Published online Jan 15, 2026. doi: 10.4251/wjgo.v18.i1.111357
Revised: August 15, 2025
Accepted: November 24, 2025
Published online: January 15, 2026
Processing time: 198 Days and 11.3 Hours
This review comprehensively summarized the potential of artificial intelligence (AI) in the management of esophageal cancer. It highlighted the significance of AI-assisted endoscopy in Japan where endoscopy is central to both screening and diagnosis. For the clinical adaptation of AI, several challenges remain for its effective translation. The establishment of high-quality clinical databases, such as the National Clinical Database and Japan Endoscopy Database in Japan, which covers almost all cases of esophageal cancer, is essential for validating multimodal AI models. This requires rigorous external validation using diverse datasets, including those from different endoscope manufacturers and image qualities. Furthermore, endoscopists’ skills significantly affect diagnostic accuracy, sug
Core Tip: This review detailed how artificial intelligence (AI) mitigates operator dependence in the endoscopic diagnosis of esophageal squamous cell carcinoma by comparing the sensitivity and specificity of innovative deep learning models with those of expert endoscopists. This further highlights the use of large-scale repositories, such as the National Clinical Database and Japan Endoscopy Database, for robust AI training and validation. Multimodal AI using big databases proposes a multi-institutional or multi-vendor AI strategy in Japan. Finally, we outlined future directions for real-time endoscopic support and the integration of clinical outcomes into next-generation AI-driven endoscopy.
- Citation: Kurisaki K, Kobayashi S, Akashi T, Nakao Y, Fukumoto M, Tasaki K, Adachi T, Eguchi S, Kanetaka K. Opportunities and challenges of artificial intelligence-assisted endoscopy and high-quality data for esophageal squamous cell carcinoma. World J Gastrointest Oncol 2026; 18(1): 111357
- URL: https://www.wjgnet.com/1948-5204/full/v18/i1/111357.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v18.i1.111357
Esophageal squamous cell carcinoma (ESCC) is often detected at an advanced stage because of its absence of symptoms. Although the 5-year survival rate for early stage-ESCC exceeds 85%, for advanced disease it is < 20%[1].
Screening endoscopy plays a central role in the early detection of ESCC, particularly in high-risk populations, such as Japanese males with a history of smoking or alcohol consumption[2-4]. A multicenter study reported that linked color imaging, an advanced image-enhanced endoscopy mode, improved the detection rate of upper gastrointestinal neoplasms, highlighting its importance in surveillance[5]. Furthermore, novel image-enhanced technologies, such as texture and color enhancement imaging, are expected to improve detection rates by enhancing the visibility of early esophageal cancer through the optimization of mucosal structure and tone[6-8]. Narrow band imaging (NBI) plays a pivotal role in the detailed examination of the detected lesions. Combining NBI with magnifying observation to analyze the microvascular patterns of the mucosal surface, based on the classification by the Japan Esophageal Society, is an established method for accurately predicting the extent and invasion depth of superficial squamous cell carcinoma[9-11]. Thus, recent endoscopic technologies have significantly contributed to the early diagnosis of esophageal cancer by enhancing both the detection capability in screening and the qualitative diagnostic performance in detailed examinations[12].
In recent years artificial intelligence (AI) techniques, particularly deep learning-based image analysis, have advanced rapidly and stimulated vigorous medical research. The applications of AI in endoscopic diagnosis are expanding and are pivotal for the early detection of esophageal cancer. Therefore, this review aimed to summarize the current status, challenges, and prospects of AI-assisted endoscopy in the management of esophageal cancer. AI offers a practical solution to overcome the long-standing challenges of operator-dependent diagnostic accuracy and the difficulty in disseminating expert-level skills, thereby promising a paradigm shift in ESCC management.
To clarify the current status of AI in the management of ESCC, we conducted a comprehensive search on PubMed for studies published between 2019 and 2025. The results are classified and summarized in three tables. Table 1 details key clinical studies on diagnostic performance while Table 2 focuses on the technical and algorithmic aspects of AI models. Table 3 provides an overview of recent review articles and meta-analyses.
| Ref. | Year | Primary outcomes | Secondary outcomes | Patients studied | Type of data | Study design |
| Zhao et al[65] | 2019 | Feasibility of CAD-based IPCL classification | Observer agreement and pixel-level/Lesion-level accuracy | 219 patients (30 with inflammation, 24 with low-grade intraepithelial neoplasia, 165 with early esophageal cancer); 185 lesions selected for analysis | NBI-ME images with histological findings | Retrospective study |
| Fukuda et al[18] | 2020 | NBI-based CAD system for ESCC diagnosis | Observer agreement, AI speed, and detection time | Training: 1544 pathological SCC lesions and 458 non-cancer/normal tissues; 354 video clips for testing | NBI/BLI endoscopic images and videos with pathology assessment | Evaluation study using retrospectively collected data |
| Shimamoto et al[66] | 2020 | AI system for invasion depth estimation in superficial ESCC | AI vs expert endoscopists on same validation videos | 909 patients for training dataset; 102 videos of superficial ESCC cases used for validation | WLI/NBI/BLI endoscopic media with invasion depth pathology | Retrospective training; independent video-based validation |
| Shiroma et al[67] | 2021 | AI detection capability of T1 ESCC in EGD videos | Real-time AI-assisted detection: Performance and comparison with endoscopists | 8428 images (training set); 144 EGD videos (validation set); 40 patients (validation set) | Retrospective EGD data: WLI/NBI videos and images | Retrospective study |
| Ikenoyama et al[68] | 2021 | LVL prediction from non-stained images and cancer risk assessment | Comparison of AI performance with experienced endoscopists | 595 patients (6634 training images); 72 patients (667 independent validation images) | WLI/NBI endoscopic images and clinical data (ESCC, HNSCC, Lugol staining) | Retrospective study |
| Li et al[69] | 2021 | Early ESCC CAD system and comparison with WLI-based models | Reduced missed diagnoses and unnecessary biopsies | Training: 235 cases (abnormal NM-NBI images), 412 cases (normal images); Validation: 284 cases (202 abnormal, 82 normal) | NM-NBI/WLI images, patient/Lesion data, and histology | 4-phase observational retrospective study with endoscopist assessment |
| Waki et al[70] | 2021 | AI performance in ESCC detection under simulated oversight | AI vs endoscopists: Sensitivity gain, specificity loss with AI support | Training: 1376 superficial ESCC cases (17336 images); 196 non-cancerous cases (2916 images); Testing: 52 superficial ESCC cases (1459 images); 47 non-cancerous lesions (1168 images) | NBI/BLI/WLI images and videos of superficial ESCC, benign lesions, and normal esophagus | DL-based AI system: Retrospective development with partial prospective validation |
| Meng et al[71] | 2022 | CAD performance metrics (AUROC, accuracy, sensitivity, specificity) | CAD vs endoscopists: Diagnostic performance and predictive values | 837 patients (training); 323 patients (test) | Non-magnified WLI/NBI images and histology-confirmed SESCC/HGIN data | Retrospective diagnostic accuracy |
| Tajiri et al[72] | 2022 | AI-based ESCC vs non-ESCC classification under simulated conditions | AI vs endoscopists: Subgroup accuracy by pathology and lesion size | Training: 1433 superficial ESCC cases (25048 images); 410 non-cancerous esophageal lesions (8557 images); Testing: 123 superficial ESCC cases (3370 images); 107 non-cancerous lesions (2075 images) | ME/non-ME endoscopic images and videos (WLI, NBI, BLI) | DL-AI system development in simulated setting (retrospective/prospective mixed design) |
| Feng et al[16] | 2023 | AI diagnostic accuracy for superficial ESCC detection | AI diagnostic support and cancer feature recognition | 1283 patients (training); 319 (internal validation); 905 (external validation); total 2507 patients, 9663 images | WLI images (Olympus/Fujifilm) and SESCC-confirmed clinical data | Retrospective diagnostic study (partly prospectively registered) |
| Yuan et al[22] | 2023 | Real-time detection and delineation of early ESCC using a novel AI system | Accurate classification of image modalities | Not available | WLI/NBI videos and Lugol-stained ME images with histology-confirmed clinical data | Pilot study (video demonstration) |
| Wang et al[73] | 2023 | IPCL-based early ESCC localization and classification | Performance metrics and model comparison | 246 patients (2887 ME-BLI images); 81 patients (493 ME-NBI images) | Magnified ME-BLI/NBI images and histology-confirmed ESCC data | Pilot retrospective image collection study |
| Wang et al[74] | 2024 | YOLO-HSI integration for early ESCC detection | HSI vs RGB models: Classification of normal, dysplasia, and ESCC | 16 patients (7 with esophageal SCC, 9 with squamous dysplasia, 10 healthy controls); training dataset: 1836 images | WLI/NBI images converted to HSI with pathology evaluation | Not clearly described; development and evaluation study |
| Nakao et al[75] | 2024 | AI support for non-experts in ESCC detection | ESCC detection rate, observation time, and adverse events | 320 patients included in analysis (AI group: 152; control group: 168) | WLI/NBI/Lugol endoscopic media with histopathology and clinical data | Prospective, single-center, exploratory randomized controlled trial |
| Aoyama et al[20] | 2025 | AI model for superficial ESCC detection from endoscopic videos | Subgroup analysis and comparison with endoscopist performance | Training data: 280 cases (140 with lesions, 140 without); Test data: 115 cases (57 with lesions, 58 without) | NBI endoscopic videos with histopathological diagnosis | Prospective data, retrospective analysis design |
| Ma et al[76] | 2025 | Diagnostic performance for ESN classification | iCLE vs experts and non-experts: Diagnostic performance and support evaluation | 2803 patients (for iCLE training/validation); 226 patients (image test); additional patients for video recognition testing (prospective, total N not shown) | pCLE video/image data with histopathological gold standard | Prospective diagnostic study |
| Ref. | Year | Primary outcomes | Secondary outcomes | Patients studied | Type of data | Study design |
| Everson et al[24] | 2019 | Abnormal IPCL classification metrics and prediction time | CNN feature visualization, eCAM analysis, and future AI insights | 17 individuals (10 with ESCN, 7 with normal esophageal squamous epithelium) | HD ME-NBI videos/images (.png) with matched histopathology | Proof-of-concept study |
| Zhao et al[65] | 2019 | Feasibility of CAD-based IPCL classification | Observer agreement and pixel-level/Lesion-level accuracy | 219 patients (30 with inflammation, 24 with low-grade intraepithelial neoplasia, 165 with early esophageal cancer); 185 lesions selected for analysis | NBI-ME images with histological findings | Retrospective study |
| Horie et al[77] | 2019 | CNN-based AI for esophageal cancer detection | Detection of small/superficial lesions and predictive values | Training: 8428 images from 384 patients; Test: 47 patients with 49 cancer lesions | WLI/NBI endoscopic images with histopathology | Retrospective study |
| Ohmori et al[17] | 2020 | Image analysis system for ESCC detection and classification | Comparison of diagnostic performance between AI system and expert endoscopists | 135 patients for validation; training data included 804 histologically confirmed superficial esophageal SCC lesions | WLI/NBI images (with/without magnification) and histological data | Retrospective training; prospective external validation |
| Guo et al[78] | 2020 | Real-time AI for early ESCC and precancerous lesion detection | Frame-based/Lesion-based sensitivity/specificity and normal video evaluation | Training data: 191 cases of precancerous/early ESCC; 358 cases of non-cancerous lesions; validation data: 100 endoscopic videos from 41 patients with ESCC and 30 normal controls | NBI images/videos with histological confirmation | Development study with retrospective training and delayed validation dataset |
| Tang et al[79] | 2021 | Diagnostic performance of the DCNN model | Evaluation of DCNN support and agreement with endoscopists | 1078 patients (for training and cross-validation); 243 patients (for independent internal and external validation) | WLI endoscopic images and retrospective clinical data | Multicenter diagnostic retrospective study |
| Uema et al[80] | 2021 | CNN system for microvessel classification in superficial ESCC | CNN vs endoscopists in microvessel classification, agreement, and diagnostic support | 336 patients with 393 SESCC lesions; 1777 training images; 617 validation images | Trimmed ME-NBI images of SESCC and clinical data | Retrospective single-center study |
| Liu et al[44] | 2022 | Development of AI model to detect and delineate early ESCC under WLI endoscopy | Detection and delineation performance | 1239 patients (13083 images for training/testing); 262 patients (1479 internal test images); 96 patients (648 external test images) | WLI images with confirmed early ESCC clinical data | Retrospective study |
| Yuan et al[21] | 2022 | AI-based IPCL subtype prediction for early ESCC | AI validation, diagnostic support, and comparison with endoscopists | 685 patients (training/validation); 176 patients (ER validation dataset) | ME-NBI images and confirmed precancerous/superficial ESCC data | Retrospective multicenter study |
| Zhao et al[81] | 2022 | Effectiveness of AI-DEN system for early EC diagnosis | Diagnostic accuracy and speed comparison | 300 patients suspected of having esophageal cancer; Training group: 200 patients (148 with early cancer, 52 with benign disease); Test group: 100 patients (92 with early cancer, 8 with benign disease) | NBI-DEN images with pathology results | Retrospective study |
| Tani et al[31] | 2023 | Diagnostic accuracy of AI system vs endoscopists | Diagnostic accuracy and safety outcomes | 388 patients (registered); 380 patients (underwent endoscopy); 237 lesions (evaluated) | Endoscopic images (WLI, NBI/BLI) and real-time clinical data | Single-center prospective single-arm non-inferiority trial |
| Zhang et al[45] | 2023 | Interpretable AI-IDPS for ESCC invasion depth prediction | AI-IDPS vs DL models and endoscopists; trust in AI predictions | 581 patients for training; validation using 196 images and 33 continuously collected videos | ME-NBI images/videos with pathology reports and endoscopist feedback | Multicenter retrospective study with crossover validation by endoscopists |
| Wang et al[74] | 2024 | YOLO-HSI integration for early ESCC detection | HSI vs RGB models: Classification of normal, dysplasia, and ESCC | 16 patients (7 with esophageal SCC, 9 with squamous dysplasia, 10 healthy controls); training dataset: 1836 images | WLI/NBI images converted to HSI with pathology evaluation | Not clearly described; development and evaluation study |
| Nakao et al[75] | 2024 | AI support for non-experts in ESCC detection | ESCC detection rate, observation time, and adverse events | 320 patients included in analysis (AI group: 152; control group: 168) | WLI/NBI/Lugol endoscopic media with histopathology and clinical data | Prospective, single-center, exploratory randomized controlled trial |
| Aoyama et al[20] | 2025 | AI model for superficial ESCC detection from endoscopic videos | Subgroup analysis and comparison with endoscopist performance | Training data: 280 cases (140 with lesions, 140 without); Test data: 115 cases (57 with lesions, 58 without) | NBI endoscopic videos with histopathological diagnosis | Prospective data, retrospective analysis design |
| Ma et al[76] | 2025 | Diagnostic performance for ESN classification | iCLE vs experts and non-experts: Diagnostic performance and support evaluation | 2803 patients (for iCLE training/validation); 226 patients (image test); additional patients for video recognition testing (prospective, total N not shown) | pCLE video/image data with histopathological gold standard | Prospective diagnostic study |
| Ref. | Year | Primary outcomes | Secondary outcomes | Patients studied | Type of data | Study design |
| Zhang et al[63] | 2020 | Not applicable | Not applicable | Not applicable | Not applicable | Mini-review |
| Syed et al[82] | 2020 | Summary of literature using DL to identify esophageal tumors | DL/CNN overview, current limitations, and future directions | Not applicable (not focused on individual study data; 21 relevant studies reviewed) | Endoscopic images with histology-confirmed clinical data | Mentored review |
| Huang et al[83] | 2020 | Not applicable | Not applicable | Not applicable | Not applicable | Mini-review |
| Lazăr et al[84] | 2020 | ML model comparison for endoscopic esophageal lesion assessment | AI in care quality, workflow efficiency, and physician support | Not applicable (review of multiple studies) | Review of AI in endoscopic assessment of esophageal disease | Review |
| Namikawa et al[85] | 2020 | Review of studies using CNNs in gastrointestinal endoscopy | AI in GI polyp/cancer detection and capsule endoscopy | Not applicable | Not applicable | Review article |
| Zhang et al[1] | 2021 | Diagnostic accuracy of AI-assisted models | AI vs endoscopists: Pooled accuracy and subgroup performance | Not applicable | Endoscopic media and histology-confirmed clinical data | Meta-analysis |
| Liu et al[44] | 2021 | Not applicable | Not applicable | Not applicable | Not applicable | Mini-review |
| Ma et al[13] | 2022 | CNN-AI for early EC detection from endoscopy | Heterogeneity and bias analysis (I2, meta-regression, Deeks’ plot) | Meta-analysis of 7 studies; number of patients and images varied across studies | Meta-analysis of WLE/NBI-based studies with histological confirmation | Meta-analysis |
| Nagao et al[64] | 2022 | AI in upper GI: Current status, clinical integration, and future outlook | AI in H. pylori infection diagnosis, gastric anatomy, and upper GI cancer detection | Not applicable (review of multiple studies) | CNN-based AI for diagnosis of GI and pharyngeal cancers | Review |
| Tokat et al[86] | 2022 | Overview of the applicability of AI technology in upper gastrointestinal endoscopy | Clinical AI challenges and applications in GI oncology | Not applicable | WLE/NBI/ME-NBI images with clinical and histological data | Review article |
| Guidozzi et al[14] | 2023 | AI in endoscopic diagnosis of esophageal cancer | AI vs endoscopists, tumor types, and bias risk (QUADAS-2) | 1590 patients for ESCC diagnosis (14 studies); 478 patients for EAC diagnosis (9 studies) | English studies using endoscopic images/videos | Systematic review and meta-analysis |
| Pan et al[87] | 2023 | AI in early ESCC endoscopic diagnosis: Current status | Discussion of limitations and future prospects of AI in ESCC diagnosis | Not applicable (review of multiple studies) | Review of DL models for ESCC diagnosis | Review |
| Tao et al[32] | 2024 | AI vs experts: Accuracy in early EC and depth diagnosis | AI vs endoscopists: Performance, invasion depth diagnosis, and bias assessment | Not applicable | Meta-analysis of 19 studies with heterogeneous sample sizes | Meta-analysis |
| Kikuchi et al[88] | 2024 | Recent studies on endoscopic AI for GI tumors | Discussion on the future outlook of AI in gastroenterology | Not applicable | Published studies on DL-based endoscopic AI | Review |
| Yan et al[33] | 2025 | Not applicable | Not applicable | Not applicable | Not applicable | Review article |
| Shahzil et al[89] | 2025 | Lesion color contrast, visibility, and GI detection rate | Lesion visibility and analysis of serrated lesions and adenomas | 16634 individuals (data integrated from 17 studies) | Clinical and endoscopic image data | Systematic review and meta-analysis |
Recent meta-analyses collectively demonstrated high diagnostic accuracy of AI-assisted endoscopy for early ESCC detection (Table 3). A meta-analysis of convolutional neural network systems reported a pooled sensitivity of 0.90 [95% confidence interval (CI): 0.82-0.94], specificity of 0.91 (95%CI: 0.79-0.96), and area under the curve of 0.95 (95%CI: 0.93-0.97) for early ESCC detection[13]. Another meta-analysis reported a pooled sensitivity of 91.2% (84.3%-95.2%) and specificity of 80.0% (64.3%-89.9%)[14]. These robust findings are supported by numerous individual clinical studies (Table 1).
An AI system combined white light imaging (WLI) and NBI images in an internal validation set with a sensitivity of 96.64%, specificity of 95.35%, and accuracy of 91.75%; for early ESCC. The accuracy, sensitivity, and specificity of early ESCC detection were 85.7%, 92.6%, and 80.0%, respectively[15,16]. For non-magnified NBI/blue-laser imaging, the same system yielded a sensitivity of 100%, specificity of 63%, and accuracy of 77%; for WLI the corresponding values were 90%, 76%, and 81%, respectively[17]. In a video-based evaluation, expert endoscopists achieved a sensitivity of 79%, specificity of 72%, and accuracy of 75%, whereas the AI model attained a sensitivity of 91%, specificity of 51%, and accuracy of 63% (a higher sensitivity but lower specificity and overall accuracy)[18]. These data suggest that AI can extract pixel-level patterns imperceptible to the human eye, maximizing diagnostic yield from various imaging modalities. Given that the interobserver agreement among endoscopists for early ESCC is approximately 70%, AI can serve as a valuable tool to enhance diagnostic consistency[19].
Beyond its standalone capabilities AI shows great potential for enhancing the skills of endoscopists. With AI assistance the sensitivity and accuracy of general endoscopists increased from 57.4% to 66.5% and from 68.6% to 75.9%, respectively. Among experts the sensitivity increased from 59.1% to 70.0%, and the accuracy increased from 72.1% to 79.3%[20]. AI has markedly enhanced the ability of less experienced endoscopists to classify intrapapillary capillary loop patterns and predict invasion depth[21] (Table 2). AI systems detect and precisely delineate very small flat lesions (approximately 3 mm) that are often missed visually regardless of WLI/NBI use, magnification, or iodine staining[22,23]. The mean processing time to detect and outline a lesion on a single image was 17 milliseconds for AI and 92 seconds for an expert endoscopist. Video diagnosis can be performed at 26-37 milliseconds per frame, and some systems can scan an entire cancer video within 1 second, making it sufficiently fast for real-time use. Several solutions have been integrated into endoscopic processors that do not require additional monitoring[16,18,22,24].
Despite these promising results several challenges must be addressed for successful clinical integration. The black-box nature of deep learning models is a significant barrier to their clinical adoption in endoscopy as it limits transparency and clinician trust[25]. Explainable AI (XAI) techniques are being actively developed to address this challenge by providing interpretable justifications for AI outputs[26]. In endoscopic diagnosis methods such as gradient-weighted class activation mapping generate heat maps that visually indicate the image regions that influence the AI decision, allowing clinicians to critically assess the focus and reasoning of the model[27]. Studies have demonstrated that XAI not only improves diagnostic support but also enhances the trust and acceptance of endoscopists. The ENDOANGEL-ED system, which incorporates explainability, was shown to significantly increase both diagnostic accuracy and clinician confidence compared with traditional deep learning models while also improving endoscopist performance in multireader studies[27]. Beyond diagnostic support XAI has educational value in visualizing expert lesion recognition patterns, which can be leveraged for training and skill development[28,29].
Thus, the integration of XAI is foundational for building clinician trust, meeting regulatory and ethical requirements, and facilitating the safe and effective incorporation of AI into routine endoscopic practice[28,30]. Some studies have failed to demonstrate the non-inferiority of AI systems compared with human endoscopists[31]. Furthermore, the creation of AI models requires large, high-quality datasets; insufficient volume, poor quality, and sampling bias remain major obstacles[1]. False positives during AI diagnosis are an additional concern[32]. Future priorities include large multicenter validation studies, optimization of algorithms for real-time video processing, and proactive resolution of ethical and legal issues from a patient-centered perspective[33].
The National Clinical Database (NCD) and Japan Endoscopy Database (JED) are pivotal national registries in Japan that provide a foundation for quality improvement, research, and policies in surgical and endoscopic medicine[34,35]. Established in 2010 and closely linked to the surgical board certification system, the NCD is a nationwide web-based registry that captures > 95% of surgical procedures in Japan[34,36]. It enables comprehensive tracking of surgical outcomes, risk adjustment, and institutional benchmarking[37,38]. By providing feedback to participating hospitals and facilitating national and international comparisons, the NCD drives continuous quality improvement and evidence-based policies in Japanese surgical care[34,36,39]. Its vast dataset has also illuminated key clinical trends, such as an aging patient population and a shift towards minimally invasive surgery[37,40,41].
The JED, initiated by the Japan Gastroenterological Endoscopy Society, is a prospective project aimed at becoming one of the world’s largest endoscopy databases[35]. It standardizes terminology and data collection for all endoscopic pro
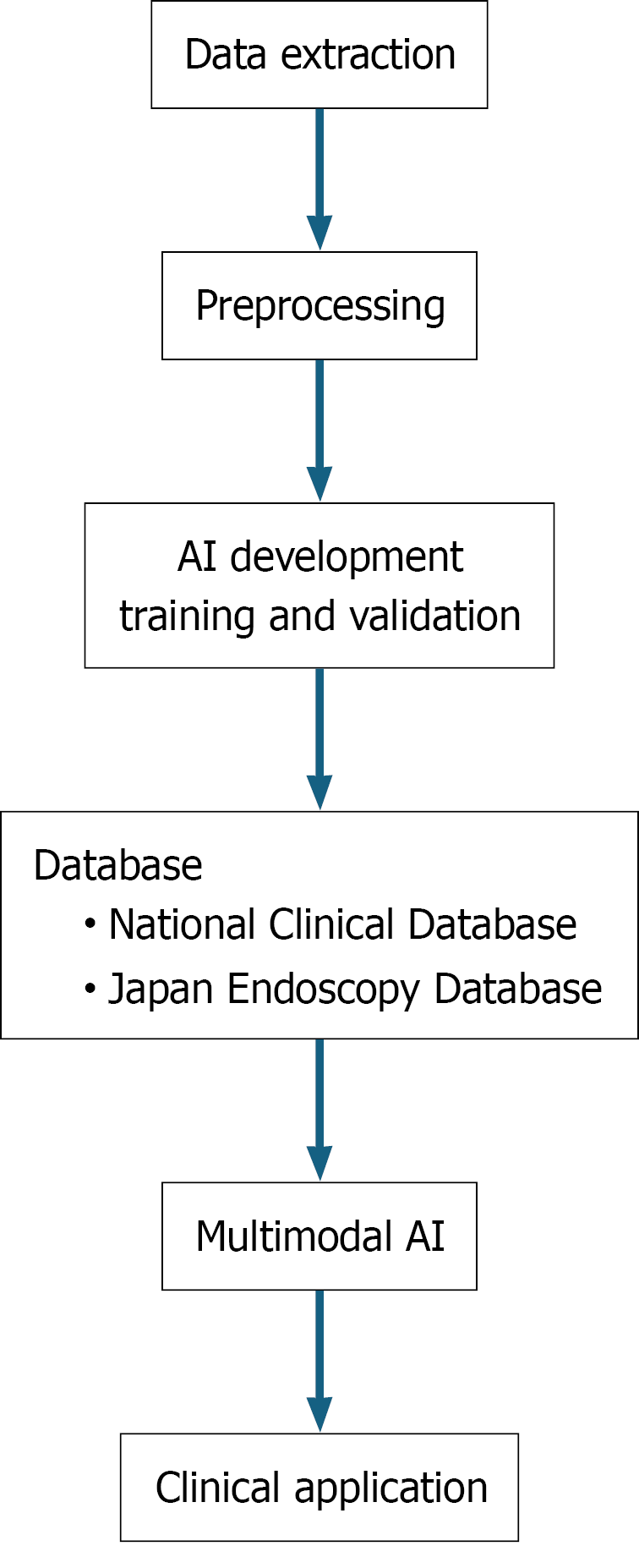
Building robust and generalizable AI models requires large-scale heterogeneous datasets to mitigate data bias, which is a challenge often faced by single-institution studies (Figure 1)[44,45]. Japan’s national registries, the NCD and JED, provide a decisive solution because of their scale, quality, and standardized structure. JED provides an unparalleled platform for the development of AI in endoscopic diagnosis[35,42,43]. It collects endoscopic images and videos under standardized protocols and a wealth of associated metadata, including histopathological diagnoses, specific endoscope models used, and experience level of the endoscopists[35]. This structured dataset is invaluable in all phases of AI development. High-quality images linked to definitive pathological results ensure the quality of both model training and internal validation. Comprehensive surgical and pathological data from the NCD are ideal resources for creating and validating AI-powered risk models and outcome-prediction tools for perioperative management[36,39,46].
The goal of these registries is to enable robust external validation. By reflecting real-world clinical heterogeneity through multicenter, multivendor, and multioperator data, these studies allow for a true assessment of the robustness and generalizability of multimodal AI[47]. This provides Japanese researchers with a significant competitive advantage in developing and deploying clinically reliable and scalable multimodal AI applications globally[48,49].
Despite their immense potential leveraging large-scale databases such as the NCD and JED for routine AI integration presents several key challenges. A primary hurdle lies in the data infrastructure: Incomplete data interoperability across different hospital systems complicates harmonization, whereas issues of data quality and completeness, such as missing entries, can undermine the reliability of AI models[35,42]. Furthermore, ensuring real-world generalizability is critical as models trained on national data must perform robustly across heterogeneous clinical practices and patient populations without overfitting[48-51]. The successful clinical adoption of these tools also depends on their seamless integration into existing workflows, a process often hindered by concerns over the interpretability of black-box models and clinician trust[52]. Finally, navigating the complex regulatory, ethical, and governance landscapes, including data privacy and liability, presents a significant barrier that requires ongoing collaboration among clinicians, regulators, and industry[48,50,53]. Systematically addressing these multifaceted challenges is essential to unlock the full potential of national databases for developing clinically impactful AI technologies. The goal is to create multimodal AI models that are not only accurate but also fair, robust, and seamlessly integrable into diverse clinical environments.
Ethical and legal concerns hinder the development of AI. The black-box nature of AI and the uncertainty regarding the legal responsibility for diagnostic errors are critical barriers to social acceptance and full incorporation into screening programs. The clinical integration of AI raises complex legal questions regarding liability in the event of diagnostic errors[54]. In the Japanese legal context, current regulations do not explicitly define the legal status or liability of AI-assisted diagnostic tools. Although the prevailing view holds that physicians retain their ultimate responsibility under the Medical Practitioners Act, the practical application of this principle in AI-supported settings remains ambiguous, particularly in cases of misdiagnosis or omission of relevant information. Although the principles that AI serves as a supportive tool and that the ultimate diagnostic responsibility rests with the clinician are widely accepted, its application in specific clinical scenarios is not straightforward[55].
Consider a false-positive case in which the AI flags a lesion, but the clinician, based on expert judgment, correctly identifies it as benign, thus avoiding an unnecessary biopsy. The current consensus in the medical literature emphasizes that robust, externally validated clinical evaluations, including prospective studies and randomized trials, when feasible, are essential before AI tools can be integrated into practice[56]. While the clinician’s decision is medically sound, a record of overruling an AI alert could potentially increase litigation risk if a different lesion is missed. Conversely, in a false-negative scenario both the AI and clinician fail to detect a lesion[57]. In such cases if an AI system is proven to have a performance level equivalent to or exceeding that of an average specialist, a new legal landscape may emerge in which a clinician’s failure to consult the AI or their overreliance[57] on it could be considered a breach of the standard of care[57]. The rapid use of AI models in clinical settings has outpaced the establishment of standardized evaluation criteria, leading to variability in performance assessments and challenges in ensuring safety, efficacy, and equity across diverse patient populations[58].
Ethical and legal challenges extend beyond liability. For instance, the issue of incidental findings in which AI flags a suspicious lesion in an adjacent area outside the primary focus of examination, such as the pharynx, during esophageal screening, presents a new ethical dilemma[59]. Clear guidelines regarding the extent of a clinician’s obligation to investigate incidental and out-of-specialty findings have not yet been established. Although no high-profile legal cases have emerged in Japan regarding incidental AI findings, international reports, particularly in radiology, have raised concerns that the failure to investigate AI-flagged abnormalities, even in adjacent or non-target areas, could lead to legal liability. For instance, in the United Kingdom and United States, legal commentaries have highlighted that overlooking AI-generated alerts may be considered a breach of the standard of care, especially when such alerts pertain to clinically significant but incidental findings[57-59].
Furthermore, ethical frameworks emphasize the need for transparent data stewardship, clear governance, and mechanisms for patient empowerment and consent management[60]. Legal frameworks in Japan, such as the Next Generation Medical Infrastructure Act, provide a basis for the secondary use of medical data but face challenges in practical implementation, including underutilization and regulatory complexity[61]. Establishing protocols for the quality management of updated AI models and ensuring transparent communication of any changes to the clinical community are equally important. Building societal consensus and regulatory frameworks to address these issues is the key to sustainable development and safe implementation of AI technology[1,44,45].
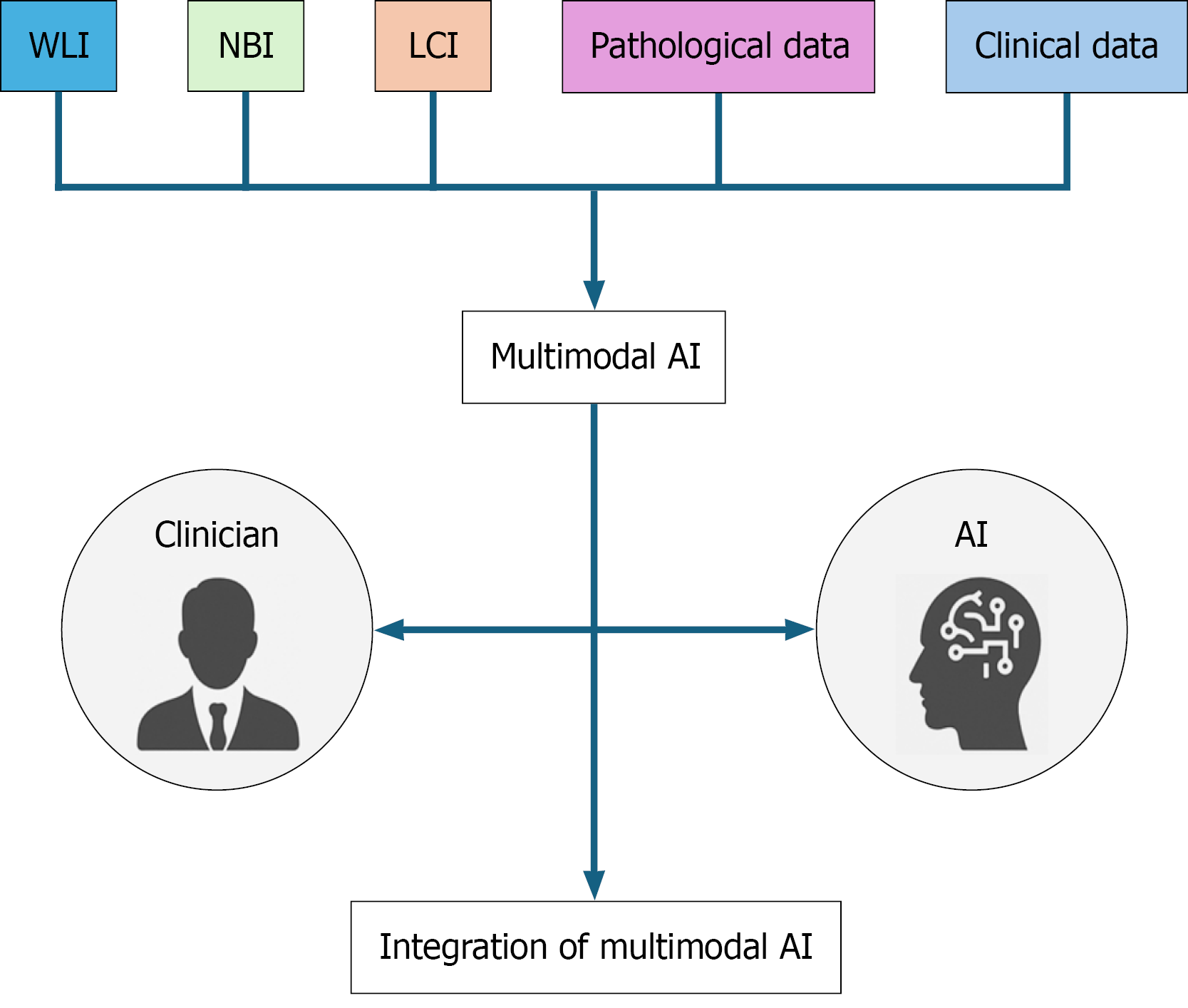
Recent advancements in image-enhanced endoscopy have led to a multimodal AI approach that integrates complementary information from diverse sources (Figure 2). By simultaneously inputting images of the same lesion captured in multiple modes, such as WLI, NBI, and linked color imaging, and training AI to analyze these features cohesively, such systems may achieve a level of robustness and diagnostic accuracy that surpass that of any single modality AI. This multimodal strategy has the potential to become the standard treatment for AI-assisted endoscopic diagnosis. However, most current multimodal AI systems remain in the experimental or proof-of-concept phase and have not yet been implemented in routine clinical workflows.
Beyond lesion detection and border delineation, multimodal AI, which integrates pathological, clinical, and genomic data, is attracting attention as a next-generation platform for personalized medicine, enabling depth-of-invasion prediction, treatment-response assessment, and therapeutic decision-making. Ideally, AI should serve as an adjunct, elevating the diagnostic ability of less-experienced physicians to the expert level, thereby standardizing care. To this end AI should not serve as a replacement for clinicians but as a tool for intelligence augmentation, enhancing their inherent skills.
Recent studies, including those involving generative AI, have consistently shown that human-AI collaboration yields superior outcomes in tasks ranging from diagnosis to text generation, surpassing the performance of either humans or AI working alone[55,62]. This collaborative paradigm is poised to contribute significantly to endoscopic diagnosis by improving accuracy, standardizing care, and reducing physicians’ cognitive loads[20,23,63,64]. AI implementation must be accompanied by continuous updates to the legal framework, insurance policies, and physician-training programs. Only through a holistic ecosystem spanning technical robustness, legal clarity, and societal consensus can AI truly augment clinical care in a safe and sustainable manner.
Advances in AI have potential benefits not only for the early detection of esophageal cancer but also for highly specialized tasks such as selecting treatment strategies and evaluating therapeutic efficacy. Multimodal AI and AI-based educational systems are expected to become future standards of care, further advancing personalized healthcare. As AI technology continues to evolve, exclusive dependency on it should be avoided. Continuous refinement of clinical expertise, knowledge, and ethical standards is indispensable. Given the inherent uncertainty of medicine, we must remain vigilant and adaptable.
However, limitations such as dataset bias, regulatory uncertainty, and the lack of long-term outcome evidence remain significant barriers to clinical implementation. Addressing these challenges will require sustained interdisciplinary collaboration among clinicians, data scientists, engineers, and ethicists. Such collaborations are essential to ensure the safe, effective, and ethically sound integration of AI into real-world clinical practice.
Ultimately, the goal of integrating AI is not to replace clinicians but to enhance their capabilities, standardize the quality of care, and disseminate expertise. This review provided a roadmap for clinicians and researchers to address opportunities and challenges, ensuring that AI is leveraged to achieve meaningful improvements in the care of patients with ESCC.
We thank the reviewers for their comments, which have helped us improve the manuscript. Open Evidence (https://openevidence.com), Gemini 2.5pro (https://gemini.google.com/), and GPT-4o and o3-pro (https://chat.openai.com/) were used for the search and draft editing, respectively.
| 1. | Codipilly DC, Qin Y, Dawsey SM, Kisiel J, Topazian M, Ahlquist D, Iyer PG. Screening for esophageal squamous cell carcinoma: recent advances. Gastrointest Endosc. 2018;88:413-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 204] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 2. | di Pietro M, Canto MI, Fitzgerald RC. Endoscopic Management of Early Adenocarcinoma and Squamous Cell Carcinoma of the Esophagus: Screening, Diagnosis, and Therapy. Gastroenterology. 2018;154:421-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 3. | Yang X, Suo C, Zhang T, Yin X, Man J, Yuan Z, Chen H, Yu J, Jin L, Chen X, Lu M, Ye W. A nomogram for screening esophageal squamous cell carcinoma based on environmental risk factors in a high-incidence area of China: a population-based case-control study. BMC Cancer. 2021;21:343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Nezu Y, Manabe N, Yoda Y, Haruma K. Effectiveness of screening endoscopy for esophageal squamous cell carcinoma in Japanese males. United European Gastroenterol J. 2022;10:868-873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 5. | Ono S, Kawada K, Dohi O, Kitamura S, Koike T, Hori S, Kanzaki H, Murao T, Yagi N, Sasaki F, Hashiguchi K, Oka S, Katada K, Shimoda R, Mizukami K, Suehiro M, Takeuchi T, Katsuki S, Tsuda M, Naito Y, Kawano T, Haruma K, Ishikawa H, Mori K, Kato M; LCI-FIND Trial Group. Linked Color Imaging Focused on Neoplasm Detection in the Upper Gastrointestinal Tract : A Randomized Trial. Ann Intern Med. 2021;174:18-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 6. | Dobashi A, Ono S, Furuhashi H, Futakuchi T, Tamai N, Yamauchi T, Suka M, Sumiyama K. Texture and Color Enhancement Imaging Increases Color Changes and Improves Visibility for Squamous Cell Carcinoma Suspicious Lesions in the Pharynx and Esophagus. Diagnostics (Basel). 2021;11:1971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 7. | Abe S, Yamazaki T, Hisada IT, Makiguchi ME, Yoshinaga S, Sato T, Nonaka S, Suzuki H, Oda I, Saito Y. Visibility of early gastric cancer in texture and color enhancement imaging. DEN Open. 2022;2:e46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 8. | Tamai N, Horiuchi H, Matsui H, Furuhashi H, Kamba S, Dobashi A, Sumiyama K. Visibility evaluation of colorectal lesion using texture and color enhancement imaging with video. DEN Open. 2022;2:e90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 9. | Oyama T, Inoue H, Arima M, Momma K, Omori T, Ishihara R, Hirasawa D, Takeuchi M, Tomori A, Goda K. Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: magnifying endoscopic classification of the Japan Esophageal Society. Esophagus. 2017;14:105-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 243] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 10. | Tanaka I, Hirasawa D, Saito H, Matsuda T, Nakahori M, Maeda Y, Okuzono T, Suzuki K, Igarashi K, Nawata Y, Ito S, Unno S, Chonan A. The sub-classification of type B2 vessels according to the magnifying endoscopic classification of the Japan Esophageal Society. Dig Endosc. 2020;32:49-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Yoshida M, Mori K, Urabe Y, Hirasawa D, Sasaki F, Takeuchi M, Kadota T, Yoshio T, Yoshinaga S, Kitamura Y, Ohno K, Ono Y, Igarashi K, Takahashi H, Ishihara R. Evaluating the usefulness of considering the size and morphological type of type B2 vessel area based on Japan Esophageal Society classification in estimating tumor invasion depth in superficial esophageal squamous cell carcinomas: study protocol for a prospective observational study (Japan BEES study). BMC Gastroenterol. 2024;24:50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 12. | Akashi T, Yamaguchi N, Isomoto H. Recent Advances in Gastrointestinal Cancer Endoscopic Diagnosis and Treatment: Focusing on Older Adults. Intern Med. 2025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Ma H, Wang L, Chen Y, Tian L. Convolutional neural network-based artificial intelligence for the diagnosis of early esophageal cancer based on endoscopic images: A meta-analysis. Saudi J Gastroenterol. 2022;28:332-340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 14. | Guidozzi N, Menon N, Chidambaram S, Markar SR. The role of artificial intelligence in the endoscopic diagnosis of esophageal cancer: a systematic review and meta-analysis. Dis Esophagus. 2023;36:doad048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 15. | Liu W, Yuan X, Guo L, Pan F, Wu C, Sun Z, Tian F, Yuan C, Zhang W, Bai S, Feng J, Hu Y, Hu B. Artificial Intelligence for Detecting and Delineating Margins of Early ESCC Under WLI Endoscopy. Clin Transl Gastroenterol. 2022;13:e00433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (1)] |
| 16. | Feng Y, Liang Y, Li P, Long Q, Song J, Li M, Wang X, Cheng CE, Zhao K, Ma J, Zhao L. Artificial intelligence assisted detection of superficial esophageal squamous cell carcinoma in white-light endoscopic images by using a generalized system. Discov Oncol. 2023;14:73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 17. | Ohmori M, Ishihara R, Aoyama K, Nakagawa K, Iwagami H, Matsuura N, Shichijo S, Yamamoto K, Nagaike K, Nakahara M, Inoue T, Aoi K, Okada H, Tada T. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc. 2020;91:301-309.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (1)] |
| 18. | Fukuda H, Ishihara R, Kato Y, Matsunaga T, Nishida T, Yamada T, Ogiyama H, Horie M, Kinoshita K, Tada T. Comparison of performances of artificial intelligence versus expert endoscopists for real-time assisted diagnosis of esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2020;92:848-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (1)] |
| 19. | Hatta W, Koike T, Ogata Y, Kondo Y, Ara N, Uno K, Asano N, Imatani A, Masamune A. Comparison of Magnifying Endoscopy with Blue Light Imaging and Narrow Band Imaging for Determining the Invasion Depth of Superficial Esophageal Squamous Cell Carcinoma by the Japanese Esophageal Society's Intrapapillary Capillary Loop Classification. Diagnostics (Basel). 2021;11:1941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Aoyama N, Nakajo K, Sasabe M, Inaba A, Nakanishi Y, Seno H, Yano T. Effects of artificial intelligence assistance on endoscopist performance: Comparison of diagnostic performance in superficial esophageal squamous cell carcinoma detection using video-based models. DEN Open. 2026;6:e70083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (1)] |
| 21. | Yuan XL, Liu W, Liu Y, Zeng XH, Mou Y, Wu CC, Ye LS, Zhang YH, He L, Feng J, Zhang WH, Wang J, Chen X, Hu YX, Zhang KH, Hu B. Artificial intelligence for diagnosing microvessels of precancerous lesions and superficial esophageal squamous cell carcinomas: a multicenter study. Surg Endosc. 2022;36:8651-8662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Yuan X, Zeng X, He L, Ye L, Liu W, Hu Y, Hu B. Artificial intelligence for detecting and delineating a small flat-type early esophageal squamous cell carcinoma under multimodal imaging. Endoscopy. 2023;55:E141-E142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Zhou Y, Liu RD, Gong H, Yuan XL, Hu B, Huang ZY. Multimodal artificial intelligence system for detecting a small esophageal high-grade squamous intraepithelial neoplasia: A case report. World J Gastrointest Endosc. 2025;17:101233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 24. | Everson M, Herrera L, Li W, Luengo IM, Ahmad O, Banks M, Magee C, Alzoubaidi D, Hsu HM, Graham D, Vercauteren T, Lovat L, Ourselin S, Kashin S, Wang HP, Wang WL, Haidry RJ. Artificial intelligence for the real-time classification of intrapapillary capillary loop patterns in the endoscopic diagnosis of early oesophageal squamous cell carcinoma: A proof-of-concept study. United European Gastroenterol J. 2019;7:297-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (1)] |
| 25. | Poon AIF, Sung JJY. Opening the black box of AI-Medicine. J Gastroenterol Hepatol. 2021;36:581-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (1)] |
| 26. | Ge Z, Wang B, Chang J, Yu Z, Zhou Z, Zhang J, Duan Z. Using deep learning and explainable artificial intelligence to assess the severity of gastroesophageal reflux disease according to the Los Angeles Classification System. Scand J Gastroenterol. 2023;58:596-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 27. | Dong Z, Wang J, Li Y, Deng Y, Zhou W, Zeng X, Gong D, Liu J, Pan J, Shang R, Xu Y, Xu M, Zhang L, Zhang M, Tao X, Zhu Y, Du H, Lu Z, Yao L, Wu L, Yu H. Explainable artificial intelligence incorporated with domain knowledge diagnosing early gastric neoplasms under white light endoscopy. NPJ Digit Med. 2023;6:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 28. | Auzine MM, Heenaye-Mamode Khan M, Baichoo S, Gooda Sahib N, Bissoonauth-Daiboo P, Gao X, Heetun Z. Development of an ensemble CNN model with explainable AI for the classification of gastrointestinal cancer. PLoS One. 2024;19:e0305628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 29. | Mascarenhas M, Mendes F, Martins M, Ribeiro T, Afonso J, Cardoso P, Ferreira J, Fonseca J, Macedo G. Explainable AI in Digestive Healthcare and Gastrointestinal Endoscopy. J Clin Med. 2025;14:549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 30. | Nazir S, Dickson DM, Akram MU. Survey of explainable artificial intelligence techniques for biomedical imaging with deep neural networks. Comput Biol Med. 2023;156:106668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 65] [Reference Citation Analysis (0)] |
| 31. | Tani Y, Ishihara R, Inoue T, Okubo Y, Kawakami Y, Matsueda K, Miyake M, Yoshii S, Shichijo S, Kanesaka T, Yamamoto S, Takeuchi Y, Higashino K, Uedo N, Michida T, Kato Y, Tada T. A single-center prospective study evaluating the usefulness of artificial intelligence for the diagnosis of esophageal squamous cell carcinoma in a real-time setting. BMC Gastroenterol. 2023;23:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 32. | Tao Y, Fang L, Qin G, Xu Y, Zhang S, Zhang X, Du S. Efficiency of endoscopic artificial intelligence in the diagnosis of early esophageal cancer. Thorac Cancer. 2024;15:1296-1304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Yan SY, Fu XY, Yang Y, Jia LY, Liang JW, Li YH, Yan LL, Zhou Y, Zhou XB, Li SW, Mao XL. Artificial intelligence in early screening for esophageal squamous cell carcinoma. Best Pract Res Clin Gastroenterol. 2025;75:102004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 34. | Miyata H, Gotoh M, Hashimoto H, Motomura N, Murakami A, Tomotaki A, Hirahara N, Ono M, Ko C, Iwanaka T. Challenges and prospects of a clinical database linked to the board certification system. Surg Today. 2014;44:1991-1999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 35. | Matsuda K, Tanaka K, Fujishiro M, Saito Y, Ohtsuka K, Oda I, Katada C, Kato M, Kida M, Kobayashi K, Hoteya S, Horimatsu T, Kodashima S, Matsuda T, Muto M, Yamamoto H, Ryozawa S, Iwakiri R, Kutsumi H, Miyata H, Kato M, Haruma K, Fujimoto K, Uemura N, Kaminishi M, Tajiri H. Design paper: Japan Endoscopy Database (JED): A prospective, large database project related to gastroenterological endoscopy in Japan. Dig Endosc. 2018;30:5-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 36. | Gotoh M, Miyata H, Hashimoto H, Wakabayashi G, Konno H, Miyakawa S, Sugihara K, Mori M, Satomi S, Kokudo N, Iwanaka T. National Clinical Database feedback implementation for quality improvement of cancer treatment in Japan: from good to great through transparency. Surg Today. 2016;46:38-47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 107] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 37. | Murakami K, Akutsu Y, Miyata H, Toh Y, Toyozumi T, Kakeji Y, Seto Y, Matsubara H. Essential risk factors for operative mortality in elderly esophageal cancer patients registered in the National Clinical Database of Japan. Esophagus. 2023;20:39-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 38. | Yamamoto H, Nashimoto A, Miyashiro I, Miyata H, Toh Y, Gotoh M, Kodera Y, Kakeji Y, Seto Y. Impact of a board certification system and adherence to the clinical practice guidelines for gastric cancer on risk-adjusted surgical mortality after distal and total gastrectomy in Japan: a questionnaire survey of departments registered in the National Clinical Database. Surg Today. 2024;54:459-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 39. | Anazawa T, Miyata H, Gotoh M. Cancer registries in Japan: National Clinical Database and site-specific cancer registries. Int J Clin Oncol. 2015;20:5-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 40. | Ito S, Takahashi A, Ueno H, Takiguchi S, Kajiwara Y, Kakeji Y, Eguchi S, Goi T, Saiura A, Sasaki A, Takeuchi H, Tanaka C, Hashimoto M, Hiki N, Horiguchi A, Matsuda S, Mizushima T, Yamamoto H, Kitagawa Y, Shirabe K. Annual report on National Clinical Database 2021 for gastroenterological surgery in Japan. Ann Gastroenterol Surg. 2025;9:32-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 41. | Kajiwara Y, Takahashi A, Ueno H, Kakeji Y, Hasegawa H, Eguchi S, Goi T, Saiura A, Sasaki A, Takiguchi S, Takeuchi H, Tanaka C, Hashimoto M, Hiki N, Horiguchi A, Matsuda S, Mizushima T, Marubashi S, Gotoh M, Konno H, Yamamoto H, Miyata H, Seto Y, Kitagawa Y; National Clinical Database. Annual report on National Clinical Database 2020 for gastroenterological surgery in Japan. Ann Gastroenterol Surg. 2023;7:367-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 62] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 42. | Kodashima S, Tanaka K, Matsuda K, Fujishiro M, Saito Y, Ohtsuka K, Oda I, Katada C, Kato M, Kida M, Kobayashi K, Hoteya S, Horimatsu T, Matsuda T, Muto M, Yamamoto H, Ryozawa S, Iwakiri R, Kutsumi H, Miyata H, Kato M, Haruma K, Fujimoto K, Uemura N, Kaminishi M, Tajiri H. First progress report on the Japan Endoscopy Database project. Dig Endosc. 2018;30:20-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 43. | Saito Y, Kodashima S, Matsuda T, Matsuda K, Fujishiro M, Tanaka K, Kobayashi K, Katada C, Horimatsu T, Muto M, Ohtsuka K, Oda I, Kato M, Kida M, Hoteya S, Yamamoto H, Ryozawa S, Iwakiri R, Kutsumi H, Kato M, Haruma K, Fujimoto K, Iishi H, Ogata H, Uemura N, Kaminishi M, Tajiri H, Inoue H. Current status of diagnostic and therapeutic colonoscopy in Japan: The Japan Endoscopic Database Project. Dig Endosc. 2022;34:144-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 44. | Liu Y. Artificial intelligence-assisted endoscopic detection of esophageal neoplasia in early stage: The next step? World J Gastroenterol. 2021;27:1392-1405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (1)] |
| 45. | Zhang L, Luo R, Tang D, Zhang J, Su Y, Mao X, Ye L, Yao L, Zhou W, Zhou J, Lu Z, Zhang M, Xu Y, Deng Y, Huang X, He C, Xiao Y, Wang J, Wu L, Li J, Zou X, Yu H. Human-Like Artificial Intelligent System for Predicting Invasion Depth of Esophageal Squamous Cell Carcinoma Using Magnifying Narrow-Band Imaging Endoscopy: A Retrospective Multicenter Study. Clin Transl Gastroenterol. 2023;14:e00606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 46. | Kakeji Y, Yamamoto H, Ueno H, Eguchi S, Endo I, Sasaki A, Takiguchi S, Takeuchi H, Hashimoto M, Horiguchi A, Masaki T, Marubashi S, Yoshida K, Miyata H, Konno H, Gotoh M, Kitagawa Y, Mori M, Seto Y. Development of gastroenterological surgery over the last decade in Japan: analysis of the National Clinical Database. Surg Today. 2021;51:187-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 47. | Okagawa Y, Abe S, Yamada M, Oda I, Saito Y. Artificial Intelligence in Endoscopy. Dig Dis Sci. 2022;67:1553-1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 48. | Parasa S, Wallace M, Bagci U, Antonino M, Berzin T, Byrne M, Celik H, Farahani K, Golding M, Gross S, Jamali V, Mendonca P, Mori Y, Ninh A, Repici A, Rex D, Skrinak K, Thakkar SJ, van Hooft JE, Vargo J, Yu H, Xu Z, Sharma P. Proceedings from the First Global Artificial Intelligence in Gastroenterology and Endoscopy Summit. Gastrointest Endosc. 2020;92:938-945.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 49. | Sumiyama K, Futakuchi T, Kamba S, Matsui H, Tamai N. Artificial intelligence in endoscopy: Present and future perspectives. Dig Endosc. 2021;33:218-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 50. | Christou CD, Tsoulfas G. Challenges involved in the application of artificial intelligence in gastroenterology: The race is on! World J Gastroenterol. 2023;29:6168-6178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 7] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 51. | Du RC, Ouyang YB, Hu Y. Research trends on artificial intelligence and endoscopy in digestive diseases: A bibliometric analysis from 1990 to 2022. World J Gastroenterol. 2023;29:3561-3573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (2)] |
| 52. | Kuo CY, Chiu HM. Application of artificial intelligence in gastroenterology: Potential role in clinical practice. J Gastroenterol Hepatol. 2021;36:267-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 53. | Shung DL, Sung JJY. Challenges of developing artificial intelligence-assisted tools for clinical medicine. J Gastroenterol Hepatol. 2021;36:295-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 54. | Cestonaro C, Delicati A, Marcante B, Caenazzo L, Tozzo P. Defining medical liability when artificial intelligence is applied on diagnostic algorithms: a systematic review. Front Med (Lausanne). 2023;10:1305756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 55. | Chadebecq F, Lovat LB, Stoyanov D. Artificial intelligence and automation in endoscopy and surgery. Nat Rev Gastroenterol Hepatol. 2023;20:171-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 58] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 56. | Park SH, Han K, Jang HY, Park JE, Lee JG, Kim DW, Choi J. Methods for Clinical Evaluation of Artificial Intelligence Algorithms for Medical Diagnosis. Radiology. 2023;306:20-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 72] [Reference Citation Analysis (0)] |
| 57. | Anderson T, Torreggiani WC, Munk PL, Mallinson PI. The impact of the introduction of artificial intelligence in radiology and its potential legal implications in the UK and Ireland. BJR Open. 2020;2:20200030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 58. | Shah NH, Halamka JD, Saria S, Pencina M, Tazbaz T, Tripathi M, Callahan A, Hildahl H, Anderson B. A Nationwide Network of Health AI Assurance Laboratories. JAMA. 2024;331:245-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 76] [Reference Citation Analysis (0)] |
| 59. | Korfiatis P, Kline TL, Meyer HM, Khalid S, Leiner T, Loufek BT, Blezek D, Vidal DE, Hartman RP, Joppa LJ, Missert AD, Potretzke TA, Taubel JP, Tjelta JA, Callstrom MR, Williamson EE. Implementing Artificial Intelligence Algorithms in the Radiology Workflow: Challenges and Considerations. Mayo Clin Proc Digit Health. 2025;3:100188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 60. | Williams M, Karim W, Gelman J, Raza M. Ethical data acquisition for LLMs and AI algorithms in healthcare. NPJ Digit Med. 2024;7:377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 61. | Yoshida M, Tanaka K, Yamamoto R. An Examination of Japan's Legal System and Issues Related to Promoting the Use of Medical Data. Stud Health Technol Inform. 2025;327:1019-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 62. | Zöller N, Berger J, Lin I, Fu N, Komarneni J, Barabucci G, Laskowski K, Shia V, Harack B, Chu EA, Trianni V, Kurvers RHJM, Herzog SM. Human-AI collectives most accurately diagnose clinical vignettes. Proc Natl Acad Sci U S A. 2025;122:e2426153122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 63. | Zhang YH, Guo LJ, Yuan XL, Hu B. Artificial intelligence-assisted esophageal cancer management: Now and future. World J Gastroenterol. 2020;26:5256-5271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 40] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (2)] |
| 64. | Nagao S, Tani Y, Shibata J, Tsuji Y, Tada T, Ishihara R, Fujishiro M. Implementation of artificial intelligence in upper gastrointestinal endoscopy. DEN Open. 2022;2:e72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 65. | Zhao YY, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 66. | Shimamoto Y, Ishihara R, Kato Y, Shoji A, Inoue T, Matsueda K, Miyake M, Waki K, Kono M, Fukuda H, Matsuura N, Nagaike K, Aoi K, Yamamoto K, Inoue T, Nakahara M, Nishihara A, Tada T. Real-time assessment of video images for esophageal squamous cell carcinoma invasion depth using artificial intelligence. J Gastroenterol. 2020;55:1037-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (1)] |
| 67. | Shiroma S, Yoshio T, Kato Y, Horie Y, Namikawa K, Tokai Y, Yoshimizu S, Yoshizawa N, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Akazawa N, Akiyama J, Tada T, Fujisaki J. Ability of artificial intelligence to detect T1 esophageal squamous cell carcinoma from endoscopic videos and the effects of real-time assistance. Sci Rep. 2021;11:7759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (1)] |
| 68. | Ikenoyama Y, Yoshio T, Tokura J, Naito S, Namikawa K, Tokai Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Katayama N, Tada T, Fujisaki J. Artificial intelligence diagnostic system predicts multiple Lugol-voiding lesions in the esophagus and patients at high risk for esophageal squamous cell carcinoma. Endoscopy. 2021;53:1105-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 69. | Li B, Cai SL, Tan WM, Li JC, Yalikong A, Feng XS, Yu HH, Lu PX, Feng Z, Yao LQ, Zhou PH, Yan B, Zhong YS. Comparative study on artificial intelligence systems for detecting early esophageal squamous cell carcinoma between narrow-band and white-light imaging. World J Gastroenterol. 2021;27:281-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (1)] |
| 70. | Waki K, Ishihara R, Kato Y, Shoji A, Inoue T, Matsueda K, Miyake M, Shimamoto Y, Fukuda H, Matsuura N, Ono Y, Yao K, Hashimoto S, Terai S, Ohmori M, Tanaka K, Kato M, Shono T, Miyamoto H, Tanaka Y, Tada T. Usefulness of an artificial intelligence system for the detection of esophageal squamous cell carcinoma evaluated with videos simulating overlooking situation. Dig Endosc. 2021;33:1101-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 71. | Meng QQ, Gao Y, Lin H, Wang TJ, Zhang YR, Feng J, Li ZS, Xin L, Wang LW. Application of an artificial intelligence system for endoscopic diagnosis of superficial esophageal squamous cell carcinoma. World J Gastroenterol. 2022;28:5483-5493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (2)] |
| 72. | Tajiri A, Ishihara R, Kato Y, Inoue T, Matsueda K, Miyake M, Waki K, Shimamoto Y, Fukuda H, Matsuura N, Egawa S, Yamaguchi S, Ogiyama H, Ogiso K, Nishida T, Aoi K, Tada T. Utility of an artificial intelligence system for classification of esophageal lesions when simulating its clinical use. Sci Rep. 2022;12:6677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 73. | Wang J, Long Q, Liang Y, Song J, Feng Y, Li P, Sun W, Zhao L. AI-assisted identification of intrapapillary capillary loops in magnification endoscopy for diagnosing early-stage esophageal squamous cell carcinoma: a preliminary study. Med Biol Eng Comput. 2023;61:1631-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 74. | Wang YK, Karmakar R, Mukundan A, Men TC, Tsao YM, Lu SC, Wu IC, Wang HC. Computer-aided endoscopic diagnostic system modified with hyperspectral imaging for the classification of esophageal neoplasms. Front Oncol. 2024;14:1423405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 75. | Nakao E, Yoshio T, Kato Y, Namikawa K, Tokai Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Kurihara N, Ishizuka N, Ishihara R, Tada T, Fujisaki J. Randomized controlled trial of an artificial intelligence diagnostic system for the detection of esophageal squamous cell carcinoma in clinical practice. Endoscopy. 2025;57:210-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 76. | Ma T, Liu GQ, Guo J, Ji R, Shao XJ, Li YQ, Li Z, Zuo XL. Artificial intelligence-aided optical biopsy improves the diagnosis of esophageal squamous neoplasm. World J Gastroenterol. 2025;31:104370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 77. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 285] [Article Influence: 40.7] [Reference Citation Analysis (4)] |
| 78. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (1)] |
| 79. | Tang D, Wang L, Jiang J, Liu Y, Ni M, Fu Y, Guo H, Wang Z, An F, Zhang K, Hu Y, Zhan Q, Xu G, Zou X. A Novel Deep Learning System for Diagnosing Early Esophageal Squamous Cell Carcinoma: A Multicenter Diagnostic Study. Clin Transl Gastroenterol. 2021;12:e00393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 80. | Uema R, Hayashi Y, Tashiro T, Saiki H, Kato M, Amano T, Tani M, Yoshihara T, Inoue T, Kimura K, Iwatani S, Sakatani A, Yoshii S, Tsujii Y, Shinzaki S, Iijima H, Takehara T. Use of a convolutional neural network for classifying microvessels of superficial esophageal squamous cell carcinomas. J Gastroenterol Hepatol. 2021;36:2239-2246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 81. | Zhao Z, Li M, Liu P, Yu J, Zhao H. Efficacy of Digestive Endoscope Based on Artificial Intelligence System in Diagnosing Early Esophageal Carcinoma. Comput Math Methods Med. 2022;2022:9018939. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 82. | Syed T, Doshi A, Guleria S, Syed S, Shah T. Artificial Intelligence and Its Role in Identifying Esophageal Neoplasia. Dig Dis Sci. 2020;65:3448-3455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 83. | Huang LM, Yang WJ, Huang ZY, Tang CW, Li J. Artificial intelligence technique in detection of early esophageal cancer. World J Gastroenterol. 2020;26:5959-5969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 84. | Lazăr DC, Avram MF, Faur AC, Goldiş A, Romoşan I, Tăban S, Cornianu M. The Impact of Artificial Intelligence in the Endoscopic Assessment of Premalignant and Malignant Esophageal Lesions: Present and Future. Medicina (Kaunas). 2020;56:364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 85. | Namikawa K, Hirasawa T, Yoshio T, Fujisaki J, Ozawa T, Ishihara S, Aoki T, Yamada A, Koike K, Suzuki H, Tada T. Utilizing artificial intelligence in endoscopy: a clinician's guide. Expert Rev Gastroenterol Hepatol. 2020;14:689-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 86. | Tokat M, van Tilburg L, Koch AD, Spaander MCW. Artificial Intelligence in Upper Gastrointestinal Endoscopy. Dig Dis. 2022;40:395-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 87. | Pan Y, He L, Chen W, Yang Y. The current state of artificial intelligence in endoscopic diagnosis of early esophageal squamous cell carcinoma. Front Oncol. 2023;13:1198941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 88. | Kikuchi R, Okamoto K, Ozawa T, Shibata J, Ishihara S, Tada T. Endoscopic Artificial Intelligence for Image Analysis in Gastrointestinal Neoplasms. Digestion. 2024;105:419-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 89. | Shahzil M, Kashif TB, Jamil Z, Khaqan MA, Munir L, Amjad Z, Faisal MS, Chaudhary AJ, Ali H, Khan S, Enofe I. Assessing the effectiveness of texture and color enhancement imaging versus white-light endoscopy in detecting gastrointestinal lesions: A systematic review and meta-analysis. DEN Open. 2026;6:e70128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/