Published online Dec 15, 2025. doi: 10.4251/wjgo.v17.i12.114037
Revised: October 2, 2025
Accepted: November 6, 2025
Published online: December 15, 2025
Processing time: 92 Days and 17.1 Hours
Early recurrence is an important factor affecting the prognosis of hepatocellular carcinoma (HCC), but its preoperative prediction remains challenging.
To explore the value of a multimodal interpretable fusion model combining computed tomography (CT) habitat imaging (HI), radiomics, and clinical features in predicting early recurrence of HCC and analyze its correlation with patho
The 191 HCC patients were categorized into early recurrence and non-early recurrence groups based on postoperative follow-up outcomes, and randomly divided into training and testing sets in a 7:3 ratio. Based on CT arterial phase and clinical data, the habitat model, radiomics model, clinical model, and fusion model were constructed and compared for their predictive ability in early recurrence of HCC. For the optimal model, SHapley Additive exPlanations (SHAP) analysis was performed to evaluate the contribution of different features in the model, and the correlation between HI and radiomics features with tumor microvascular invasion (MVI), Ki67 expression, GPC-3 expression, and pathological grading was analyzed.
The fusion model demonstrated the best performance in predicting early recurrence of HCC, achieving the area under the curve of 0.933 on the validation set. The decision curve analysis curve indicated that the fusion model yielded the highest clinical net benefit. SHAP analysis provided valuable insights into explaining the fusion model's prediction of early HCC recurrence. Correlation analysis revealed significant associations between certain radiomics and Habitat features and pathological indicators such as MVI and Ki-67 expression in HCC.
An interpretable fusion model integrating clinical, radiomic, and habitat features can assist clinicians in identifying early postoperative recurrence of HCC, offering significant potential for prognosis prediction and clinical mana
Core Tip: This study explores an interpretable fusion model that combines clinical, radiomics, and habitat features based on enhanced arterial-phase computed tomography images of the liver to predict early recurrence after hepatocellular carcinoma (HCC) surgery. The model outperforms traditional radiomics, clinical, and habitat models. It can help clinicians identify early recurrence of HCC after surgery.
- Citation: Huang LH, Fang YJ, Zheng XJ, Huang C, Li CL, Yu B, Huang MJ, Qin SJ, Huang DY, Lu DW. Application of multimodal fusion technology in early recurrence prediction and pathological analysis of hepatocellular carcinoma. World J Gastrointest Oncol 2025; 17(12): 114037
- URL: https://www.wjgnet.com/1948-5204/full/v17/i12/114037.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i12.114037
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths globally. Its insidious onset and rapid progression result in most patients being diagnosed at an advanced stage, leading to a poor prognosis[1]. Although radical surgical resection is the main curative treatment for early-stage HCC, the early recurrence rate within two years post-surgery remains as high as 30%-50%, which becomes a critical bottleneck limiting long-term survival[2,3]. Current clinical prognostic indicators, such as alpha-fetoprotein (AFP) and tumor size, can only reflect partial biological characteristics of the tumor and are limited in capturing tumor heterogeneity, reducing the accuracy and clinical utility of preoperative early recurrence risk prediction[4,5]. Therefore, developing imaging tools capable of non-invasive, precise assessment of HCC recurrence risk holds significant clinical importance for optimizing postoperative follow-up strategies and guiding individualized interventions.
Imageomics technology offers a novel approach to quantifying tumor biological behavior by extracting high-throughput texture, morphological, and functional features from medical images, demonstrating potential in HCC diagnosis and prognostic assessment[6,7]. However, traditional radiomics models often treat the tumor as a homo
This study aims to construct a multimodal fusion model based on CT-enhanced arterial-phase images, integrating clinical features, traditional radiomics features, and HI features, and compare its performance with that of single-modality models (clinical model, radiomics model, HI model) to verify its advantages in predicting early postoperative recurrence of HCC. At the same time, SHapley Additive exPlanations (SHAP) analysis will be used to analyze the contribution of key predictive features in the model and explore the correlation between HI and radiomics features and MVI, Ki-67, GPC-3 expression, and pathological grading. The aim is to provide new imaging biomarkers for non-invasive assessment of early recurrence of HCC and promote the application of radiology in precision prognostic management of HCC.
This study was approved by the Ethics Committee of the Affiliated Hospital of Youjiang Medical University for Nationalities (approval No. YYFY-LL-2024-038). In accordance with the Helsinki Declaration, retrospective studies are exempt from the requirement to obtain informed consent from participants.
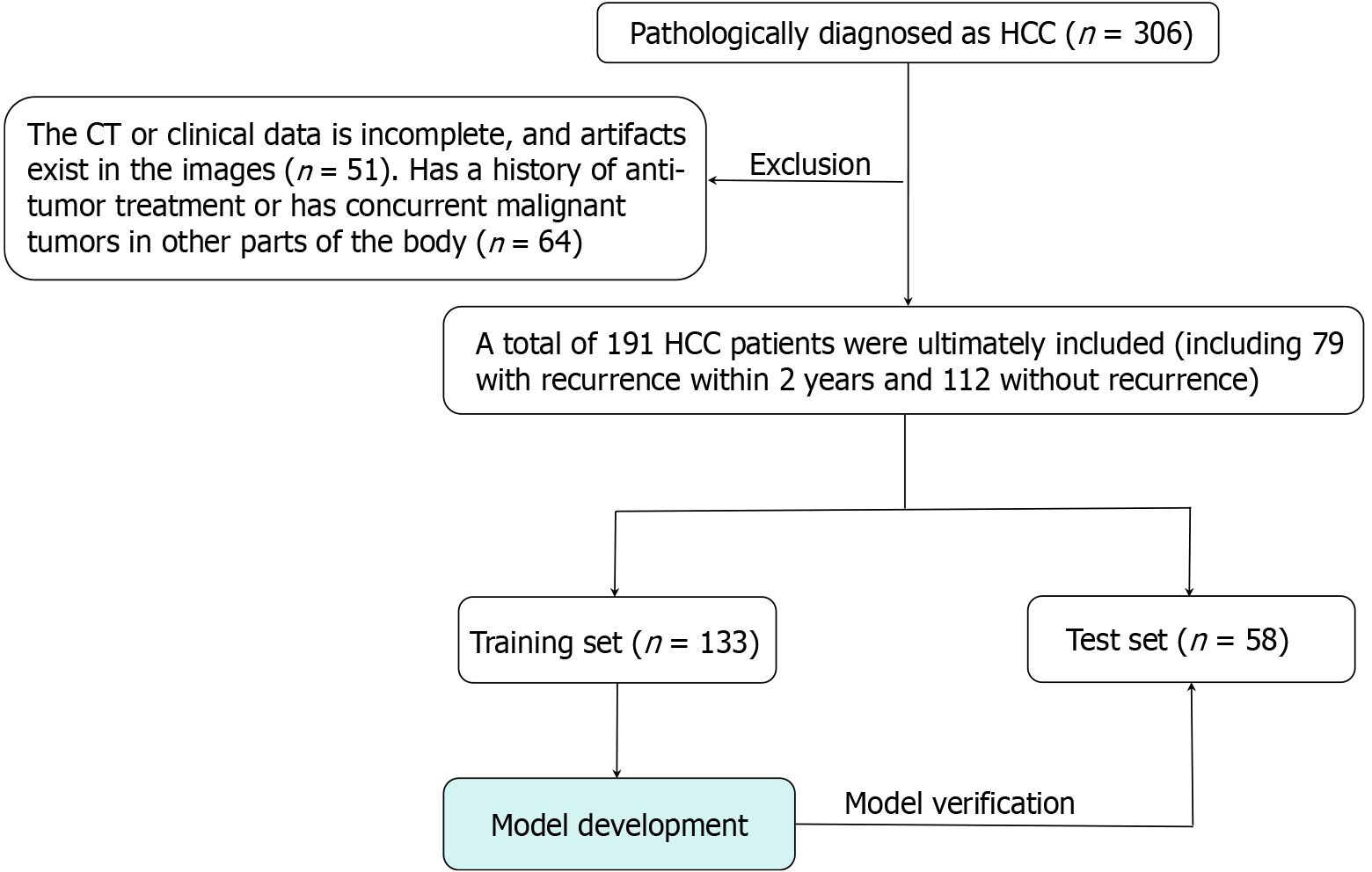
This retrospective study included 191 patients with HCC who were treated at the Affiliated Hospital of Youjiang Medical University for Nationalities from January 2019 to August 2023. Based on postoperative follow-up data, the study subjects were first categorized into an early recurrence group (n = 79) and a non-early recurrence group (n = 112). Subsequently, using random allocation, all patients were further divided into a training set (n = 133) and a test set (n = 58) at a ratio of 7:3.
Inclusion criteria: (1) Patients undergoing radical resection with postoperative pathological confirmation of HCC, possessing comprehensive clinical and follow-up data; and (2) Within one week prior to surgery, perform a contrast-enhanced CT scan of the liver concurrently with the corresponding laboratory tests.
Exclusion criteria: (1) CT images with artifacts during the arterial phase; and (2) Patients with prior antitumor treatments (radiotherapy, chemotherapy, interventional therapy) or concomitant malignancies in other sites.
All patients underwent follow-up at 3-month intervals for the first 2 years post-surgery, then every 6 months thereafter. Each follow-up included enhanced upper abdominal CT or magnetic resonance imaging (MRI) and tumor marker testing.
Clinical and laboratory parameters included age, gender, body mass index, alcohol consumption history, hepatitis B surface antigen, AFP, albumin, aspartate aminotransferase, and alanine aminotransferase.
This study referred to previous findings and included four pathological indicators closely related to postoperative recurrence of HCC: MVI[12], Ki-67[13], GPC-3[14], and pathological grading[15], in order to enhance the interpretability of the results. Among these, MVI was categorized into three levels: M0, M1, and M2. Low expression of Ki-67 (< 10%-20%) suggested slow tumor proliferation, better differentiation, and a favorable prognosis, while high expression (≥ 30%-50%) indicated active proliferation, poor differentiation, strong invasiveness, and a higher risk of recurrence[16]. GPC-3 was considered positive when tumor cells showed brown-yellow granular staining in the cytoplasm/membrane, with positive cells ≥ 10%, and negative otherwise[17]. Pathological grading used the Edmondson-Steiner system[18], with differentiation gradually decreasing from grade I to grade IV, accompanied by increasing cellular atypia, nuclear-to-cytoplasm ratio, and a shift in arrangement from regular trabecular to loose and disordered.
During the follow-up within 2 years after surgery for HCC patients, the discovery of new tumors in the liver or extrahepatic regions through imaging studies like ultrasound, CT, or MRI can diagnose early recurrence[18]. The diagnosis of tumor recurrence requires joint evaluation by a hepatobiliary surgeon and a radiologist. During the evaluation, both doctors rely solely on the designated clinical data and imaging findings, without access to any irrelevant information. Based on follow-up outcomes: Cases experiencing tumor recurrence within 2 years post-surgery were classified into the recurrence group; those without recurrence were assigned to the non-recurrence group.
The CT scanning method involved a scanning range from the diaphragm to the inferior margin of the liver. Contrast-enhanced scanning used a high-pressure injector with an 18G intravenous catheter to inject non-ionic iodinated contrast agent at 1.5-2.0 mL/kg at a rate of 3.0-3.5 mL/s, followed by immediate injection of 20-50 mL of saline to flush the catheter. After the bolus injection of the contrast agent, the aortic concentration reached the threshold [120 Hounsfield unit (HU)], triggering the arterial phase scan automatically after a 10-second delay (Table 1 showed the scanning parameters for each device).
| Devices | Revolution aca (GE) | Ingenuity core 64 (Philips) | Revolution (GE) |
| Layer thickness (mm) | 5 | 5 | 5 |
| Layer interval (mm) | 5 | 5 | 5 |
| Tube voltage (kV) | 120 | 120 | 120 |
| Tube current (mA) | 50 | 30 | 50 |
| Matrix | 512 × 512 | 512 × 512 | 512 × 512 |
| Threshold of ROI (HU) | 100 | 150 | 120 |
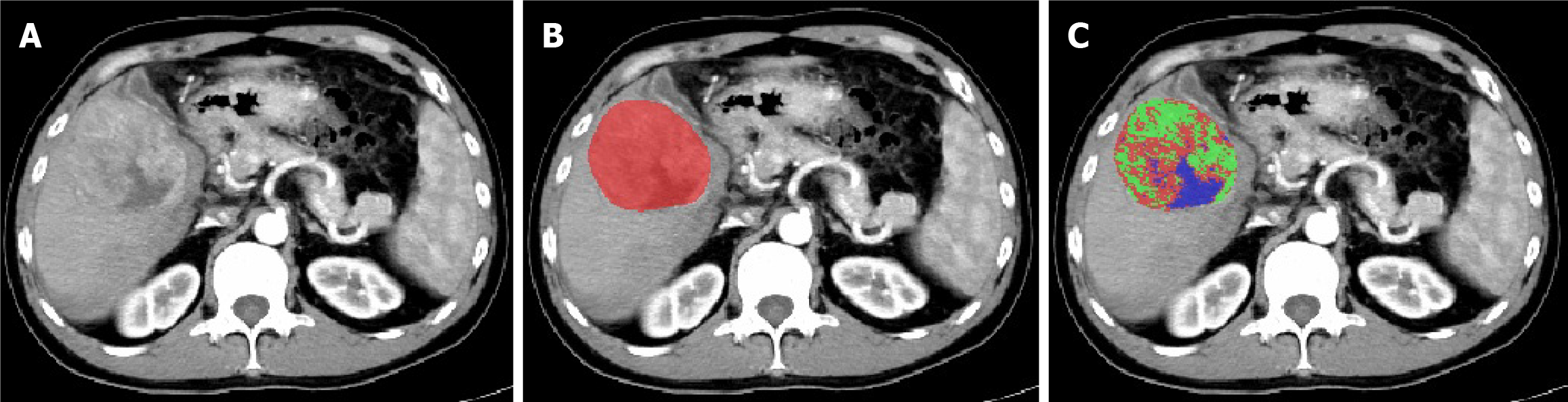
Download all patients' liver CT-enhanced arterial phase images in transverse DICOM format and convert them to NIFTI format for storage. Preprocessing of CT image datasets: First, calibrate the voxel spacing of all images to 1 mm × 1 mm × 1 mm. Then, standardize the window width and window level to 259 HU and 40 HU, respectively. Using ITK-SNAP 3.8.0 software, a radiologist with over 10 years of experience independently performed manual slice-by-slice segmentation of tumor regions in the CT arterial phase images (Figure 1). These segmentations were then cross-checked one-by-one by a senior-level physician (the study workflow is illustrated in Figure 2).
Radiomics analysis was performed using Pyradiomics (version 3.0.1) to extract 1834 radiomic features from the CT-enhanced arterial phase images, including tumor shape features, first-order features, texture features, and wavelet features. The texture features were derived from the Gray Level Co-occurrence Matrix, Gray Level Size Zone Matrix, Gray Level Run Length Matrix, Neighbouring Gray Tone Difference Matrix, and Gray Level Dependence Matrix.
The K-means algorithm, one of the most widely used methods, worked by iteratively finding the result that minimized the sum of the squared distances between each data point and its nearest K cluster centers, dividing the data points into different clusters such that the similarity within the same cluster was high, and the similarity between different clusters was low. Due to its high computational efficiency and strong interpretability, it was particularly suitable for high-dimensional and large-scale data[19]. In this study, we used the K-means clustering algorithm from the scikit-learn library, performing clustering analysis on the voxel signal intensities within the tumor based on arterial phase liver CT enhancement images. The number of clusters (k) ranged from 2 to 5, and through optimization using the Calinski-Harabasz index, k = 3 was determined as the optimal number of clusters. After clustering, the PyRadiomics module was used to extract features from each sub-region, and by calculating the mean of the sub-region features, 5502 representative features (1834 × 3) were obtained.
Feature selection and model construction involved selecting features with good reproducibility and low redundancy by first applying Z-score normalization to the training and testing set features. Significant features were selected using t-tests or Mann-Whitney U tests (P < 0.05), and distinctive features with a Pearson correlation coefficient > 0.9 were selectively excluded to reduce multicollinearity. The least absolute shrinkage and selection operator (LASSO) algorithm was then used to construct a penalty function λ, shrinking some regression coefficients to zero to select stable features, with the optimal λ value determined by 10-fold cross-validation using the scikit-learn library, completing the feature optimization. Based on the selected features, clinical, radiomics, habitat radiomics, and fusion models were constructed. Model construction was carried out using three machine learning algorithms: RandomForest, ExtraTrees, and multi-layer perceptron (MLP), with the models constructed from the corresponding optimal features being defined as target models. The synthetic minority oversampling technique was combined with oversampling to address the class imbalance in the training set, while the validation set maintained the original sample distribution. The model's robustness is ensured through hyperparameter tuning via 5-fold cross-validation combined with grid search.
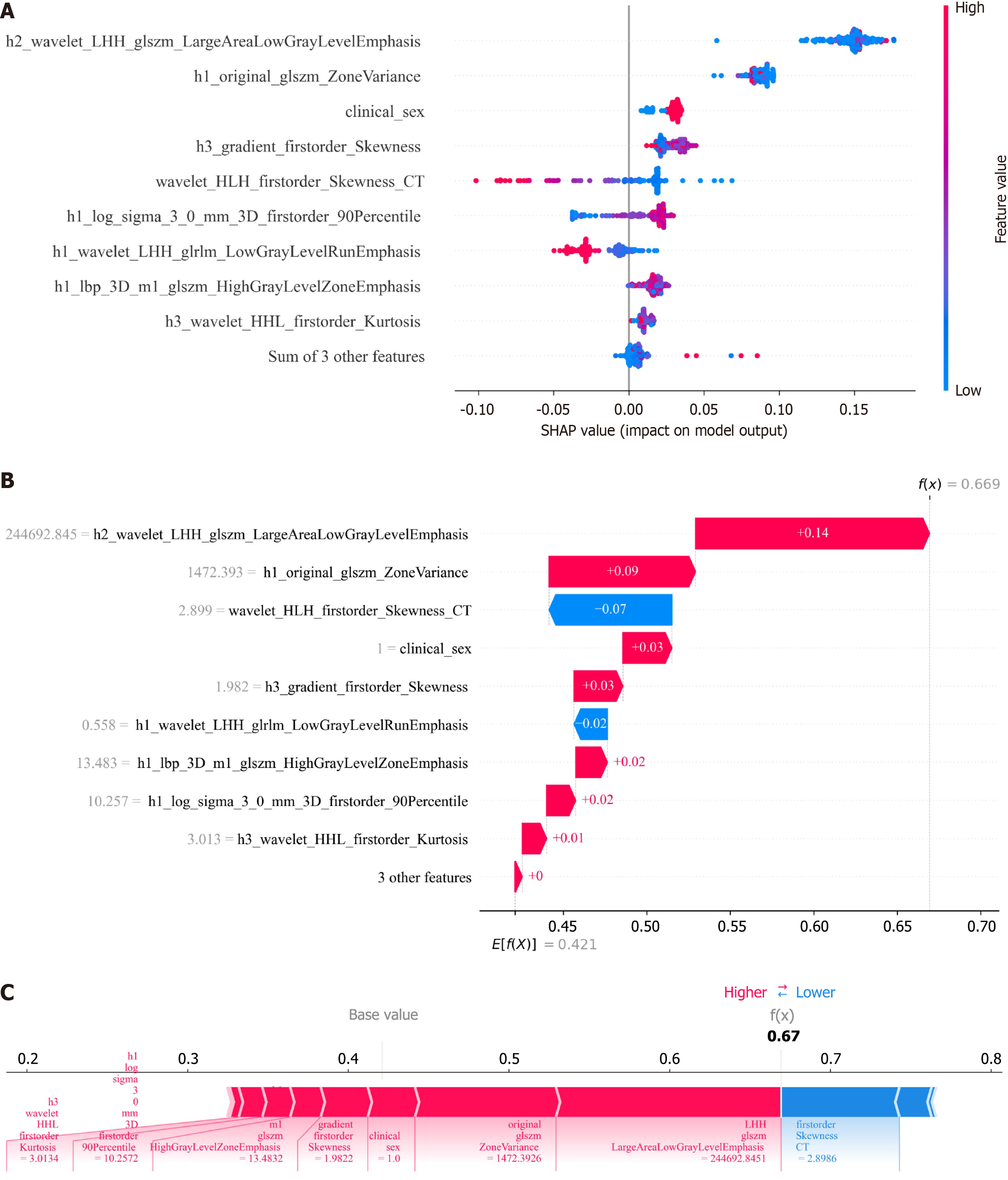
Model explanation: In this study, the SHAP package in Python was used for an in-depth analysis of the fusion model, quantifying the importance of each feature to visualize the model's decision logic. SHAP calculates the importance value for each feature and intuitively presents its impact on the model's output in numerical form[19]. Specifically, features with higher average absolute SHAP values play a more crucial role in the model's prediction process. Additionally, using the SHAP summary plot, the degree of feature impact can be presented visually, helping researchers quickly understand the relationship between features and prediction results, providing scientific evidence for model optimization and feature selection.
This study conducted data processing and model construction in a Python 3.7.12 environment: Statistical analysis was performed using Statsmodels 0.13.2, while machine learning algorithms were implemented with Scikit-learn 1.0.2. Continuous variables underwent intergroup comparisons via t-tests or Mann-Whitney U tests based on normality. Model diagnostic performance was evaluated using receiver operating characteristic curves, with clinical utility validated through decision curve analysis. For the model with optimal performance, Pearson correlation analysis was performed in R software (version 4.2.2, http://www.Rproject.org) to investigate the association among the selected features and MVI, Ki67 expression, GPC-3 expression, and pathological grade in HCC.
The patients' baseline clinical characteristics are shown in Table 2. Training set analysis revealed significant differences in AFP levels (P = 0.012) and gender (P = 0.016) between the group with early HCC recurrence and the group without early recurrence. No statistically significant differences were observed for other clinical indicators between the two groups (P > 0.05). Therefore, AFP and sex were selected for clinical model building.
| Clinical feature | Training set (n = 133) | Validation set (n = 58) | ||||
| No early recurrence (n = 77) | Early recurrence (n = 56) | P value | No early recurrence (n = 35) | Early recurrence (n = 23) | P value | |
| Age (year) | 54.49 ± 10.24 | 51.04 ± 10.88 | 0.063 | 50.46 ± 10.31 | 50.17 ± 9.73 | 0.917 |
| BMI (kg/m2) | 22.26 ± 3.46 | 22.00 ± 2.54 | 0.632 | 21.71 ± 3.41 | 22.80 ± 2.29 | 0.183 |
| HBsAg (IU/mL) | 967.49 ± 869.13 | 1019.11 ± 726.57 | 0.496 | 1141.26 ± 1046.98 | 1141.35 ± 762.88 | 0.893 |
| AFP (ng/mL) | 369.27 ± 494.96 | 539.03 ± 519.80 | 0.012 | 523.78 ± 532.39 | 473.20 ± 539.57 | 0.975 |
| ALB (g/L) | 39.35 ± 5.21 | 39.18 ± 4.60 | 0.842 | 41.55 ± 7.36 | 38.83 ± 4.59 | 0.069 |
| AST (IU/L) | 82.31 ± 135.98 | 51.94 ± 27.49 | 0.758 | 64.54 ± 69.10 | 68.47 ± 71.76 | 0.169 |
| ALT (IU/L) | 75.34 ± 118.98 | 50.77 ± 37.14 | 0.531 | 62.21 ± 77.61 | 74.23 ± 98.27 | 0.474 |
| Sex, n (%) | 0.016 | 0.08 | ||||
| Female | 17 (22.08) | 3 (5.36) | 9 (25.71) | 1 (4.35) | ||
| Male | 60 (77.92) | 53 (94.64) | 26 (74.29) | 22 (95.65) | ||
| Drinking, n (%) | 0.173 | 1 | ||||
| No | 54 (70.13) | 32 (57.14) | 24 (68.57) | 15 (65.22) | ||
| Yes | 23 (29.87) | 24 (42.86) | 11 (31.43) | 8 (34.78) | ||
In terms of physical characteristics, the average Ki-67 expression index among 191 HCC patients was 0.23 (standard deviation 0.21); the MVI incidence rate was 35.08% (67/191). Among the MVI-positive patients, 37 were in the low-risk group (M1), and 30 were in the high-risk group (M2). The pathological grade distribution was as follows: 13 cases (6.81%) at Edmondson-Steiner grade I, 82 cases (42.93%) at grade II, 73 cases (38.22%) at grade III, and 23 cases (12.04%) at grade IV. GPC-3 expression was positive in 163 cases and negative in 28 cases. Table 2 provides more detailed information about all the patients.
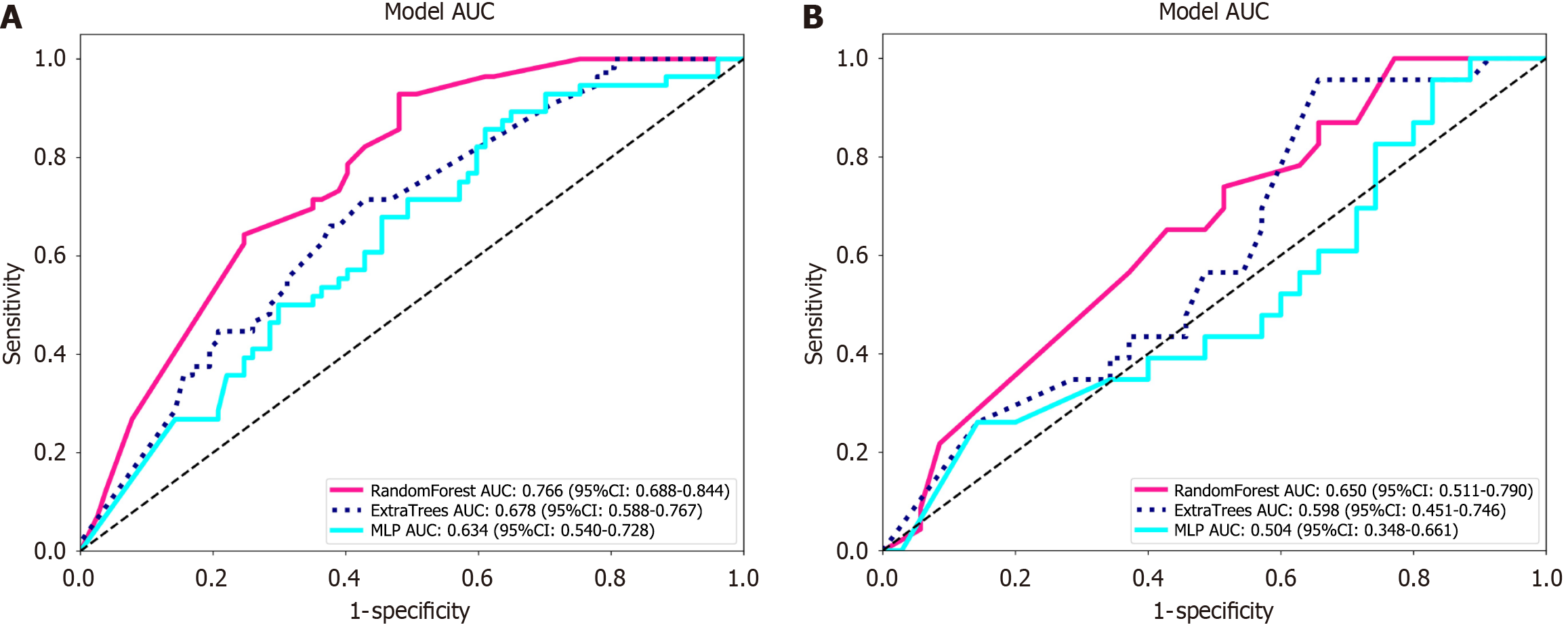
Among the three models constructed with AFP and gender, RandomForest, ExtraTrees, and MLP, the RandomForest model demonstrated the best overall predictive performance, with test set accuracy, area under the curve (AUC), sensitivity, and specificity of 0.517, 0.650, 0.870, and 0.286, respectively. Therefore, it was chosen as the clinical model (Table 3 and Figure 3).
| Dataset | Model name | Accuracy | AUC | 95%CI | Sensitivity | Specificity |
| Training | RandomForest | 0.677 | 0.766 | 0.6884-0.8443 | 0.893 | 0.519 |
| Test | RandomForest | 0.517 | 0.650 | 0.5106-0.7900 | 0.870 | 0.286 |
| Training | ExtraTrees | 0.632 | 0.678 | 0.5881-0.7671 | 0.661 | 0.61 |
| Test | ExtraTrees | 0.534 | 0.598 | 0.4506-0.7457 | 0.696 | 0.429 |
| Training | MLP | 0.579 | 0.634 | 0.5398-0.7281 | 0.839 | 0.39 |
| Test | MLP | 0.466 | 0.504 | 0.3482-0.6605 | 0.913 | 0.171 |
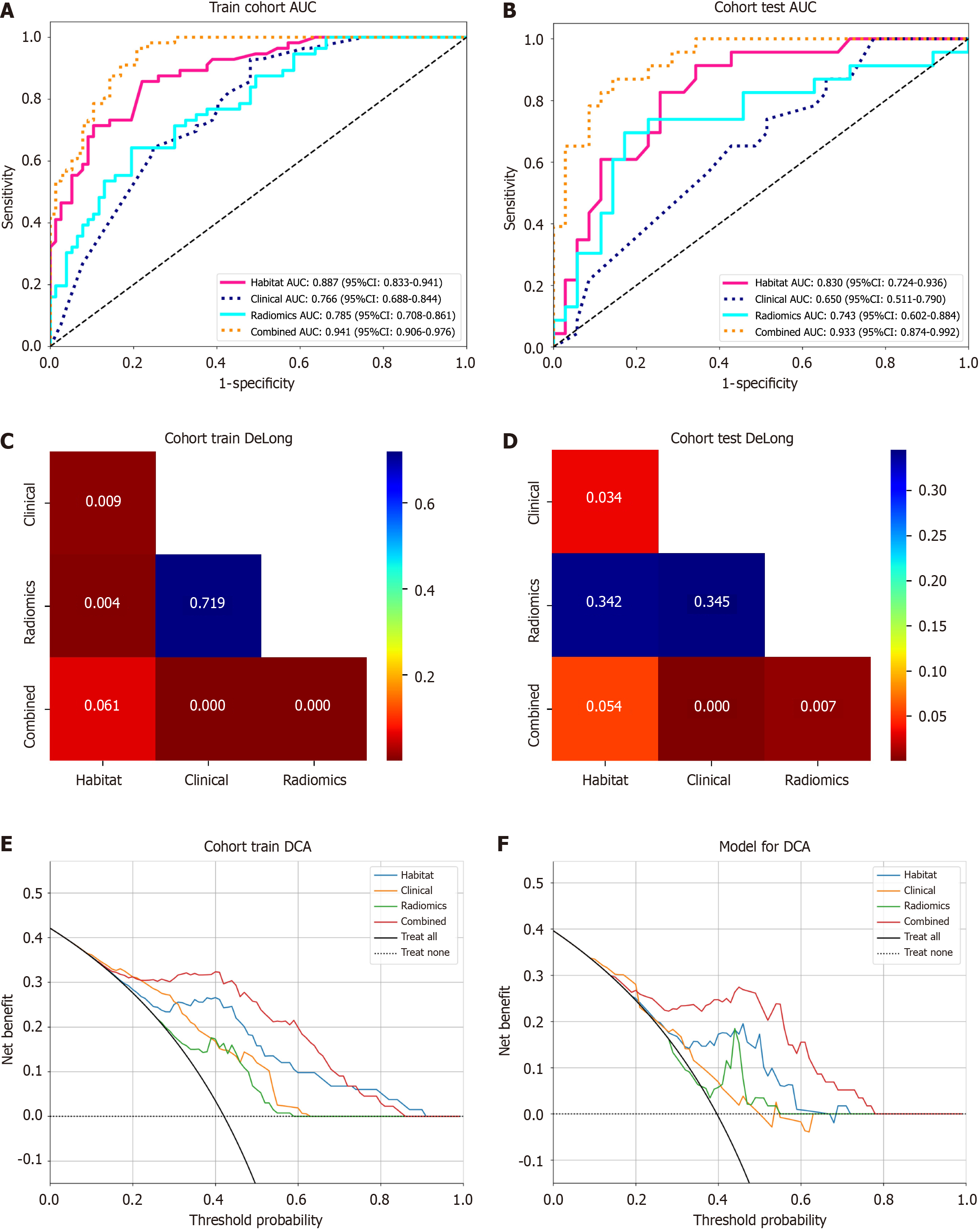
After performing t-test, Mann-Whitney U test, Pearson correlation analysis, and LASSO regression for dimensionality reduction, 9 radiomics features were selected to construct the radiomics prediction model, 10 habitat features were used to build the habitat prediction model, and 12 fusion features were used to construct the fusion model. The predictive metrics of different models were shown in Table 4 and Figure 4, and the results indicated that the fusion model had the best overall predictive performance. The Delong test (Figure 4) showed that in the test set, the fusion model's AUC (0.933) was significantly better than that of the clinical model (0.650) and radiomics model (0.743) (all P < 0.05); compared with the habitat model (0.830), the fusion model's AUC did not show a statistically significant difference in both the training and test sets (P > 0.05), but the decision curve analysis (Figure 4) indicated that the fusion model had higher clinical net benefit.
| Dataset | Model | Accuracy | AUC | 95%CI | Sensitivity | Specificity |
| Training | Clinical | 0.677 | 0.766 | 0.6884-0.8443 | 0.893 | 0.519 |
| Test | Clinical | 0.517 | 0.650 | 0.5106-0.7900 | 0.870 | 0.286 |
| Training | Radiomics | 0.729 | 0.785 | 0.7083-0.8608 | 0.625 | 0.805 |
| Test | Radiomics | 0.759 | 0.743 | 0.6018-0.8840 | 0.652 | 0.829 |
| Training | Habitat | 0.774 | 0.887 | 0.8328-0.9413 | 0.732 | 0.805 |
| Test | Habitat | 0.741 | 0.830 | 0.7239-0.9357 | 0.87 | 0.657 |
| Training | Combined | 0.857 | 0.941 | 0.9058-0.9764 | 0.946 | 0.792 |
| Test | Combined | 0.845 | 0.933 | 0.8735-0.9923 | 0.826 | 0.857 |
Further analyze the contribution levels and directional effects of the features within the best-performing ensemble model using SHAP values. The results indicated that h2_wavelet_LHH_glszm_LargeAreaLowGrayLevelEmphasis, gender, h3_gradient_firstorder_Skewness, h1_lbp_3D_m1_glszm_HighGrayLevelZoneEmphasis, and h3_wave
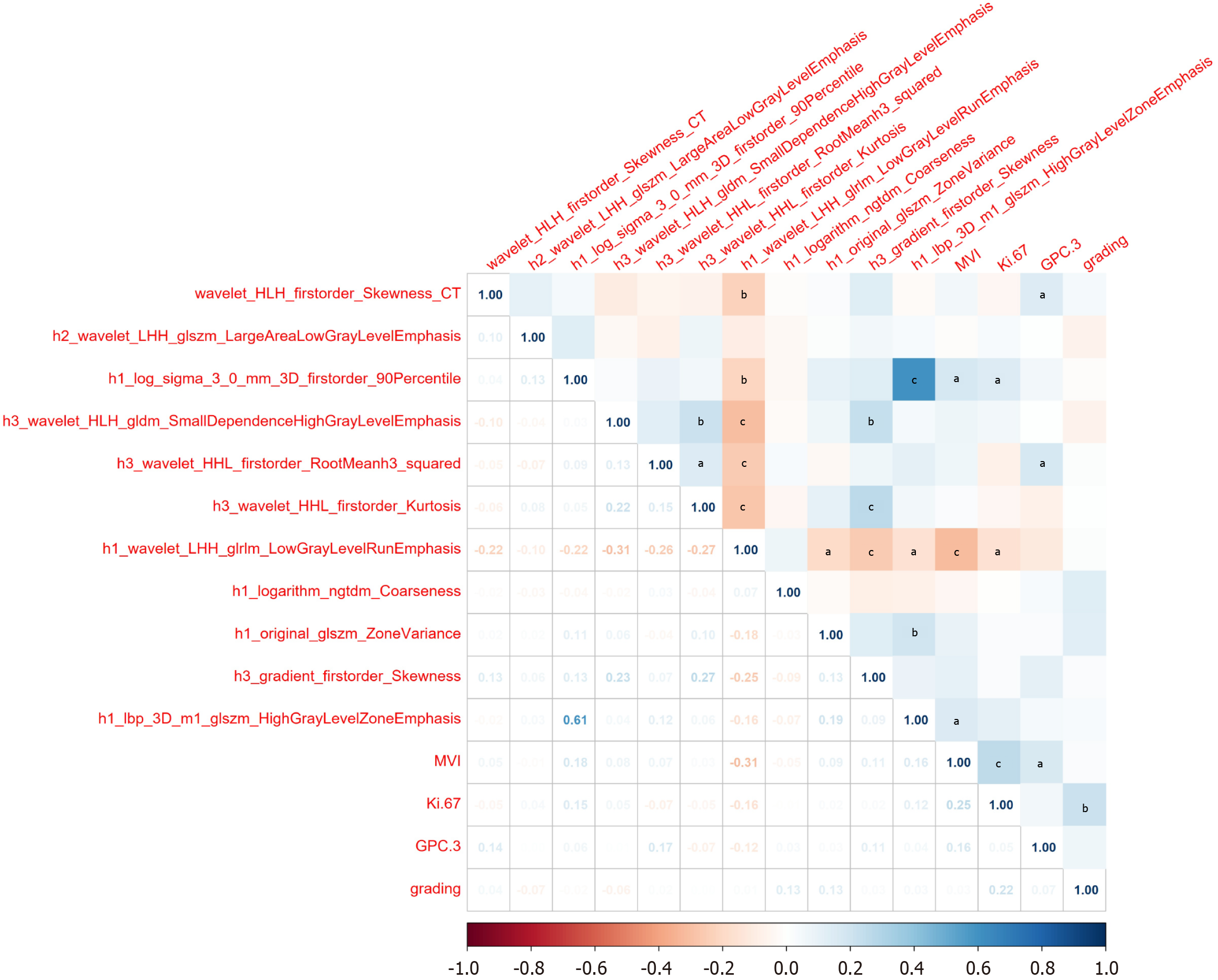
To further explore the potential biological significance of imagingomics/habitat features, we conducted an in-depth analysis of the correlations between the selected features and MVI, Ki-67 exposure, GPC-3 expression, and tumor grade. The results are presented in Table 5 and Figure 6. Correlation analysis showed that wavelet_HLH_firstorder_Skew
| MVI | Ki-67 | GPC-3 | Grading | |||||
| r value1 | P value | r value1 | P value | r value1 | P value | r value1 | P value | |
| wavelet_HLH_firstorder_Skewness_CT | 0.0535 | 0.4625 | -0.0461 | 0.5267 | 0.1425 | 0.0492 | 0.0441 | 0.5451 |
| h2_wavelet_LHH_glszm_LargeAreaLowGrayLevelEmphasis | -0.0148 | 0.8389 | 0.0354 | 0.6271 | 0.0042 | 0.9535 | -0.0664 | 0.3613 |
| h1_log_sigma_3_0_mm_3D_firstorder_90Percentile | 0.1792 | 0.0131 | 0.1462 | 0.0435 | 0.0572 | 0.4319 | -0.0196 | 0.7883 |
| h3_wavelet_HLH_gldm_SmallDependenceHighGrayLevelEmphasis | 0.0758 | 0.2975 | 0.0514 | 0.4805 | 0.0077 | 0.9154 | -0.0605 | 0.4056 |
| h3_wavelet_HHL_firstorder_RootMeanh3_squared | 0.0687 | 0.3452 | -0.0729 | 0.3163 | 0.1703 | 0.0185 | 0.0198 | 0.786 |
| h3_wavelet_HHL_firstorder_Kurtosis | 0.028 | 0.7007 | -0.0464 | 0.5238 | -0.0733 | 0.3133 | 0.001 | 0.9891 |
| h1_wavelet_LHH_glrlm_LowGrayLevelRunEmphasis | -0.3128 | < 0.001 | -0.1574 | 0.0297 | -0.1179 | 0.1042 | 0.0141 | 0.8467 |
| h1_logarithm_ngtdm_Coarseness | -0.0491 | 0.5004 | -0.0097 | 0.8940 | 0.0301 | 0.6797 | 0.1327 | 0.0672 |
| h1_original_glszm_ZoneVariance | 0.0926 | 0.2024 | 0.0223 | 0.7596 | 0.0289 | 0.6918 | 0.1272 | 0.0796 |
| h3_gradient_firstorder_Skewness | 0.1075 | 0.1389 | 0.0235 | 0.7474 | 0.113 | 0.1195 | 0.0261 | 0.7201 |
| h1_lbp_3D_m1_glszm_HighGrayLevelZoneEmphasis | 0.159 | 0.0281 | 0.1189 | 0.1013 | 0.0419 | 0.5651 | 0.0321 | 0.6594 |
In this study, we developed a fusion model based on CT arterial phase images, alongside habitat models, radiomics models, and clinical models. We compared their predictive capabilities for early recurrence of HCC and assessed the relevance of screened features to lesion parameters. A results show that the fusion model demonstrated the highest predictive performance. The results indicated that the fusion model had the best predictive performance. Some of the fused features showed significant correlations with MVI, Ki-67, and GPC-3 expression in HCC. The study results demonstrate strong innovation and clinical value.
The fusion model demonstrated significant advantages in predicting early recurrence of HCC, achieving a verification set AUC of 0.933. This performance markedly surpassed that of the habitat model (AUC = 0.830), traditional radiomics model (AUC = 0.743), and clinical model (AUC = 0.650). This advantage is similar to previous results of fusion models based on CT or MRI[20-23], further confirming the scientific validity of "multi-feature fusion enhancing prediction accuracy". At the same time, decision curve analysis shows that it provides greater net clinical benefit at various thresholds, which together confirm the clinical utility of the fusion model, significantly surpassing other single-type models. This advantage may stem from multi-dimensional support: (1) In terms of feature comprehensiveness, the fusion model does not rely on a single information source but innovatively integrates clinical features (such as gender, AFP levels), traditional radiomics features (such as gray-level co-occurrence matrix, texture statistical parameters), and HI features (such as tumor sub-region blood flow, cell density distribution). Compared to clinical models that rely solely on limited clinical indicators and cannot reflect tumor heterogeneity[24], and traditional radiomics models that depend on manually designed texture features and are susceptible to subjective bias and functional redundancy[24,25], the fusion model, through multi-modal feature complementarity, achieves a "panoramic" depiction of tumor biological behaviors (such as invasiveness, proliferative activity) and individual patient differences, capturing richer recurrence-related information, laying the foundation for high-precision prediction; and (2) The unique value of habitat features adds to the advantage. The habitat model quantifies tumor heterogeneity non-invasively by dividing the tumor into sub-regions with specific physiological states (such as blood flow, necrosis, and edema)[11], and tumor heterogeneity is a core driver of HCC invasion and recurrence[26]. This is also the key reason why the habitat model (AUC = 0.830) outperforms traditional radiomics and clinical models. The fusion model further integrates habitat features with other features, essentially adding clinical risk stratification, microscopic texture features, and other dimensions on top of "heterogeneity assessment", which further amplifies the ability to identify recurrence risk, breaking the limitations of single-model predictions.
SHAP analysis is an important method for understanding the decision-making mechanism of ensemble models. The results show that the five features: H2_wavelet_LHH_glszm_LargeAreaLowGrayLevelEmphasis, gender, h3_gradient_firstorder_Skewness, h1_lbp_3D_m1_glszm_HighGrayLevelZoneEmphasis, and h3_wavelet_HHL_first
The relevance of features to disease mechanisms provides the fusion model with superior biological rationale. Existing research indicates that MVI serves as a crucial pathological biomarker for assessing tumor aggressiveness and migratory potencyl[28], and the expression level of KI-67 is closely associated with tumor cell proliferation activity[31]. Furthermore, HCC patients with higher MVI grades and Ki-67 expression indices have a significantly increased risk of postoperative recurrence[12,32]. This study found that some fusion features are directly associated with key pathological parameters of HCC: For example, h1_log_sigma_3_0_mm_3D_firstorder_90Percentile shows a significant positive correlation with MVI and Ki-67 expression, while h1_wavelet_LHH_glrlm_LowGrayLevelRunEmphasis shows a significant negative correlation with both MVI and Ki-67 expression. This suggests that HCC with high h1_log_sigma_3_0_mm_3D_firstorder_90Percentile values and low h1_wavelet_LHH_glrlm_LowGrayLevelRunEmphasis values have higher aggressiveness and tumor cell active multiplication, making these patients more likely to experience short-term recurrence. This aligns with the impact direction of these characteristics on the model as shown in SHAP analysis. wavelet_HLH_firstorder_Skewness_CT (r = 0.1425, P = 0.0492) and h3_wavelet_HHL_first
In conclusion, the fusion model demonstrates significant advantages in predicting early recurrence of HCC, due to the comprehensiveness of feature dimensions, the uniqueness of habitat features, and the correlation with pathological mechanisms. It not only provides a high-precision prediction tool for clinical use but also offers a new direction for HCC recurrence research and individualized treatment through the association of "imaging features - pathological mechanisms". However, this study also has several limitations: (1) This study is a retrospective, single-center investigation lacking external validation. Furthermore, the lack of statistically significant improvement in the AUC of the fusion model may be attributed to insufficient statistical power due to the small sample size. Future research should expand the sample size and incorporate data from multicenter studies to enhance the generalizability and reliability of the findings; (2) The research process relies on the physician's experience and skill. When manually delineating lesions, subjective bias is unavoidable. In the future, a combined manual and semi-automated delineation approach will be adopted, where physicians only refine blurred boundaries. This aims to minimize subjective bias and enhance the accuracy of lesion delineation; (3) When extracting ITH using HI, this study exclusively utilized arterial phase images from contrast-enhanced CT scans, whose precision remains significantly limited. Future efforts will optimize the HI algorithm by integrating multimodal imaging to enhance ITH extraction accuracy. Large-scale studies will correlate HI habitat maps with histopathological and genomic data to elucidate the biological significance of each subregion, providing theoretical support for understanding tumor characteristics; and (4) This study did not concurrently collect pathological slide data (e.g., tumor necrosis fraction, microvascular density measurements) from enrolled patients. It relied solely on imaging features to infer correlations with pathological indicators, lacking direct validation. Future work will integrate pathological slides to more thoroughly explore the relationship between “imaging features and pathological mechanisms”.
This study, based on CT arterial-phase images and combining clinical features, radiomics features, and HI characteristics, constructed a fusion model that has significant advantages in predicting early recurrence of HCC. Through SHAP analysis and correlation studies, the model's decision logic and the biological significance of key features were revealed. Despite its limitations, this multi-feature fusion framework provides an innovative approach for early recurrence assessment of HCC. It lays the foundation for clinical translation and has the potential to advance the application of imaging in precise diagnosis and treatment of HCC. It is expected to help improve the level of patient prognosis management.
We would like acknowledge all funding organizations and patients of this study.
| 1. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 12107] [Article Influence: 6053.5] [Reference Citation Analysis (6)] |
| 2. | Singal AG, Kanwal F, Llovet JM. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat Rev Clin Oncol. 2023;20:864-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 508] [Article Influence: 169.3] [Reference Citation Analysis (2)] |
| 3. | Calderaro J, Petitprez F, Becht E, Laurent A, Hirsch TZ, Rousseau B, Luciani A, Amaddeo G, Derman J, Charpy C, Zucman-Rossi J, Fridman WH, Sautès-Fridman C. Intra-tumoral tertiary lymphoid structures are associated with a low risk of early recurrence of hepatocellular carcinoma. J Hepatol. 2019;70:58-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 343] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 4. | Ganesan P, Kulik LM. Hepatocellular Carcinoma: New Developments. Clin Liver Dis. 2023;27:85-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 291] [Article Influence: 97.0] [Reference Citation Analysis (1)] |
| 5. | van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 588] [Cited by in RCA: 845] [Article Influence: 140.8] [Reference Citation Analysis (0)] |
| 6. | Zhang YP, Zhang XY, Cheng YT, Li B, Teng XZ, Zhang J, Lam S, Zhou T, Ma ZR, Sheng JB, Tam VCW, Lee SWY, Ge H, Cai J. Artificial intelligence-driven radiomics study in cancer: the role of feature engineering and modeling. Mil Med Res. 2023;10:22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 81] [Reference Citation Analysis (0)] |
| 7. | Wei J, Jiang H, Gu D, Niu M, Fu F, Han Y, Song B, Tian J. Radiomics in liver diseases: Current progress and future opportunities. Liver Int. 2020;40:2050-2063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 8. | Woznicki P, Laqua FC, Al-Haj A, Bley T, Baeßler B. Addressing challenges in radiomics research: systematic review and repository of open-access cancer imaging datasets. Insights Imaging. 2023;14:216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 9. | Li S, Dai Y, Chen J, Yan F, Yang Y. MRI-based habitat imaging in cancer treatment: current technology, applications, and challenges. Cancer Imaging. 2024;24:107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 45] [Reference Citation Analysis (0)] |
| 10. | Aminu M, Yadav D, Hong L, Young E, Edelkamp P Jr, Saad M, Salehjahromi M, Chen P, Sujit SJ, Chen MM, Sabloff B, Gladish G, de Groot PM, Godoy MCB, Cascone T, Vokes NI, Zhang J, Brock KK, Daver N, Woodman SE, Tawbi HA, Sheshadri A, Lee JJ, Jaffray D, D Code Team, Wu CC, Chung C, Wu J. Habitat Imaging Biomarkers for Diagnosis and Prognosis in Cancer Patients Infected with COVID-19. Cancers (Basel). 2022;15:275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Yang Y, Han Y, Zhao S, Xiao G, Guo L, Zhang X, Cui G. Spatial heterogeneity of edema region uncovers survival-relevant habitat of Glioblastoma. Eur J Radiol. 2022;154:110423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 12. | Xu XF, Diao YK, Zeng YY, Li C, Li FW, Sun LY, Wu H, Lin KY, Yao LQ, Wang MD, Zhang CW, Lau WY, Shen F, Yang T. Association of severity in the grading of microvascular invasion with long-term oncological prognosis after liver resection for early-stage hepatocellular carcinoma: a multicenter retrospective cohort study from a hepatitis B virus-endemic area. Int J Surg. 2023;109:841-849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 13. | Zhang L, Xiao Y, Dong M, Li M, Chen H, Wang J. Three-dimensional MR elastography-based stiffness for assessing the status of Ki67 proliferation index and Cytokeratin-19 in hepatocellular carcinoma. Eur Radiol. 2025;35:4722-4735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 14. | Tehrani HA, Zangi M, Fathi M, Vakili K, Hassan M, Rismani E, Hossein-Khannazer N, Vosough M. GPC-3 in hepatocellular carcinoma; A novel biomarker and molecular target. Exp Cell Res. 2025;444:114391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 15. | Fuster-Anglada C, Mauro E, Ferrer-Fàbrega J, Caballol B, Sanduzzi-Zamparelli M, Bruix J, Fuster J, Reig M, Díaz A, Forner A. Histological predictors of aggressive recurrence of hepatocellular carcinoma after liver resection. J Hepatol. 2024;81:995-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 54] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 16. | Zhao YF, Xiong X, Chen K, Tang W, Yang X, Shi ZR. Evaluation of the Therapeutic Effect of Adjuvant Transcatheter Arterial Chemoembolization Based on Ki67 After Hepatocellular Carcinoma Surgery. Front Oncol. 2021;11:605234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 17. | Kaseb AO, Hassan M, Lacin S, Abdel-Wahab R, Amin HM, Shalaby A, Wolff RA, Yao J, Rashid A, Vennapusa B, Feng J, Ohtomo T. Evaluating clinical and prognostic implications of Glypican-3 in hepatocellular carcinoma. Oncotarget. 2016;7:69916-69926. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 18. | Brandão ABM, Rodriguez S, Marroni CA, Junior AMF, Fernandes MV, Mucenic M. Performance of eight predictive models for hepatocellular carcinoma recurrence after liver transplantation: A comparative study. Ann Hepatol. 2024;29:101184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 19. | Kazerouni AS, Hormuth DA 2nd, Davis T, Bloom MJ, Mounho S, Rahman G, Virostko J, Yankeelov TE, Sorace AG. Quantifying Tumor Heterogeneity via MRI Habitats to Characterize Microenvironmental Alterations in HER2+ Breast Cancer. Cancers (Basel). 2022;14:1837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 20. | Huang YH, Shi ZY, Zhu T, Zhou TH, Li Y, Li W, Qiu H, Wang SQ, He LF, Wu ZY, Lin Y, Wang Q, Gu WC, Gu CC, Song XY, Zhou Y, Guan DG, Wang K. Longitudinal MRI-Driven Multi-Modality Approach for Predicting Pathological Complete Response and B Cell Infiltration in Breast Cancer. Adv Sci (Weinh). 2025;12:e2413702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 21. | Sui C, Su Q, Chen K, Tan R, Wang Z, Liu Z, Xu W, Li X. (18)F-FDG PET/CT-based habitat radiomics combining stacking ensemble learning for predicting prognosis in hepatocellular carcinoma: a multi-center study. BMC Cancer. 2024;24:1457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 22. | Liu HF, Wang M, Lu YJ, Wang Q, Lu Y, Xing F, Xing W. CEMRI-Based Quantification of Intratumoral Heterogeneity for Predicting Aggressive Characteristics of Hepatocellular Carcinoma Using Habitat Analysis: Comparison and Combination of Deep Learning. Acad Radiol. 2024;31:2346-2355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 23. | Qin Y, Zhang LG, Zhou X, Song C, Wu Y, Tang M, Ling Z, Wang J, Cai H, Peng Z, Feng ST. Explainable Fusion Model for Predicting Postoperative Early Recurrence in Hepatocellular Carcinoma Using Gadoxetic Acid-Enhanced MRI Habitat Imaging. Acad Radiol. 2025;32:5162-5172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 24. | Huynh BN, Groendahl AR, Tomic O, Liland KH, Knudtsen IS, Hoebers F, van Elmpt W, Malinen E, Dale E, Futsaether CM. Head and neck cancer treatment outcome prediction: a comparison between machine learning with conventional radiomics features and deep learning radiomics. Front Med (Lausanne). 2023;10:1217037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 25. | Meng Y, Yang Y, Hu M, Zhang Z, Zhou X. Artificial intelligence-based radiomics in bone tumors: Technical advances and clinical application. Semin Cancer Biol. 2023;95:75-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 26. | Ma M, Liu R, Wen C, Xu W, Xu Z, Wang S, Wu J, Pan D, Zheng B, Qin G, Chen W. Predicting the molecular subtype of breast cancer and identifying interpretable imaging features using machine learning algorithms. Eur Radiol. 2022;32:1652-1662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 27. | Wei Y, Pei W, Qin Y, Su D, Liao H. Preoperative MR imaging for predicting early recurrence of solitary hepatocellular carcinoma without microvascular invasion. Eur J Radiol. 2021;138:109663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 28. | Zhang ZH, Jiang C, Qiang ZY, Zhou YF, Ji J, Zeng Y, Huang JW. Role of microvascular invasion in early recurrence of hepatocellular carcinoma after liver resection: A literature review. Asian J Surg. 2024;47:2138-2143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 30] [Reference Citation Analysis (0)] |
| 29. | Wu Y, Ye Z, Yang T, Yao S, Chen J, Yin T, Song B. Preoperative prediction of early recurrence in hepatocellular carcinoma using simultaneous multislice diffusion kurtosis imaging. Eur Radiol. 2025;35:7398-7409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 30. | Zhong X, Salahuddin Z, Chen Y, Woodruff HC, Long H, Peng J, Xie X, Lin M, Lambin P. An Interpretable Radiomics Model Based on Two-Dimensional Shear Wave Elastography for Predicting Symptomatic Post-Hepatectomy Liver Failure in Patients with Hepatocellular Carcinoma. Cancers (Basel). 2023;15:5303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 31. | Andrés-Sánchez N, Fisher D, Krasinska L. Physiological functions and roles in cancer of the proliferation marker Ki-67. J Cell Sci. 2022;135:jcs258932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 32. | Xia TY, Zhou ZH, Meng XP, Zha JH, Yu Q, Wang WL, Song Y, Wang YC, Tang TY, Xu J, Zhang T, Long XY, Liang Y, Xiao WB, Ju SH. Predicting Microvascular Invasion in Hepatocellular Carcinoma Using CT-based Radiomics Model. Radiology. 2023;307:e222729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 151] [Reference Citation Analysis (1)] |
| 33. | Kim YH, Kang JS. Expression of glypican-3 in mouse embryo stem cells and its derived hepatic lineage cells treated with diethylnitrosamine in vitro. Asian Pac J Cancer Prev. 2013;14:6341-6345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |