vitro | HDAC1, TGFB, SATB2, ADAM10, MYC, and MAPK1 | Cell proliferation, invasion and poor survival | Ishikawa et al[160] |
| miR-519d-3p | Downregulated | In vitro | TROAP | Apoptosis, cell proliferation, migration, and invasion | Ye and Lv[161] |
| miRNA-31 | Upregulated | In vitro | STK40 | NF-κB signaling pathway and invasion | Zhu and Xue[162] |
| miR-200a | Upregulated | In vitro | PTEN | Cell proliferation, migration and invasion | Li et al[163] |
| miRNA-552 | Upregulated | In vitro | PTEN | Poor prognosis | Im et al[164] |
| miRNA-552 | Upregulated | In vitro and in vivo | ADAM28 | Cell proliferation, migration and tumor growth | Wang et al[165] |
| miR-592 | Upregulated | In vitro | mTOR and FOXO | Cell proliferation, migration and invasion | Pan et al[166] |
| miR-708 and miR-31 | Upregulated | In vitro | CDKN2B | Cell proliferation, invasion and apoptosis resistance | Lei et al[167] |
| miR-25 | Upregulated | In vitro and in vivo | SIRT6 | Metastasis through inhibited | Wang et al[168] |
| miR-130b-3p | Upregulated | In vitro and in vivo | CHD9 | Cell proliferation and tumor growth | Song et al[169] |
| miRNA-221 | Upregulated | In vitro and in vivo | TP53BP2 | Cell proliferation through TP53 inhibition | Ali et al[124] |
The tumor-suppressive miR’s miR-145-5p and miR-16-5p inhibit PI3K/AKT signaling, reduce mesenchymal marker expression (N-cadherin and vimentin), restore epithelial markers such as E-cadherin, and suppress proliferation, migra
Conversely, miR-21-3p and miR-21-5p inhibit apoptosis by upregulating multidrug resistance proteins (multidrug resistance-1, multidrug resistance protein 1), enhancing antioxidant defenses (glutathione, superoxide dismutase, glu
MiR-145-5p represses EMT- and CSC-associated transcription factors (OCT4, SOX2, and KLF4), thereby inhibiting spheroid formation and stem-like properties[35,44,85]. miR-16-5p suppresses HMGA2 and FOX-1 expression, limiting EMT and metastatic potential[37,63].
In contrast, miR-21-5p promotes CSC traits via Wnt/β-catenin activation and TGF-βR2 suppression, leading to nuclear β-catenin accumulation and upregulation of epithelial cell adhesion molecule and catenins[75,80,83,84,86]. miR-21-3p enhances EMT, migration, and invasion, whereas miR-199a-3p reduces mesenchymal marker expression under hypoxic conditions, highlighting context-dependent regulation[70-73]. Collectively, these miR’s establish a dynamic balance that dictates CRC invasiveness and recurrence.
Angiogenesis and microenvironmental modulation are critical for tumor progression. miR-145-5p inhibits VEGF, HIF-1α, and N-RAS, reducing neovascularization. miR-16-5p suppresses VEGFA expression, impairing vascular support for tumors. In contrast, miR-21-5p enhances angiogenesis by repressing KRIT1 and activating β-catenin signaling, promoting vascular permeability and neovascularization[31,42]. miR-199a-3p regulates endothelial inflammation and barrier function, indirectly influencing tumor angiogenesis and metastatic niche formation[36,87].
Therapy resistance arises from the combined effects of these miRNAs on DNA repair, survival signaling, EMT, stemness, and drug efflux. miR-145-5p modulates the 5-FU response via the ATF4/miR-145/HDAC4/p53 axis and RAD18-mediated DNA repair, while reducing cetuximab resistance by targeting RREB1 and Ras/MAPK signaling[44,88,89]. miR-16-5p enhances chemotherapy and radiotherapy sensitivity by targeting KRAS, BCL2, and the PI3K/AKT/mTOR pathway, and its low expression predicts poor response. miR-199b-3p contributes to cetuximab resistance via Wnt/β-catenin signaling and CRIM1 regulation; its inhibition restores drug sensitivity both in vitro and in vivo[90-93]. miR-21-3p promotes cisplatin and paclitaxel resistance by enhancing drug efflux, antioxidant defense, and survival signaling[86]. miR-21-5p mediates resistance to 5-FU, oxaliplatin, and radiation by repressing SATB1, PDCD4, PTEN, and TIMP3[77,79,80,83,94,95]. Contextually, miR-21-5p can sensitize cells to chemoradiotherapy in rectal cancer, illustrating its dual role as both a resistance mediator and a therapeutic target.
Collectively, miR-145-5p, miR-16-5p, miR-199a-3p, miR-21-3p, and miR-21-5p form a highly interconnected regulatory network[43-51,79,80]. Their coordinated modulation of PI3K/AKT, Ras/MAPK, Wnt/β-catenin, apoptosis, DNA repair, EMT, stemness, and angiogenesis underlies CRC progression, metastasis, and therapy resistance[53-84]. Therapeutic strategies targeting these miRs, individually or in combination[31-38,66], offer potential to improve chemosensitivity[39-42,76,79,85], radiosensitivity[43-51,79,80], and overall clinical outcomes, emphasizing the importance of understanding miR crosstalk[53-84,89] and pathway integration in precision oncology[73-100]. Table 4 provides an overview of the miRs linked to treatment response. Below are selected examples highlighting specific miRs and the molecular mechanisms by which they influence therapeutic outcomes in CRC.
| miR | Mechanism | Study model | Target | Response therapy | Ref. |
| miR-153-5p | Overexpression | In vitro | BCL-2 | Sensibilize oxaliplatin | He et al[170] |
| miR-145-5p | Decreased | In vitro | BIRC5, Fli-1 | Sensibilize, 5-FU, oxaliplatin | Xie et al[171] |
| miR-1451 | Overexpression | In vitro and in vivo | SNAI1, HDAC4 and ATF4 | Sensibilize radiotherapy and 5-FU | Zhao et al[88]; Zhu et al[172] |
| miR-150-5p | Overexpression | In vitro and in vivo | BIRC5, CASP7, VEGFA | Anti-VEGF | Slattery et al[173]; Chen et al[174] |
| miR-195-5p | Expression | In vivo | GDPD5 | Sensibilize 5-FU | Feng et al[175] |
| Overexpression | In vitro | BIRC5, BCL-2, YAP | Sensibilize, doxorrubicin and oxaliplatin | Qu et al[176]; Poel et al[177] | |
| miR20b-5p1 | Expression | In vitro and in vivo | CTSS, ADAM9, EGFR, CCND1/CDK4/FOXM1 axis | Sensibilize 5-FU | Fu et al[178]; Yang et al[179] |
| miR21-3p | Overexpression | In vitro | MDR1 and MRP1 | Cisplatin resistance | Dong et al[86] |
| miR21-5p | Overexpression | In vitro | SATB1, PTEN, MSH2, PDCD4 | Chemoresistance (oxaliplatin) | Chen et al[94] |
| miR497-5p | Overexpression | In vitro and in vivo | KSR1, BCL-2, IGF1-R | Sensibilize 5-FU, oxaliplatin | Poel et al[177]; Wang et al[180] |
| miR-17-5p | Overexpression | In vitro and in vivo | MFN2, vimentin STAT3, E2F1, HMGA2, SOX4, TWIST1, and EGFR | Resistance oxaliplatin, irinotecan, and fluorouracil | Kim et al[26]; Sun et al[181] |
| miR-199b-3p; miR-199a-5p | Overexpression | In vitro and in vivo | CRIM1 | Resistance cetuximab, sensitive cetuximab | Kim et al[26]; Han et al[93]; Mussnich et al[182] |
| miR-124 | Overexpression | In vitro and in vivo | PRRX1 | Sensitive radiotherapy (inhibition PRRX1) | Zhang et al[183] |
| miR-1226-5p | Overexpression | In vitro | IRF1 | Resistance radiotherapy | Choi et al[184] |
| miR-7-5p | Downregulated | In vitro and in vivo | KLF4 | Resistance radiotherapy | Shang et al[185] |
| miR-16-5p | Downregulated | In vitro | FOXK, PI3K/AKT/mTOR | Resistance radiotherapy | Mousavikia et al[91] |
| miR-423-5p | Downregulated | In vitro | BCL-2 | Resistance radiotherapy | Shang et al[186] |
In CRC, the interaction between tumor cells and the TME is orchestrated through a dynamic exchange of signals mediated by EVs, cytokines, growth factors, and non-coding RNA[101-103]. EVs including exosomes, micro-vesicles, and apoptotic bodies act as critical carriers of miR’s, thereby regulating gene expression and sustaining tumor stromal communication. TAM-derived EVs often promote malignancy by delivering miR-21, miR-155, and miR-105, which enhance angiogenesis, immune evasion, and tumor growth[20,21]. In the immune evasion context, miR-934 and miR-106b-5p inhibit M1 macrophage polarization through the upregulation of miR-19a-3p and downregulation of miR-155[18,104-107]. CAFs also display distinct miR signatures, with overexpression of miR-345-5p, miR-17-5p, miR-20a-5p, miR-122-5p, miR-93-5p, and miR-590-3p, together with reduced levels of miR-200b-3p, contributing to stromal remodeling[108-113].
Tumor cells further evade immune surveillance by expressing miR-24 and miR-218, which suppress NK cell cyto
Currently, most clinical trials (n = 11) evaluating miRNAs in CRC have focused on their roles as biomarkers and predictors of tumor stage. These studies, registered in the United States National Library of Medicine (https://clinicaltrials.gov/), highlight the diagnostic and prognostic potential of miRs. Therapeutically, two main strategies are being pursued: (1) The use of miR mimics to restore tumor-suppressive miRs; and (2) The inhibition of oncogenic miRs using antisense oligonucleotides, locked nucleic acids (LNAs), or miR sponges[119,120]. Although viral and liposomal systems have been employed to improve mimic delivery, clinical translation has been limited by toxicity, off-target effects, and immune-related complications[121,122].
Among these mimics, MRX34 (a liposomal miR-34a mimic) represented the first in-human clinical trial, demonstrating preliminary antitumor activity in refractory solid tumors before termination due to severe immune-mediated toxicities[123]. Other candidates, such as TargomiRs (miR-16 mimic) and INT-1B3 (miR-193a-3p mimic), have shown encouraging safety and preclinical efficacy. However, their clinical progress has been slowed by modest outcomes and financial constraints. In contrast, inhibitors targeting oncogenic miRs have produced more favorable results. For example, LNA-i-miR-221 demonstrated an excellent safety profile and durable clinical benefit in CRC patients, while coboomarsen (anti-miR-155) improved safety and quality-of-life outcomes in lymphoma patients before its discontinuation for strategic reasons[124,125]. More recently, TTX-MC138 (anti-miR-10b) entered early-phase clinical trials for metastatic cancers, and lademirsen (anti-miR-21) was well tolerated in patients with Alport syndrome, although it failed to provide significant clinical benefits[126-128]. Collectively, these findings suggest that miR inhibition may represent a safer and more feasible therapeutic approach than mimic-based strategies, while highlighting the need for further optimization.
Given the limitations of conventional delivery systems, there is growing interest in developing novel, efficient, and non-cytotoxic vehicles. Cell-derived carriers, particularly exosomes, have emerged as promising delivery platforms for miRs[129]. Among these, mesenchymal stem cell (MSC)-derived exosomes are especially attractive because of their clinical applicability in gene therapy delivery systems (e.g., IL-2, interferon, and TNF-related apoptosis-inducing ligand)[130,131]. Table 5 presents selected examples of MSC-derived exosomes engineered for miRNA delivery, which may offer a potential therapeutic avenue for CRC treatment.
| Exo-miR | Cell delivery | Study model | Mechanism | Ref. |
| Exo-miR30a; miR222 | hCC-MSC | In vitro and in vivo | Growth (increase), migration and metastasis (inhibit MIA3) | Du et al[187] |
| Exo-miR4461 | hBM-MSC | In vitro | The proliferation, migration and invasion by down-regulating COPB2 (decrease) | Chen et al[159] |
| Exo-miR22-3p | hBM-MSC | In vitro | Proliferation and invasion (RAP2B/PI3K/AKT pathway) (decrease) | Wang and Lin[188] |
| Exo-miR-16-5p | hBM-MSC | In vitro and in vivo | Proliferation, invasion and migration (downregulating ITGA2) (decrease) | Xu et al[36] |
| Exo-miR431-5p | hUC-MSC | In vitro and in vivo | Progression (suppress PRDX1) (decrease) | Qu et al[189] |
| Exo-anti-miR-146b-5p ASO | hUC-MSC | In vitro and in vivo | Proliferation, migration and EMT (inhibition of Smad signaling) (decrease) | Yu et al[190] |
| Exo-miR-486-5p | hUC-MSC | In vitro | Glycolysis and cell stemness by targeting NEK2 (decrease) | Cui et al[191] |
| Exo-miR-431-5p | hUC-MSC | In vitro and in vivo | Cell growth and progression by inhibiting PRDX1 (decrease) | Qu et al[189] |
| 1Exo-miR-199a-3p | AMSCs | In vitro and in vivo | Sensitized to chemotherapeutic agents by targeting mTOR pathway | Lou et al[192] |
While the role of the microenvironment in modulating miRs and influencing CRC prognosis and treatment has been acknowledged, ongoing research continues to clarify the mechanisms underlying specific regulatory targets that are critical for understanding tumor cellular and immunological characteristics.
The present work provides an extensive examination of miR’s implicated in carcinogenesis, expressed in CRC, and regulated by molecular pathways associated with tumor progression and resistance. The continued investigation of tissues and circulating miR’s is crucial, as it represents a promising approach to developing noninvasive diagnostic tools capable of predicting treatment response and recurrence risk.
Because the functional role of each miR varies greatly, numerous miR’s, especially newly identified ones, remain under intensive study. Modern methodologies for RNA analysis, exosome isolation, and expression panel analyses have led to considerable improvements in the identification of biomarkers, predictive signatures, and therapeutic targets. A major challenge in advancing the functional understanding of miR’s in CRC is the lack of preclinical models that accurately replicate the complexity of the TME. The use of organoids, three dimensional models, and co-culture systems incor
To date, miR-based therapies have faced substantial challenges, particularly in developing strategies that ensure efficient and stable delivery of miR’s to target sites without inducing cytotoxicity. Promising results have been observed when MSCs were used as delivery vehicles; however, these strategies have thus far only been evaluated in in vitro and in vivo models, and their effectiveness in clinical settings remains to be determined.
Ultimately, integrating miR profiles with genomic and clinical data is essential for advancing personalized medical strategies for CRC. Implementing such an approach could enable the development of customized therapies tailored to the specific molecular and biological features of each patient. Achieving meaningful progress in this direction will require a multidisciplinary effort that brings together molecular biology, preclinical modeling technologies, and clinical research.
In summary, our findings emphasize the regulatory roles of miRNAs in CRC that must be interpreted within the context of TME heterogeneity. While certain mechanisms are broadly shared across diverse TMEs, others are restricted to specific molecular subtypes or cellular contexts. MiRNAs emerge as central modulators of CRC progression, acting as both oncogenic and tumor-suppressive molecules that shape key biological processes, including epithelial mesenchymal transition, stemness acquisition, and therapeutic response. Their expression profiles not only hold prognostic value but also highlight promise as biomarkers for treatment stratification. Although definite clinical validation is still lacking, innovative strategies particularly stem cell-derived exosomes for targeted delivery of miRNAs or anti-miRNAs offer a path towards more precise and personalized therapies. Continued research in this field is therefore essential to improve patient outcomes and advance our understanding of CRC biology.
We are grateful to the students of Universidad Autonoma de Nuevo Leon for their assistance in the literature analysis that contributed to this manuscript.
| 1. | Roshandel G, Ghasemi-Kebria F, Malekzadeh R. Colorectal Cancer: Epidemiology, Risk Factors, and Prevention. Cancers (Basel). 2024;16:1530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 182] [Reference Citation Analysis (1)] |
| 2. | Fadlallah H, El Masri J, Fakhereddine H, Youssef J, Chemaly C, Doughan S, Abou-Kheir W. Colorectal cancer: Recent advances in management and treatment. World J Clin Oncol. 2024;15:1136-1156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 85] [Reference Citation Analysis (0)] |
| 3. | Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 413] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 4. | Sur DG, Colceriu M, Sur G, Aldea C, Silaghi C, Samasca G, Lupan I, Căinap C, Burz C, Irimie A. MiRNAs roles in the diagnosis, prognosis and treatment of colorectal cancer. Expert Rev Proteomics. 2019;16:851-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 5. | Wu HH, Leng S, Sergi C, Leng R. How MicroRNAs Command the Battle against Cancer. Int J Mol Sci. 2024;25:5865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Marei HE. Epigenetic regulators in cancer therapy and progression. NPJ Precis Oncol. 2025;9:206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 7. | Wang Z, Wang H, Zhou S, Mao J, Zhan Z, Duan S. miRNA interplay: Mechanisms and therapeutic interventions in cancer. MedComm Oncol. 2024;3:e93. [DOI] [Full Text] |
| 8. | Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1378] [Cited by in RCA: 1687] [Article Influence: 153.4] [Reference Citation Analysis (0)] |
| 9. | Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci. 2020;21:1723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 502] [Cited by in RCA: 725] [Article Influence: 120.8] [Reference Citation Analysis (0)] |
| 10. | Tan S, Xia L, Yi P, Han Y, Tang L, Pan Q, Tian Y, Rao S, Oyang L, Liang J, Lin J, Su M, Shi Y, Cao D, Zhou Y, Liao Q. Exosomal miRNAs in tumor microenvironment. J Exp Clin Cancer Res. 2020;39:67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 144] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 11. | Savardashtaki A, Shabaninejad Z, Movahedpour A, Sahebnasagh R, Mirzaei H, Hamblin MR. miRNAs derived from cancer-associated fibroblasts in colorectal cancer. Epigenomics. 2019;11:1627-1645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 12. | Shen Z, Yu N, Zhang Y, Jia M, Sun Y, Li Y, Zhao L. The potential roles of HIF-1α in epithelial-mesenchymal transition and ferroptosis in tumor cells. Cell Signal. 2024;122:111345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 13. | Kunte DP, DelaCruz M, Wali RK, Menon A, Du H, Stypula Y, Patel A, Backman V, Roy HK. Dysregulation of microRNAs in colonic field carcinogenesis: implications for screening. PLoS One. 2012;7:e45591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Li H, Rokavec M, Jiang L, Horst D, Hermeking H. Antagonistic Effects of p53 and HIF1A on microRNA-34a Regulation of PPP1R11 and STAT3 and Hypoxia-induced Epithelial to Mesenchymal Transition in Colorectal Cancer Cells. Gastroenterology. 2017;153:505-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 122] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 15. | Nijhuis A, Thompson H, Adam J, Parker A, Gammon L, Lewis A, Bundy JG, Soga T, Jalaly A, Propper D, Jeffery R, Suraweera N, McDonald S, Thaha MA, Feakins R, Lowe R, Bishop CL, Silver A. Remodelling of microRNAs in colorectal cancer by hypoxia alters metabolism profiles and 5-fluorouracil resistance. Hum Mol Genet. 2017;26:1552-1564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 16. | Yang Y, Qu A, Wu Q, Zhang X, Wang L, Li C, Dong Z, Du L, Wang C. Prognostic value of a hypoxia-related microRNA signature in patients with colorectal cancer. Aging (Albany NY). 2020;12:35-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 17. | Zhang Y, Zhao Y, Li Q, Wang Y. Macrophages, as a Promising Strategy to Targeted Treatment for Colorectal Cancer Metastasis in Tumor Immune Microenvironment. Front Immunol. 2021;12:685978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 18. | Hu J, Huang S, Liu X, Zhang Y, Wei S, Hu X. miR-155: An Important Role in Inflammation Response. J Immunol Res. 2022;2022:7437281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 104] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 19. | Xing Y, Ruan G, Ni H, Qin H, Chen S, Gu X, Shang J, Zhou Y, Tao X, Zheng L. Tumor Immune Microenvironment and Its Related miRNAs in Tumor Progression. Front Immunol. 2021;12:624725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 20. | Hussain QM, Al-Hussainy AF, Sanghvi G, Roopashree R, Kashyap A, Anand DA, Panigrahi R, Shavazi N, Taher SG, Alwan M, Jawad M, Mushtaq H. Dual role of miR-155 and exosomal miR-155 in tumor angiogenesis: implications for cancer progression and therapy. Eur J Med Res. 2025;30:393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 21. | Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis Model Mech. 2017;10:197-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 117] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 22. | Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, Liu Q, Dou R, Xiong B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 637] [Article Influence: 91.0] [Reference Citation Analysis (1)] |
| 23. | Yin Y, Yao S, Hu Y, Feng Y, Li M, Bian Z, Zhang J, Qin Y, Qi X, Zhou L, Fei B, Zou J, Hua D, Huang Z. The Immune-microenvironment Confers Chemoresistance of Colorectal Cancer through Macrophage-Derived IL6. Clin Cancer Res. 2017;23:7375-7387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 208] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 24. | Lan J, Sun L, Xu F, Liu L, Hu F, Song D, Hou Z, Wu W, Luo X, Wang J, Yuan X, Hu J, Wang G. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res. 2019;79:146-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 508] [Article Influence: 63.5] [Reference Citation Analysis (2)] |
| 25. | Shen Z, Zhou R, Liu C, Wang Y, Zhan W, Shao Z, Liu J, Zhang F, Xu L, Zhou X, Qi L, Bo F, Ding Y, Zhao L. MicroRNA-105 is involved in TNF-α-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017;8:3213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 26. | Kim TW, Lee YS, Yun NH, Shin CH, Hong HK, Kim HH, Cho YB. MicroRNA-17-5p regulates EMT by targeting vimentin in colorectal cancer. Br J Cancer. 2020;123:1123-1130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 27. | Niu L, Yang W, Duan L, Wang X, Li Y, Xu C, Liu C, Zhang Y, Zhou W, Liu J, Zhao Q, Hong L, Fan D. Biological Implications and Clinical Potential of Metastasis-Related miRNA in Colorectal Cancer. Mol Ther Nucleic Acids. 2021;23:42-54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 28. | Ma H, Pan JS, Jin LX, Wu J, Ren YD, Chen P, Xiao C, Han J. MicroRNA-17~92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Lett. 2016;376:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 29. | Yamada NO, Heishima K, Akao Y, Senda T. Extracellular Vesicles Containing MicroRNA-92a-3p Facilitate Partial Endothelial-Mesenchymal Transition and Angiogenesis in Endothelial Cells. Int J Mol Sci. 2019;20:4406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 30. | Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, Hu J, Zhu X, Yang W, Liao W, Li G, Ding Y, Liang L. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9:5395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 383] [Cited by in RCA: 747] [Article Influence: 93.4] [Reference Citation Analysis (0)] |
| 31. | He Q, Ye A, Ye W, Liao X, Qin G, Xu Y, Yin Y, Luo H, Yi M, Xian L, Zhang S, Qin X, Zhu W, Li Y. Cancer-secreted exosomal miR-21-5p induces angiogenesis and vascular permeability by targeting KRIT1. Cell Death Dis. 2021;12:576. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 118] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 32. | Hu HY, Yu CH, Zhang HH, Zhang SZ, Yu WY, Yang Y, Chen Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol. 2019;132:470-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 33. | Wu Y, Zhang J, Li G, Wang L, Zhao Y, Zheng B, Lin F, Xie L. Exosomal miR-320d promotes angiogenesis and colorectal cancer metastasis via targeting GNAI1 to affect the JAK2/STAT3 signaling pathway. Cell Death Dis. 2024;15:913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 34. | Sychowski G, Romanowicz H, Ciesielski W, Hogendorf P, Durczyński A, Smolarz B. Diagnostic and Therapeutic Potential of Selected microRNAs in Colorectal Cancer: A Literature Review. Cancers (Basel). 2025;17:2135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 35. | Rahman MS, Ghorai S, Panda K, Santiago MJ, Aggarwal S, Wang T, Rahman I, Chinnapaiyan S, Unwalla HJ. Dr. Jekyll or Mr. Hyde: The multifaceted roles of miR-145-5p in human health and disease. Noncoding RNA Res. 2025;11:22-37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 36. | Xu Y, Shen L, Li F, Yang J, Wan X, Ouyang M. microRNA-16-5p-containing exosomes derived from bone marrow-derived mesenchymal stem cells inhibit proliferation, migration, and invasion, while promoting apoptosis of colorectal cancer cells by downregulating ITGA2. J Cell Physiol. 2019;234:21380-21394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 37. | Huang X, Xu X, Ke H, Pan X, Ai J, Xie R, Lan G, Hu Y, Wu Y. microRNA-16-5p suppresses cell proliferation and angiogenesis in colorectal cancer by negatively regulating forkhead box K1 to block the PI3K/Akt/mTOR pathway. Eur J Histochem. 2022;66:3333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Tan HY, Zheng YB, Liu J. Serum miR-199a as a potential diagnostic biomarker for detection of colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22:8657-8663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 39. | Jiao W, Leng X, Zhou Q, Wu Y, Sun L, Tan Y, Ni H, Dong X, Shen T, Liu Y, Li J. Different miR-21-3p isoforms and their different features in colorectal cancer. Int J Cancer. 2017;141:2103-2111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 40. | Ye G, Chen Y. LncRNA FAM30A Predicts Adverse Prognosis and Regulates Cellular Processes in Colorectal Cancer via Modulating miR-21-3p. Turk J Gastroenterol. 2024;35:532-538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 41. | Liu T, Liu D, Guan S, Dong M. Diagnostic role of circulating MiR-21 in colorectal cancer: a update meta-analysis. Ann Med. 2021;53:87-102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 42. | Yang Z, Zhang X, Bai X, Xi X, Liu W, Zhong W. Anti-angiogenesis in colorectal cancer therapy. Cancer Sci. 2024;115:734-751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 43. | Yuan F, Sun R, Li L, Jin B, Wang Y, Liang Y, Che G, Gao L, Zhang L. A functional variant rs353292 in the flanking region of miR-143/145 contributes to the risk of colorectal cancer. Sci Rep. 2016;6:30195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 44. | Mozammel N, Amini M, Baradaran B, Mahdavi SZB, Hosseini SS, Mokhtarzadeh A. The function of miR-145 in colorectal cancer progression; an updated review on related signaling pathways. Pathol Res Pract. 2023;242:154290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 45. | Kadkhoda S, Ghafouri-Fard S. Function of miRNA-145-5p in the pathogenesis of human disorders. Pathol Res Pract. 2022;231:153780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 46. | Ghafouri-Fard S, Khoshbakht T, Hussen BM, Abdullah ST, Taheri M, Samadian M. A review on the role of mir-16-5p in the carcinogenesis. Cancer Cell Int. 2022;22:342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 66] [Reference Citation Analysis (0)] |
| 47. | Aslan ES, Yavas C, Akcali N, Eslamkhah S, Meral G, Batur LK. The functional correlation between mir-16-5p and BIRC5 gene in colorectal cancer: integrated analysis of transcriptomics and in vitro validation. Mol Biol Rep. 2025;52:252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 48. | Zheng Y, Li Y, Wei Z, Wang Y, Liu Y, Liu F, Li X, Zhang Y. HUC-MSC-derived exosomal miR-16-5p attenuates inflammation via dual suppression of M1 macrophage polarization and Th1 differentiation. Biochem Biophys Rep. 2025;43:102078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 49. | Chai C, Song LJ, Yang B, Han SY, Li XQ, Li M. Circulating miR-199a-3p in plasma and its potential diagnostic and prognostic value in glioma. Eur Rev Med Pharmacol Sci. 2016;20:4885-4890. [PubMed] |
| 50. | Liu X, Wang X, Chai B, Wu Z, Gu Z, Zou H, Zhang H, Li Y, Sun Q, Fang W, Ma Z. miR-199a-3p/5p regulate tumorgenesis via targeting Rheb in non-small cell lung cancer. Int J Biol Sci. 2022;18:4187-4202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 51. | Báez-Vega PM, Echevarría Vargas IM, Valiyeva F, Encarnación-Rosado J, Roman A, Flores J, Marcos-Martínez MJ, Vivas-Mejía PE. Targeting miR-21-3p inhibits proliferation and invasion of ovarian cancer cells. Oncotarget. 2016;7:36321-36337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 52. | Li S, Zeng X, Ma R, Wang L. MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp Ther Med. 2018;16:2038-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 53. | Hou N, Guo Z, Zhao G, Jia G, Luo B, Shen X, Bai Y. Inhibition of microRNA-21-3p suppresses proliferation as well as invasion and induces apoptosis by targeting RNA-binding protein with multiple splicing through Smad4/extra cellular signal-regulated protein kinase signalling pathway in human colorectal cancer HCT116 cells. Clin Exp Pharmacol Physiol. 2018;45:729-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 54. | Arisan ED, Rencuzogullari O, Cieza-Borrella C, Miralles Arenas F, Dwek M, Lange S, Uysal-Onganer P. MiR-21 Is Required for the Epithelial-Mesenchymal Transition in MDA-MB-231 Breast Cancer Cells. Int J Mol Sci. 2021;22:1557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 55. | Ding B, Yao M, Fan W, Lou W. Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer. Aging (Albany NY). 2020;12:5259-5279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 56. | Yu Y, Nangia-Makker P, Farhana L, G Rajendra S, Levi E, Majumdar AP. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer. 2015;14:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 57. | Bahreini F, Saidijam M, Afshar S, Mousivand Z, Najafi R. The Effect of miR-145-5p, DANCR and NRAS Expression Levels on the Survival Rate of Colorectal Cancer Patients. Asian Pac J Cancer Prev. 2021;22:4043-4049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 58. | Wang W, Ji G, Xiao X, Chen X, Qin WW, Yang F, Li YF, Fan LN, Xi WJ, Huo Y, Wen WH, Yang AG, Wang T. Epigenetically regulated miR-145 suppresses colon cancer invasion and metastasis by targeting LASP1. Oncotarget. 2016;7:68674-68687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 59. | Sheng N, Tan G, You W, Chen H, Gong J, Chen D, Zhang H, Wang Z. MiR-145 inhibits human colorectal cancer cell migration and invasion via PAK4-dependent pathway. Cancer Med. 2017;6:1331-1340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 60. | Zhuang W, Niu T, Li Z. MicroRNA miR-145-5p regulates cell proliferation and cell migration in colon cancer by inhibiting chemokine (C-X-C motif) ligand 1 and integrin α2. Bioengineered. 2021;12:9909-9917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 61. | Thuringer D, Jego G, Berthenet K, Hammann A, Solary E, Garrido C. Gap junction-mediated transfer of miR-145-5p from microvascular endothelial cells to colon cancer cells inhibits angiogenesis. Oncotarget. 2016;7:28160-28168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 62. | Yang J, Shay C, Saba NF, Teng Y. Cancer metabolism and carcinogenesis. Exp Hematol Oncol. 2024;13:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 73] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 63. | Cai K, Yang Y, Guo ZJ, Cai RL, Hashida H, Li HX. Amentoflavone inhibits colorectal cancer epithelial-mesenchymal transition via the miR-16-5p/HMGA2/β-catenin pathway. Ann Transl Med. 2022;10:1009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 64. | Huang X, Hou Y, Weng X, Pang W, Hou L, Liang Y, Wang Y, Du L, Wu T, Yao M, Wang J, Meng X. Diethyldithiocarbamate-copper complex (CuET) inhibits colorectal cancer progression via miR-16-5p and 15b-5p/ALDH1A3/PKM2 axis-mediated aerobic glycolysis pathway. Oncogenesis. 2021;10:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 65. | Qin J, Hu S, Lou J, Xu M, Gao R, Xiao Q, Chen Y, Ding M, Pan Y, Wang S. Selumetinib overcomes ITGA2-induced 5-fluorouracil resistance in colorectal cancer. Int Immunopharmacol. 2024;137:112487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 66. | Meng W, Li Y, Chai B, Liu X, Ma Z. miR-199a: A Tumor Suppressor with Noncoding RNA Network and Therapeutic Candidate in Lung Cancer. Int J Mol Sci. 2022;23:8518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 67. | Nonaka R, Nishimura J, Kagawa Y, Osawa H, Hasegawa J, Murata K, Okamura S, Ota H, Uemura M, Hata T, Takemasa I, Mizushima T, Okuzaki D, Yamamoto H, Doki Y, Mori M. Circulating miR-199a-3p as a novel serum biomarker for colorectal cancer. Oncol Rep. 2014;32:2354-2358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 68. | Wang X, Li Y, Zhou H, Han N, Pan L, Yu C. Effect of Gambogic Acid on miR-199a-3p Expression and Cell Biological Behavior in Colorectal Cancer Cells. J Oncol. 2021;2021:5140621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 69. | Dos Santos IL, Penna KGBD, Dos Santos Carneiro MA, Libera LSD, Ramos JEP, Saddi VA. Tissue micro-RNAs associated with colorectal cancer prognosis: a systematic review. Mol Biol Rep. 2021;48:1853-1867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 70. | Hou J, Mi X, Liu N, Li X, Li XN, Yang Y, Lu X, Fang Y, Jin NY. MiR-199a/b-3p inhibits colorectal cancer cell proliferation, migration and invasion through targeting PAK4 and BCAR3. Eur J Med Res. 2022;27:121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 71. | Lin L, Chen B, Li Y. Hypoxia-Induced Downregulation of MiR-199a-3p Promotes Proliferation, Invasion and Epithelial-Mesenchymal Transition in Colorectal Cancer Cells. J Biol Regul Homeost Agents. 2022;36. [DOI] [Full Text] |
| 72. | Han Y, Kuang Y, Xue X, Guo X, Li P, Wang X, Guo X, Yuan B, Zhi Q, Zhao H. NLK, a novel target of miR-199a-3p, functions as a tumor suppressor in colorectal cancer. Biomed Pharmacother. 2014;68:497-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 73. | Urh K, Žlajpah M, Zidar N, Boštjančič E. Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis. Biomedicines. 2021;9:179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 74. | Bautista-Sánchez D, Arriaga-Canon C, Pedroza-Torres A, De La Rosa-Velázquez IA, González-Barrios R, Contreras-Espinosa L, Montiel-Manríquez R, Castro-Hernández C, Fragoso-Ontiveros V, Álvarez-Gómez RM, Herrera LA. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol Ther Nucleic Acids. 2020;20:409-420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 381] [Article Influence: 63.5] [Reference Citation Analysis (0)] |
| 75. | Le MNT, Takahi M, Ohnuma K. Auto/paracrine factors and early Wnt inhibition promote cardiomyocyte differentiation from human induced pluripotent stem cells at initial low cell density. Sci Rep. 2021;11:21426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 76. | Wu Y, Song Y, Xiong Y, Wang X, Xu K, Han B, Bai Y, Li L, Zhang Y, Zhou L. MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer. Cell Physiol Biochem. 2017;43:945-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 157] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 77. | Lin PL, Wu DW, Huang CC, He TY, Chou MC, Sheu GT, Lee H. MicroRNA-21 promotes tumour malignancy via increased nuclear translocation of β-catenin and predicts poor outcome in APC-mutated but not in APC-wild-type colorectal cancer. Carcinogenesis. 2014;35:2175-2182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 78. | Nguyen HT, Kacimi SEO, Nguyen TL, Suman KH, Lemus-Martin R, Saleem H, Do DN. MiR-21 in the Cancers of the Digestive System and Its Potential Role as a Diagnostic, Predictive, and Therapeutic Biomarker. Biology (Basel). 2021;10:417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 79. | Yu W, Zhu K, Wang Y, Yu H, Guo J. Overexpression of miR-21-5p promotes proliferation and invasion of colon adenocarcinoma cells through targeting CHL1. Mol Med. 2018;24:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 80. | Li A, Yang PM. Overexpression of miR-21-5p in colorectal cancer cells promotes self-assembly of E-cadherin-dependent multicellular tumor spheroids. Tissue Cell. 2020;65:101365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 81. | Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si K, Sun B, Chen B, Xiao Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology. 2020;18:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 486] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 82. | Jiang R, Chen X, Ge S, Wang Q, Liu Y, Chen H, Xu J, Wu J. MiR-21-5p Induces Pyroptosis in Colorectal Cancer via TGFBI. Front Oncol. 2020;10:610545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 83. | Lopes-Ramos CM, Habr-Gama A, Quevedo Bde S, Felício NM, Bettoni F, Koyama FC, Asprino PF, Galante PA, Gama-Rodrigues J, Camargo AA, Perez RO, Parmigiani RB. Overexpression of miR-21-5p as a predictive marker for complete tumor regression to neoadjuvant chemoradiotherapy in rectal cancer patients. BMC Med Genomics. 2014;7:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 84. | Chen Q, Zhou L, Ye X, Tao M, Wu J. miR-145-5p suppresses proliferation, metastasis and EMT of colorectal cancer by targeting CDCA3. Pathol Res Pract. 2020;216:152872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 85. | Cheng X, Shen T, Liu P, Fang S, Yang Z, Li Y, Dong J. mir-145-5p is a suppressor of colorectal cancer at early stage, while promotes colorectal cancer metastasis at late stage through regulating AKT signaling evoked EMT-mediated anoikis. BMC Cancer. 2022;22:1151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 86. | Dong Z, Ren L, Lin L, Li J, Huang Y, Li J. Effect of microRNA-21 on multidrug resistance reversal in A549/DDP human lung cancer cells. Mol Med Rep. 2015;11:682-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 87. | Zhang N, Li WW, Lv CM, Gao YW, Liu XL, Zhao L. miR-16-5p and miR-19b-3p prevent amyloid β-induced injury by targeting BACE1 in SH-SY5Y cells. Neuroreport. 2020;31:205-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 88. | Zhao L, Chen H, Zhang Q, Ma J, Hu H, Xu L. ATF4-mediated microRNA-145/HDAC4/p53 axis affects resistance of colorectal cancer cells to 5-fluorouracil by regulating autophagy. Cancer Chemother Pharmacol. 2022;89:595-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 89. | Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32:2576-2585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 113] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 90. | Venturutti L, Cordo Russo RI, Rivas MA, Mercogliano MF, Izzo F, Oakley RH, Pereyra MG, De Martino M, Proietti CJ, Yankilevich P, Roa JC, Guzmán P, Cortese E, Allemand DH, Huang TH, Charreau EH, Cidlowski JA, Schillaci R, Elizalde PV. MiR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene. 2016;35:6189-6202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 91. | Mousavikia SN, M Matin M, Bahreyni Tossi MT, Aghaee-Bakhtiari SH, Azimian H. Identification and modulation of a PI3K/AKT/mTOR pathway-targeting microRNA in order to increase colorectal cancer cells radiosensitivity in vitro. BMC Cancer. 2025;25:1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 92. | Ghanbarian M, Afgar A, Yadegarazari R, Najafi R, Teimoori-Toolabi L. Through oxaliplatin resistance induction in colorectal cancer cells, increasing ABCB1 level accompanies decreasing level of miR-302c-5p, miR-3664-5p and miR-129-5p. Biomed Pharmacother. 2018;108:1070-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 93. | Han H, Li Y, Qin W, Wang L, Yin H, Su B, Yuan X. miR-199b-3p contributes to acquired resistance to cetuximab in colorectal cancer by targeting CRIM1 via Wnt/β-catenin signaling. Cancer Cell Int. 2022;22:42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 94. | Chen J, Huang XF, Qiao L, Katsifis A. Insulin caused drug resistance to oxaliplatin in colon cancer cell line HT29. J Gastrointest Oncol. 2011;2:27-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 95. | Luo J, Yuan J, Yang Y, Jiang Y, Yan J, Tong Q. Special AT-rich sequence binding protein 1 promotes multidrug resistance in gastric cancer by regulation of Ezrin to alter subcellular localization of ATP-binding cassette transporters. Cancer Sci. 2023;114:1353-1364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 96. | Xia X, Yang B, Zhai X, Liu X, Shen K, Wu Z, Cai J. Prognostic role of microRNA-21 in colorectal cancer: a meta-analysis. PLoS One. 2013;8:e80426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 97. | Wan D, He S, Xie B, Xu G, Gu W, Shen C, Hu Y, Wang X, Zhi Q, Wang L. Aberrant expression of miR-199a-3p and its clinical significance in colorectal cancers. Med Oncol. 2013;30:378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 98. | Rahmati S, Moeinafshar A, Rezaei N. The multifaceted role of extracellular vesicles (EVs) in colorectal cancer: metastasis, immune suppression, therapy resistance, and autophagy crosstalk. J Transl Med. 2024;22:452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 99. | Jenike AE, Halushka MK. miR-21: a non-specific biomarker of all maladies. Biomark Res. 2021;9:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 175] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 100. | Otmani K, Rouas R, Lewalle P. OncomiRs as noncoding RNAs having functions in cancer: Their role in immune suppression and clinical implications. Front Immunol. 2022;13:913951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 101. | Pieniądz P, Pięt M, Paduch R. Characteristics of the Colorectal Cancer Microenvironment—Role in Cancer Progression and Therapeutic Possibilities. Appl Sci. 2024;14:2930. [DOI] [Full Text] |
| 102. | Yu J, Dong W, Liang J. Extracellular Vesicle-Transported Long Non-Coding RNA (LncRNA) X Inactive-Specific Transcript (XIST) in Serum is a Potential Novel Biomarker for Colorectal Cancer Diagnosis. Med Sci Monit. 2020;26:e924448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 103. | Wu H, Wei M, Jiang X, Tan J, Xu W, Fan X, Zhang R, Ding C, Zhao F, Shao X, Zhang Z, Shi R, Zhang W, Wu G. lncRNA PVT1 Promotes Tumorigenesis of Colorectal Cancer by Stabilizing miR-16-5p and Interacting with the VEGFA/VEGFR1/AKT Axis. Mol Ther Nucleic Acids. 2020;20:438-450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 104. | Fernández-Messina L, Gutiérrez-Vázquez C, Rivas-García E, Sánchez-Madrid F, de la Fuente H. Immunomodulatory role of microRNAs transferred by extracellular vesicles. Biol Cell. 2015;107:61-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 118] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 105. | Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, Zhang Z, Cai S, Xu Y, Li X, He X, Zhong X, Li G, Chen Z, Li D. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 2020;13:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 610] [Article Influence: 101.7] [Reference Citation Analysis (0)] |
| 106. | Zhu X, Guo Q, Zou J, Wang B, Zhang Z, Wei R, Zhao L, Zhang Y, Chu C, Fu X, Li X. MiR-19a-3p Suppresses M1 Macrophage Polarization by Inhibiting STAT1/IRF1 Pathway. Front Pharmacol. 2021;12:614044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 107. | Yang C, Dou R, Wei C, Liu K, Shi D, Zhang C, Liu Q, Wang S, Xiong B. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol Ther. 2021;29:2088-2107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 176] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 108. | Shi W, Liu Y, Qiu X, Yang L, Lin G. Cancer-associated fibroblasts-derived exosome-mediated transfer of miR-345-5p promotes the progression of colorectal cancer by targeting CDKN1A. Carcinogenesis. 2023;44:317-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 109. | Ghofrani-Shahpar M, Pakravan K, Razmara E, Amooie F, Mahmoudian M, Heshmati M, Babashah S. Cancer-associated fibroblasts drive colorectal cancer cell progression through exosomal miR-20a-5p-mediated targeting of PTEN and stimulating interleukin-6 production. BMC Cancer. 2024;24:400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 110. | Chen X, Liu J, Zhang Q, Liu B, Cheng Y, Zhang Y, Sun Y, Ge H, Liu Y. Exosome-mediated transfer of miR-93-5p from cancer-associated fibroblasts confer radioresistance in colorectal cancer cells by downregulating FOXA1 and upregulating TGFB3. J Exp Clin Cancer Res. 2020;39:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 112] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 111. | Chen X, Liu Y, Zhang Q, Liu B, Cheng Y, Zhang Y, Sun Y, Liu J. Exosomal miR-590-3p derived from cancer-associated fibroblasts confers radioresistance in colorectal cancer. Mol Ther Nucleic Acids. 2021;24:113-126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 112. | Yuan H, Chen B, Chai R, Gong W, Wan Z, Zheng B, Hu X, Guo Y, Gao S, Dai Q, Yu P, Tu S. Loss of exosomal micro-RNA-200b-3p from hypoxia cancer-associated fibroblasts reduces sensitivity to 5-flourouracil in colorectal cancer through targeting high-mobility group box 3. Front Oncol. 2022;12:920131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 113. | Zhang Y, Wang S, Lai Q, Fang Y, Wu C, Liu Y, Li Q, Wang X, Gu C, Chen J, Cai J, Li A, Liu S. Cancer-associated fibroblasts-derived exosomal miR-17-5p promotes colorectal cancer aggressive phenotype by initiating a RUNX3/MYC/TGF-β1 positive feedback loop. Cancer Lett. 2020;491:22-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 114. | Zhang LL, Zhang LF, Shi YB. miR-24 inhibited the killing effect of natural killer cells to colorectal cancer cells by downregulating Paxillin. Biomed Pharmacother. 2018;101:257-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 115. | Yang Q, Li J, Hu Y, Tang X, Yu L, Dong L, Chen D. MiR-218-5p Suppresses the Killing Effect of Natural Killer Cell to Lung Adenocarcinoma by Targeting SHMT1. Yonsei Med J. 2019;60:500-508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 116. | Zheng J, Wang W, Hong T, Yang S, Shen J, Liu C. Suppression of microRNA-155 exerts an anti-inflammatory effect on CD4+ T cell-mediated inflammatory response in the pathogenesis of atherosclerosis. Acta Biochim Biophys Sin (Shanghai). 2020;52:654-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 117. | Drebber U, Lay M, Wedemeyer I, Vallböhmer D, Bollschweiler E, Brabender J, Mönig SP, Hölscher AH, Dienes HP, Odenthal M. Altered levels of the onco-microRNA 21 and the tumor-supressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int J Oncol. 2011;39:409-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 118. | Farace C, Pisano A, Griñan-Lison C, Solinas G, Jiménez G, Serra M, Carrillo E, Scognamillo F, Attene F, Montella A, Marchal JA, Madeddu R. Deregulation of cancer-stem-cell-associated miRNAs in tissues and sera of colorectal cancer patients. Oncotarget. 2020;11:116-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 119. | Zhang C, Sun C, Zhao Y, Wang Q, Guo J, Ye B, Yu G. Overview of MicroRNAs as Diagnostic and Prognostic Biomarkers for High-Incidence Cancers in 2021. Int J Mol Sci. 2022;23:11389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 120. | Pagoni M, Cava C, Sideris DC, Avgeris M, Zoumpourlis V, Michalopoulos I, Drakoulis N. miRNA-Based Technologies in Cancer Therapy. J Pers Med. 2023;13:1586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 52] [Reference Citation Analysis (0)] |
| 121. | Saenz-Pipaon G, Dichek DA. Targeting and delivery of microRNA-targeting antisense oligonucleotides in cardiovascular diseases. Atherosclerosis. 2023;374:44-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 122. | Attia MS, Kijanka G, Nguyen NT, Zhang J, An H. Advances and prospects of RNA delivery nanoplatforms for cancer therapy. Acta Pharm Sin B. 2025;15:52-96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 123. | Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, Shin S, Becerra CR, Falchook G, Stoudemire J, Martin D, Kelnar K, Peltier H, Bonato V, Bader AG, Smith S, Kim S, O'Neill V, Beg MS. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer. 2020;122:1630-1637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 409] [Cited by in RCA: 689] [Article Influence: 114.8] [Reference Citation Analysis (0)] |
| 124. | Ali A, Grillone K, Ascrizzi S, Caridà G, Fiorillo L, Ciliberto D, Staropoli N, Tagliaferri P, Tassone P, Di Martino MT. LNA-i-miR-221 activity in colorectal cancer: A reverse translational investigation. Mol Ther Nucleic Acids. 2024;35:102221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (2)] |
| 125. | Anastasiadou E, Seto AG, Beatty X, Hermreck M, Gilles ME, Stroopinsky D, Pinter-Brown LC, Pestano L, Marchese C, Avigan D, Trivedi P, Escolar DM, Jackson AL, Slack FJ. Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin Cancer Res. 2021;27:1139-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 126. | Medarova Z, Robertson N, Ghosh S, Duggan S, Varkaris A. Initial clinical experience with the first-in-class anti-metastasis therapeutic TTX-MC138. J Clin Oncol. 2024;42:e15072. [DOI] [Full Text] |
| 127. | Halim A, Kim B, Kenyon E, Moore A. miR-10b as a Clinical Marker and a Therapeutic Target for Metastatic Breast Cancer. Technol Cancer Res Treat. 2025;24:15330338251339256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 128. | Gale DP, Gross O, Wang F, Esteban de la Rosa RJ, Hall M, Sayer JA, Appel G, Hariri A, Liu S, Maski M, Shen Y, Zhang Q, Iqbal S, Kowthalam MU, Lin J, Ding J; HERA Clinical Trial Group. A Randomized Controlled Clinical Trial Testing Effects of Lademirsen on Kidney Function Decline in Adults with Alport Syndrome. Clin J Am Soc Nephrol. 2024;19:995-1004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 129. | Lu Y, Huang W, Li M, Zheng A. Exosome-Based Carrier for RNA Delivery: Progress and Challenges. Pharmaceutics. 2023;15:598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 56] [Reference Citation Analysis (0)] |
| 130. | Balaraman AK, Arockia Babu M, Afzal M, Sanghvi G, M M R, Gupta S, Rana M, Ali H, Goyal K, Subramaniyan V, Wong LS, Kumarasamy V. Exosome-based miRNA delivery: Transforming cancer treatment with mesenchymal stem cells. Regen Ther. 2025;28:558-572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 131. | Ma ZJ, Yang JJ, Lu YB, Liu ZY, Wang XX. Mesenchymal stem cell-derived exosomes: Toward cell-free therapeutic strategies in regenerative medicine. World J Stem Cells. 2020;12:814-840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 132. | Coronel-Hernández J, Delgado-Waldo I, Cantú de León D, López-Camarillo C, Jacobo-Herrera N, Ramos-Payán R, Pérez-Plasencia C. HypoxaMIRs: Key Regulators of Hallmarks of Colorectal Cancer. Cells. 2022;11:1895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 133. | Lai CY, Yeh KY, Liu BF, Chang TM, Chang CH, Liao YF, Liu YW, Her GM. MicroRNA-21 Plays Multiple Oncometabolic Roles in Colitis-Associated Carcinoma and Colorectal Cancer via the PI3K/AKT, STAT3, and PDCD4/TNF-α Signaling Pathways in Zebrafish. Cancers (Basel). 2021;13:5565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 134. | Xiong B, Huang Q, Zheng H, Lin S, Xu J. Recent advances microRNAs and metabolic reprogramming in colorectal cancer research. Front Oncol. 2023;13:1165862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 135. | Soheilifar MH, Grusch M, Neghab HK, Amini R, Maadi H, Saidijam M, Wang Z. Angioregulatory microRNAs in Colorectal Cancer. Cancers (Basel). 2019;12:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 136. | Wu MY, Luo YX, Jia WX, Wang DD, Sun DL, Song J, Wang J, Niu WW, Zhang XL. miRNA-320 inhibits colitis-associated colorectal cancer by regulating the IL-6R/STAT3 pathway in mice. J Gastrointest Oncol. 2022;13:695-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 137. | Mjelle R, Kristensen AK, Tora S Solheim, Westvik GS, Elvebakken H, Hofsli E. Serum small RNAs in metastatic colorectal cancer predict response to chemotherapy and characterize high-risk patients. Mol Cancer. 2024;23:133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 138. | Gou Z, Li J, Liu J, Yang N. The hidden messengers: cancer associated fibroblasts-derived exosomal miRNAs as key regulators of cancer malignancy. Front Cell Dev Biol. 2024;12:1378302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 139. | Włodarczyk M, Maryńczak K, Burzyński J, Włodarczyk J, Basak J, Fichna J, Majsterek I, Ciesielski P, Spinelli A, Dziki Ł. The role of miRNAs in the pathogenesis, diagnosis, and treatment of colorectal cancer and colitis-associated cancer. Clin Exp Med. 2025;25:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 140. | Zhang X, Ai F, Li X, Tian L, Wang X, Shen S, Liu F. MicroRNA-34a suppresses colorectal cancer metastasis by regulating Notch signaling. Oncol Lett. 2017;14:2325-2333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 141. | Valencia-Cervantes J, Sierra-Vargas MP. Regulation of Cancer-Associated miRNAs Expression under Hypoxic Conditions. Anal Cell Pathol (Amst). 2024;2024:5523283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 142. | Rahbar Farzam O, Najafi S, Amini M, Rahimi Z, Dabbaghipour R, Zohdi O, Asemani Shahgoli G, Baradaran B, Akbari B. Interplay of miRNAs and lncRNAs in STAT3 signaling pathway in colorectal cancer progression. Cancer Cell Int. 2024;24:16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 143. | Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood. 2014;124:3748-3757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 504] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 144. | Shao C, Yang F, Miao S, Liu W, Wang C, Shu Y, Shen H. Role of hypoxia-induced exosomes in tumor biology. Mol Cancer. 2018;17:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 248] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 145. | Feng CZ, Zhong SQ, Ye SW, Zheng Z, Sun H, Zhou SH. Tumor-derived exosomal miR-425-5p and miR-135b-3p enhance colorectal cancer progression through immune suppression and vascular permeability promotion. World J Gastrointest Oncol. 2025;17:106161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (3)] |
| 146. | Wang B, Zhao L, Yang C, Lin Y, Wang S, Ye Y, Luo J, Shen Z. IDH1 K224 acetylation promotes colorectal cancer via miR-9-5p/NHE1 axis-mediated regulation of acidic microenvironment. iScience. 2023;26:107206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 147. | Yi C, Yu AM. MicroRNAs in the Regulation of Solute Carrier Proteins Behind Xenobiotic and Nutrient Transport in Cells. Front Mol Biosci. 2022;9:893846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 148. | Gregersen LH, Jacobsen A, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer. 2012;12:232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 149. | Guo L, Fu J, Sun S, Zhu M, Zhang L, Niu H, Chen Z, Zhang Y, Guo L, Wang S. MicroRNA-143-3p inhibits colorectal cancer metastases by targeting ITGA6 and ASAP3. Cancer Sci. 2019;110:805-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 150. | Adam RS, Poel D, Ferreira Moreno L, Spronck JMA, de Back TR, Torang A, Gomez Barila PM, Ten Hoorn S, Markowetz F, Wang X, Verheul HMW, Buffart TE, Vermeulen L. Development of a miRNA-based classifier for detection of colorectal cancer molecular subtypes. Mol Oncol. 2022;16:2693-2709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 151. | Gherman A, Bolundut D, Ecea R, Balacescu L, Curcean S, Dina C, Balacescu O, Cainap C. Molecular Subtypes, microRNAs and Immunotherapy Response in Metastatic Colorectal Cancer. Medicina (Kaunas). 2024;60:397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 152. | Yin Y, Yan ZP, Lu NN, Xu Q, He J, Qian X, Yu J, Guan X, Jiang BH, Liu LZ. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim Biophys Acta. 2013;1829:239-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 82] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 153. | Shen X, Jiang H, Chen Z, Lu B, Zhu Y, Mao J, Chai K, Chen W. MicroRNA-145 Inhibits Cell Migration and Invasion in Colorectal Cancer by Targeting TWIST. Onco Targets Ther. 2019;12:10799-10809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 154. | Yang Y, Li XJ, Li P, Guo XT. MicroRNA-145 regulates the proliferation, migration and invasion of human primary colon adenocarcinoma cells by targeting MAPK1. Int J Mol Med. 2018;42:3171-3180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 155. | Sathyanarayanan A, Chandrasekaran KS, Karunagaran D. microRNA-145 downregulates SIP1-expression but differentially regulates proliferation, migration, invasion and Wnt signaling in SW480 and SW620 cells. J Cell Biochem. 2018;119:2022-2035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 156. | Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li D, Lai L, Jiang BH. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. 2012;40:761-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 241] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 157. | Slattery ML, Mullany LE, Sakoda LC, Wolff RK, Samowitz WS, Herrick JS. The MAPK-Signaling Pathway in Colorectal Cancer: Dysregulated Genes and Their Association With MicroRNAs. Cancer Inform. 2018;17:1176935118766522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 158. | Yu Y, Kanwar SS, Patel BB, Oh PS, Nautiyal J, Sarkar FH, Majumdar AP. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis. 2012;33:68-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 226] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 159. | Chen HL, Li JJ, Jiang F, Shi WJ, Chang GY. MicroRNA-4461 derived from bone marrow mesenchymal stem cell exosomes inhibits tumorigenesis by downregulating COPB2 expression in colorectal cancer. Biosci Biotechnol Biochem. 2020;84:338-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 160. | Ishikawa D, Takasu C, Kashihara H, Nishi M, Tokunaga T, Higashijima J, Yoshikawa K, Yasutomo K, Shimada M. The Significance of MicroRNA-449a and Its Potential Target HDAC1 in Patients With Colorectal Cancer. Anticancer Res. 2019;39:2855-2860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 161. | Ye X, Lv H. MicroRNA-519d-3p inhibits cell proliferation and migration by targeting TROAP in colorectal cancer. Biomed Pharmacother. 2018;105:879-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 162. | Zhu Y, Xue L. miRNA-31 Increases Radiosensitivity Through Targeting a Novel Target STK40 in Colorectal Cancer Cell Lines. Int J Radiat Oncol. 2017;99:E629. [RCA] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 163. | Li Y, Sun J, Cai Y, Jiang Y, Wang X, Huang X, Yin Y, Li H. MiR-200a acts as an oncogene in colorectal carcinoma by targeting PTEN. Exp Mol Pathol. 2016;101:308-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 164. | Im J, Nam SK, Lee HS. MicroRNA-552 expression in colorectal cancer and its clinicopathological significance. J Pathol Transl Med. 2021;55:125-131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 165. | Wang J, Li H, Wang Y, Wang L, Yan X, Zhang D, Ma X, Du Y, Liu X, Yang Y. MicroRNA-552 enhances metastatic capacity of colorectal cancer cells by targeting a disintegrin and metalloprotease 28. Oncotarget. 2016;7:70194-70210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 166. | Pan Z, Xie R, Song W, Gao C. MicroRNA‑592 promotes cell proliferation, migration and invasion in colorectal cancer by directly targeting SPARC. Mol Med Rep. 2021;23:261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 167. | Lei SL, Zhao H, Yao HL, Chen Y, Lei ZD, Liu KJ, Yang Q. Regulatory roles of microRNA-708 and microRNA-31 in proliferation, apoptosis and invasion of colorectal cancer cells. Oncol Lett. 2014;8:1768-1774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 168. | Wang S, Zhang Z, Gao Q. Transfer of microRNA-25 by colorectal cancer cell-derived extracellular vesicles facilitates colorectal cancer development and metastasis. Mol Ther Nucleic Acids. 2021;23:552-564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 169. | Song D, Zhang Q, Zhang H, Zhan L, Sun X. MiR-130b-3p promotes colorectal cancer progression by targeting CHD9. Cell Cycle. 2022;21:585-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 170. | He Y, Zhang L, Tan F, Wang LF, Liu DH, Wang RJ, Yin XZ. MiR-153-5p promotes sensibility of colorectal cancer cells to oxaliplatin via targeting Bcl-2-mediated autophagy pathway. Biosci Biotechnol Biochem. 2020;84:1645-1651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 171. | Xie L, Cui G, Li T. Long Noncoding RNA CBR3-AS1 Promotes Stem-like Properties and Oxaliplatin Resistance of Colorectal Cancer by Sponging miR-145-5p. J Oncol. 2022;2022:2260211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 172. | Zhu Y, Wang C, Becker SA, Hurst K, Nogueira LM, Findlay VJ, Camp ER. miR-145 Antagonizes SNAI1-Mediated Stemness and Radiation Resistance in Colorectal Cancer. Mol Ther. 2018;26:744-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 173. | Slattery ML, Mullany LE, Sakoda LC, Wolff RK, Samowitz WS, Herrick JS. Dysregulated genes and miRNAs in the apoptosis pathway in colorectal cancer patients. Apoptosis. 2018;23:237-250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 174. | Chen X, Xu X, Pan B, Zeng K, Xu M, Liu X, He B, Pan Y, Sun H, Wang S. miR-150-5p suppresses tumor progression by targeting VEGFA in colorectal cancer. Aging (Albany NY). 2018;10:3421-3437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 175. | Feng C, Zhang L, Sun Y, Li X, Zhan L, Lou Y, Wang Y, Liu L, Zhang Y. GDPD5, a target of miR-195-5p, is associated with metastasis and chemoresistance in colorectal cancer. Biomed Pharmacother. 2018;101:945-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 176. | Qu J, Zhao L, Zhang P, Wang J, Xu N, Mi W, Jiang X, Zhang C, Qu J. MicroRNA-195 chemosensitizes colon cancer cells to the chemotherapeutic drug doxorubicin by targeting the first binding site of BCL2L2 mRNA. J Cell Physiol. 2015;230:535-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 177. | Poel D, Boyd LNC, Beekhof R, Schelfhorst T, Pham TV, Piersma SR, Knol JC, Jimenez CR, Verheul HMW, Buffart TE. Proteomic Analysis of miR-195 and miR-497 Replacement Reveals Potential Candidates that Increase Sensitivity to Oxaliplatin in MSI/P53wt Colorectal Cancer Cells. Cells. 2019;8:1111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 178. | Fu Q, Cheng J, Zhang J, Zhang Y, Chen X, Luo S, Xie J. miR-20b reduces 5-FU resistance by suppressing the ADAM9/EGFR signaling pathway in colon cancer. Oncol Rep. 2017;37:123-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 179. | Yang H, Lin J, Jiang J, Ji J, Wang C, Zhang J. miR-20b-5p functions as tumor suppressor microRNA by targeting cyclinD1 in colon cancer. Cell Cycle. 2020;19:2939-2954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 180. | Wang L, Jiang CF, Li DM, Ge X, Shi ZM, Li CY, Liu X, Yin Y, Zhen L, Liu LZ, Jiang BH. MicroRNA-497 inhibits tumor growth and increases chemosensitivity to 5-fluorouracil treatment by targeting KSR1. Oncotarget. 2016;7:2660-2671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 181. | Sun K, Chen L, Li Y, Huang B, Yan Q, Wu C, Lai Q, Fang Y, Cai J, Liu Y, Chen J, Wang X, Zhu Y, Dong S, Tan J, Li A, Liu S, Zhang Y. METTL14-dependent maturation of pri-miR-17 regulates mitochondrial homeostasis and induces chemoresistance in colorectal cancer. Cell Death Dis. 2023;14:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 182. | Mussnich P, Rosa R, Bianco R, Fusco A, D'Angelo D. MiR-199a-5p and miR-375 affect colon cancer cell sensitivity to cetuximab by targeting PHLPP1. Expert Opin Ther Targets. 2015;19:1017-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 183. | Zhang Y, Zheng L, Huang J, Gao F, Lin X, He L, Li D, Li Z, Ding Y, Chen L. MiR-124 Radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One. 2014;9:e93917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 184. | Choi JY, Seok HJ, Lee DH, Kwon J, Shin US, Shin I, Bae IH. miR-1226-5p is involved in radioresistance of colorectal cancer by activating M2 macrophages through suppressing IRF1. J Transl Med. 2024;22:980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 185. | Shang Y, Zhu Z, Zhang Y, Ji F, Zhu L, Liu M, Deng Y, Lv G, Li D, Zhou Z, Lu B, Fu CG. MiR-7-5p/KLF4 signaling inhibits stemness and radioresistance in colorectal cancer. Cell Death Discov. 2023;9:42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 186. | Shang Y, Wang L, Zhu Z, Gao W, Li D, Zhou Z, Chen L, Fu CG. Downregulation of miR-423-5p Contributes to the Radioresistance in Colorectal Cancer Cells. Front Oncol. 2020;10:582239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 187. | Du Q, Ye X, Lu SR, Li H, Liu HY, Zhai Q, Yu B. Exosomal miR-30a and miR-222 derived from colon cancer mesenchymal stem cells promote the tumorigenicity of colon cancer through targeting MIA3. J Gastrointest Oncol. 2021;12:52-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 188. | Wang Y, Lin C. Exosomes miR-22-3p Derived from Mesenchymal Stem Cells Suppress Colorectal Cancer Cell Proliferation and Invasion by Regulating RAP2B and PI3K/AKT Pathway. J Oncol. 2021;2021:3874478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 189. | Qu M, Li J, Hong Z, Jia F, He Y, Yuan L. The role of human umbilical cord mesenchymal stem cells-derived exosomal microRNA-431-5p in survival and prognosis of colorectal cancer patients. Mutagenesis. 2022;37:164-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 190. | Yu S, Liao R, Bai L, Guo M, Zhang Y, Zhang Y, Yang Q, Song Y, Li Z, Meng Q, Wang S, Huang X. Anticancer effect of hUC-MSC-derived exosome-mediated delivery of PMO-miR-146b-5p in colorectal cancer. Drug Deliv Transl Res. 2024;14:1352-1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 20] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 191. | Cui F, Chen Y, Wu X, Zhao W. Mesenchymal stem cell-derived exosomes carrying miR-486-5p inhibit glycolysis and cell stemness in colorectal cancer by targeting NEK2. BMC Cancer. 2024;24:1356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 192. | Lou G, Chen L, Xia C, Wang W, Qi J, Li A, Zhao L, Chen Z, Zheng M, Liu Y. MiR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. J Exp Clin Cancer Res. 2020;39:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/