Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2592
Revised: March 17, 2024
Accepted: April 22, 2024
Published online: June 15, 2024
Processing time: 164 Days and 17.5 Hours
Liver cancer (LIHC) is a malignant tumor that occurs in the liver and has a high mortality in cancer. The ING family genes were identified as tumor suppressor genes. Dysregulated expression of these genes can lead to cell cycle arrest, senescence and/or apoptosis. ING family genes are promising targets for anticancer therapy. However, their role in LIHC is still not well understood.
To have a better understanding of the important roles of ING family members in LIHC.
A series of bioinformatics approaches (including gene expression analysis, genetic alteration analysis, survival analysis, immune infiltration analysis, prediction of upstream microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) of ING1, and ING1-related gene functional enrichment analysis) was applied to study the expression profile, clinical relationship, prognostic significance and immune infiltration of ING in LIHC. The relationship between ING family genes expression and tumor associated immune checkpoints was investigated in LIHC. The molecular mechanism of ING1 mediated hepatocarcinogenesis was preliminarily discussed.
mRNA/protein expression of different ING family genes in LIHC was analyzed in different databases, showing that ING family genes were highly expressed in LIHC. In 47 samples from 366 LIHC patients, the ING family genes were altered at a rate of 13%. By comprehensively analyzing the expression, clinical pathological parameters and prognostic value of ING family genes, ING1/5 was identified. ING1/5 was related to poor prognosis of LIHC, suggesting that they may play key roles in LIHC tumorigenesis and progression. One of the target miRNAs of ING1 was identified as hsa-miR-214-3p. Two upstream lncRNAs of hsa-miR-214-3p, U91328.1, and HCG17, were identified. At the same time, we found that the expression of ING family genes was correlated with immune cell infiltration and immune checkpoint genes.
This study lays a foundation for further research on the potential mechanism and clinical value of ING family genes in the treatment and prognosis of LIHC.
Core Tip: Liver cancer (LIHC) is a malignant tumor that occurs in the liver and has a high mortality in cancer. Comprehensive research of expression, mutation, prognosis, and biological mechanisms of ING family genes in LIHC is still lacking. We studied the expression profile, clinical relationship, prognostic potential and immune infiltration of ING family genes in LIHC. The relationship between ING family genes expression and tumor associated immune checkpoints was investigated in LIHC. The molecular mechanism of ING1-mediated hepatocarcinogenesis was preliminarily discussed. Our results highlight the potential mechanism and clinical value of ING family genes in treatment and prognosis of LIHC.
- Citation: Liu SC. Comprehensive analysis of clinical and biological value of ING family genes in liver cancer. World J Gastrointest Oncol 2024; 16(6): 2592-2609
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2592.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2592
Liver cancer (LIHC) is a malignant tumor that occurs in the liver and has a high mortality in cancer[1,2]. At present, the main methods to treat LIHC are radiotherapy, chemotherapy, surgery and liver transplantation[3,4]. Recently, although immunotherapy has made significant progress, such as programmed cell death-1 (PD-1) targeted therapy, its prognosis is poor[5]. Therefore, the discovery of new markers is of importance for individualized therapy and effective improvement of prognosis.
The ING family genes (ING1-5) were identified as tumor suppressor genes[6,7]. Dysregulated expression of these genes can lead to cell cycle arrest, senescence and/or apoptosis[6]. ING2 is highly expressed in colon cancer and induces colon cancer cells invasion through matrix-metalloproteinase-13-dependent pathway[8]. Studies have shown that knockdown of ING2 with small interfering RNA (siRNA) leads to premature aging of normal fibroblasts[9], and to apoptosis or cell-cycle arrest of various adherent cancer cells[10]. These findings indicate that ING2 plays a role in malignant trans
In this study, based on several large public databases, the expression profile, clinical relationship, prognostic significance and immune infiltration of ING family genes in LIHC were explored. The association of ING family genes expression with tumor associated immune checkpoints was investigated in LIHC. The molecular mechanism of ING1 mediated hepatocarcinogenesis was preliminarily discussed. Our results highlight the potential mechanism and clinical value of ING family genes in the treatment and prognosis of LIHC.
Using the Gene_DE module of Tumor Immune Estimation Resource 2.0 (TIMER2)[12], the expression of ING family genes in different cancers in The Cancer Genome Atlas (TCGA) project was analyzed. TIMER2 is freely available at http://timer.cistrome.org/. The University of Alabama at Birmingham Cancer (UALCAN) web-portal[13] was applied to study the expression of ING family genes in normal, primary tumor based on clinical pathological parameters, including sample types, individual cancer stages, race, sex, body weight, age, tumor histology, nodal metastasis status, TP53 mutation status and tumor grade. UALCAN is freely available at https://ualcan.path.uab.edu/. The protein expression data of the Clinical proteomic tumor analysis consortium (CPTAC) was applied to study the expression of ING proteins in normal and primary tumors tissues.
GEPIA2[14] was applied to acquire significant maps of overall survival (OS) and disease-free survival (DFS) for ING family genes. Patients with LIHC were divided into low-risk and high-risk subgroups based on low (50%) and high (50%) threshold values. The Kaplan-Meier curve was obtained by GEPIA2 survival analysis module. GEPIA2 is freely available at http://gepia2.cancer-pku.cn/.
The cBio cancer genomics portal (cBioPortal) is a publicly database that provides resources for analyzing, exploring and visualizing genomics data of cancer[15]. Based on cBioPortal, the LIHC (TCGA, Firehose Legacy) dataset containing 366 samples was selected to explore the ING family genes, and their genetic changes were evaluated. cBioPortal is available at https://www.cbioportal.org/.
The Immune-Gene module of TIMER2[12] was applied to visualize the relationship between the expression of ING family genes and the level of immune infiltration in LIHC tissues. The relationship between ING family genes expression and immune checkpoints and immune cell chemotaxis was evaluated.
The ENCORI/starBase database[16] is a comprehensive database specifically designed for RNA interactions, which can be applied to predict potential microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) that may bind to ING1 and corresponding miRNAs. The upstream binding miRNAs identification of ING1 was based on these principles: Present in at least five databases including microT, miRanda, miRmap, PicTar, TargetScan, PITA, and RNA22. Co-expression analysis of miRNA-target and RNA-RNA was performed using the Pan-Cancer module. ENCORI is available at https://rnasysu.com/encori/.
The STRING database[17] was searched with ING1 and Homo sapiens (main parameters: “experiments”, “no more than 50 interactors” in first shell, “evidence”). The experimentally validated ING1 binding proteins were obtained through the above search. Based on the dataset of LIHC tissues in TCGA, the top 100 ING1-related target genes were obtained using the similarity gene detection module of GEPIA2. Using the GEPIA2[14] correlation analysis module, pairwise gene correlation analysis between ING1 and the selected genes was performed. Functional analysis was performed through DAVID[18] to determine the biological function of genes. The P value was adjusted using the Benjamini-Hochberg method, and P < 0.05 was the critical standard. STRING is available at https://string-db.org/.
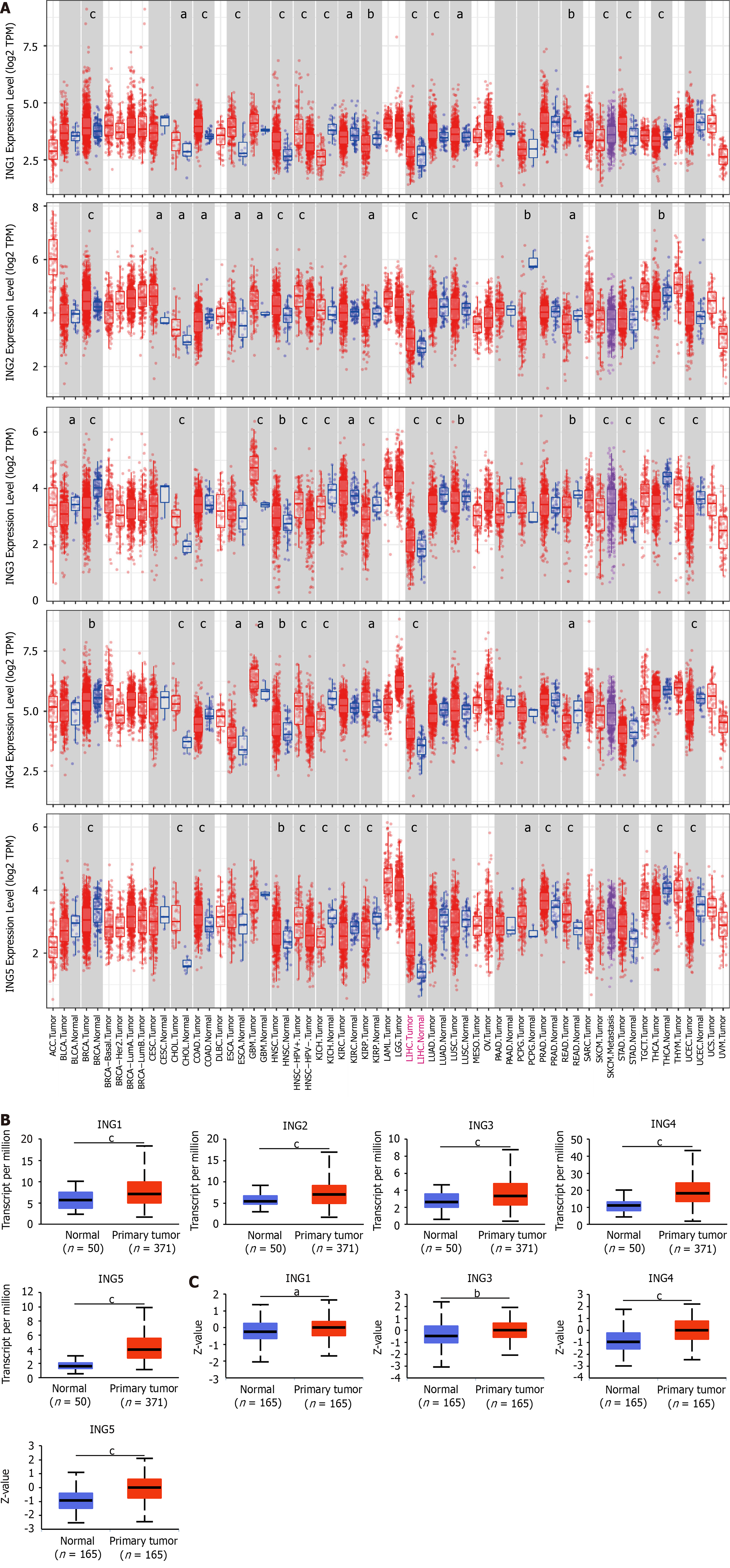
To study the expression of ING family genes in LIHC, based on multiple databases, mRNA and protein expression was studied. Analysis by TIMER2[12] showed that mRNA expression of the ING family genes was significantly upregulated in LIHC tissues (Figure 1A). ING family genes were also significantly upregulated in other cancers, such as gallbladder cancer, head and neck cancer (Figure 1A). The transcriptional level of ING family genes was further studied using the UALCAN database[13], and similar results were obtained (Figure 1B), which also showed that mRNA expressions of ING family genes was significantly upregulated in LIHC tissues. The gene expression at the protein level is closer to the original manifestation of the disease. Therefore, the protein expression data of LIHC were studied using CPTAC (Figure 1C), which showed that protein expression of ING1/3/4/5 was significantly upregulated in LIHC tissues. This further verified the reliability of these results.
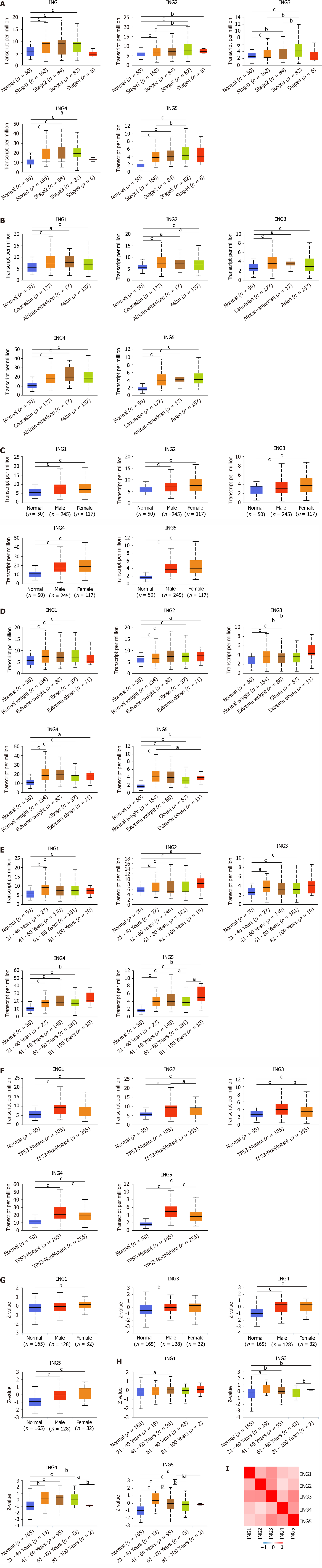
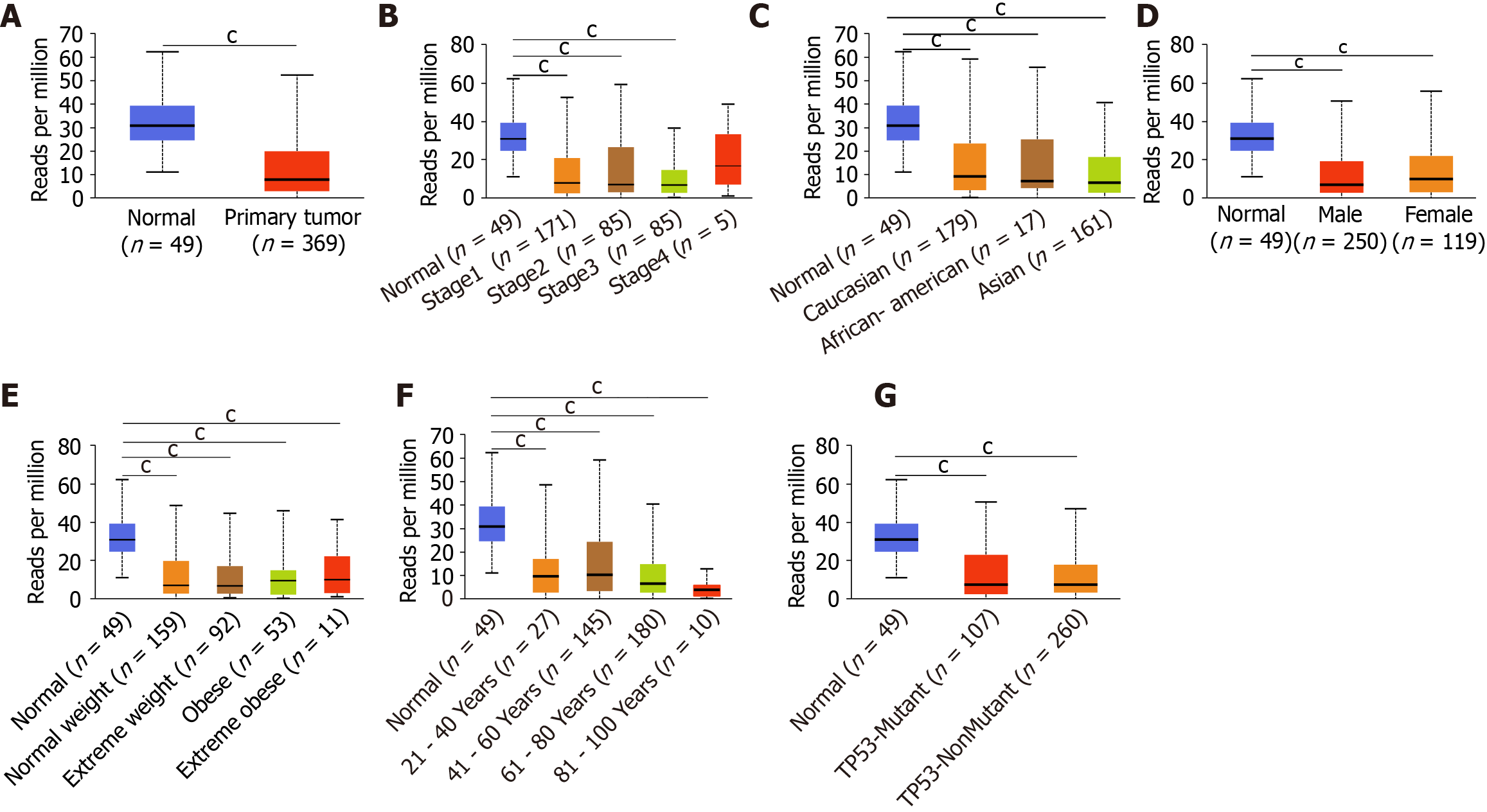
After comprehensive research of the expression patterns of various ING family genes, we further studied the correlation between mRNA expression of ING family genes and clinical pathologic characteristics such as sex, age, individual cancer stage, race, body weight, and TP53 mutation in LIHC patients. There was a significant difference in the expression of ING2/3/5 between stages 1 and 3, and a significant difference in expression of ING3 between stages 2 and 3 (Figure 2A). Between normal tissues and different pathological stages of LIHC, there were significant differences in the expression of ING family genes. There was no significant difference in the expression of ING family members among different races in LIHC (Figure 2B). There was no significant sex difference in expression of ING family members in patients with LIHC (Figure 2C). There was a significant difference in the expression of ING5 between normal weight and obese patients with LIHC (Figure 2D). There was a significant difference in the expression of ING5 between patients aged 41-60 and 61-80 years, and between ages 61-80 and 81-100 years (Figure 2E). The expression of ING2/3/4/5 was significantly different between patients with nonmutated and mutated TP53 (Figure 2F).
The protein expression data of LIHC from the CPTAC database were used to analyze the relationship between the expression of ING family members and the sex and age of patients (Figure 2G and H). There was no significant sex difference in the protein expression of ING family members in LIHC patients. There was a significant difference in the expression of ING5 between patients aged 21-40 and 41-60 years, and between 21-40 and 81-100 years. Expression of ING5 also differed significantly between patients aged 21-40 and 61-80 years. This study also evaluated the correlation between different ING family genes by analyzing their mRNA expression, showing that there was a significant positive correlation among all genes (Figure 2I).
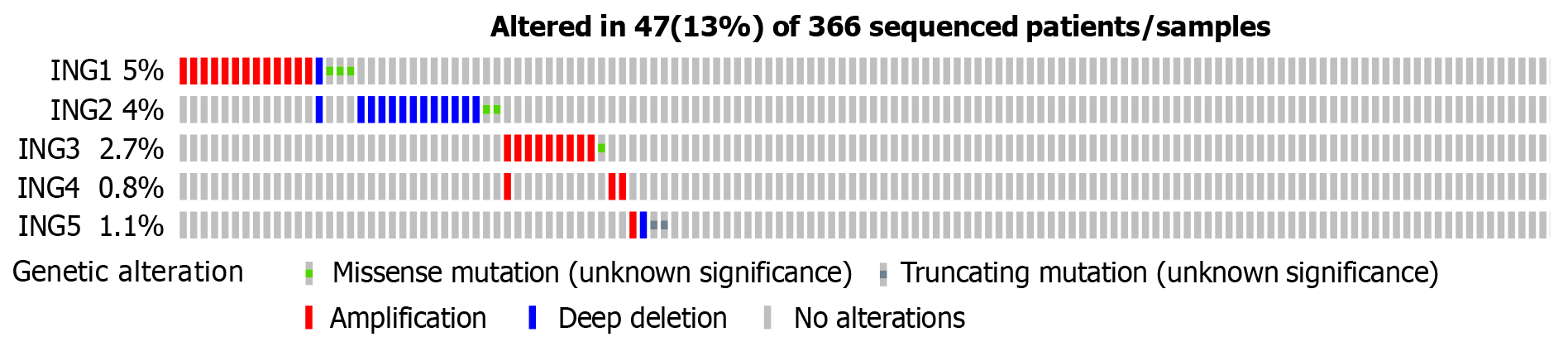
The genetic changes in the ING family genes of LIHC patients were studied using the cBioPortal website. ING family genes were altered in 47 samples from 366 patients with LIHC, accounting for a 13% alteration rate (Figure 3). According to TCGA data, the genetic change percentages of ING1-5 in LIHC were 5%, 4%, 2.7%, 0.8%, and 1.1%, respectively (Figure 3).
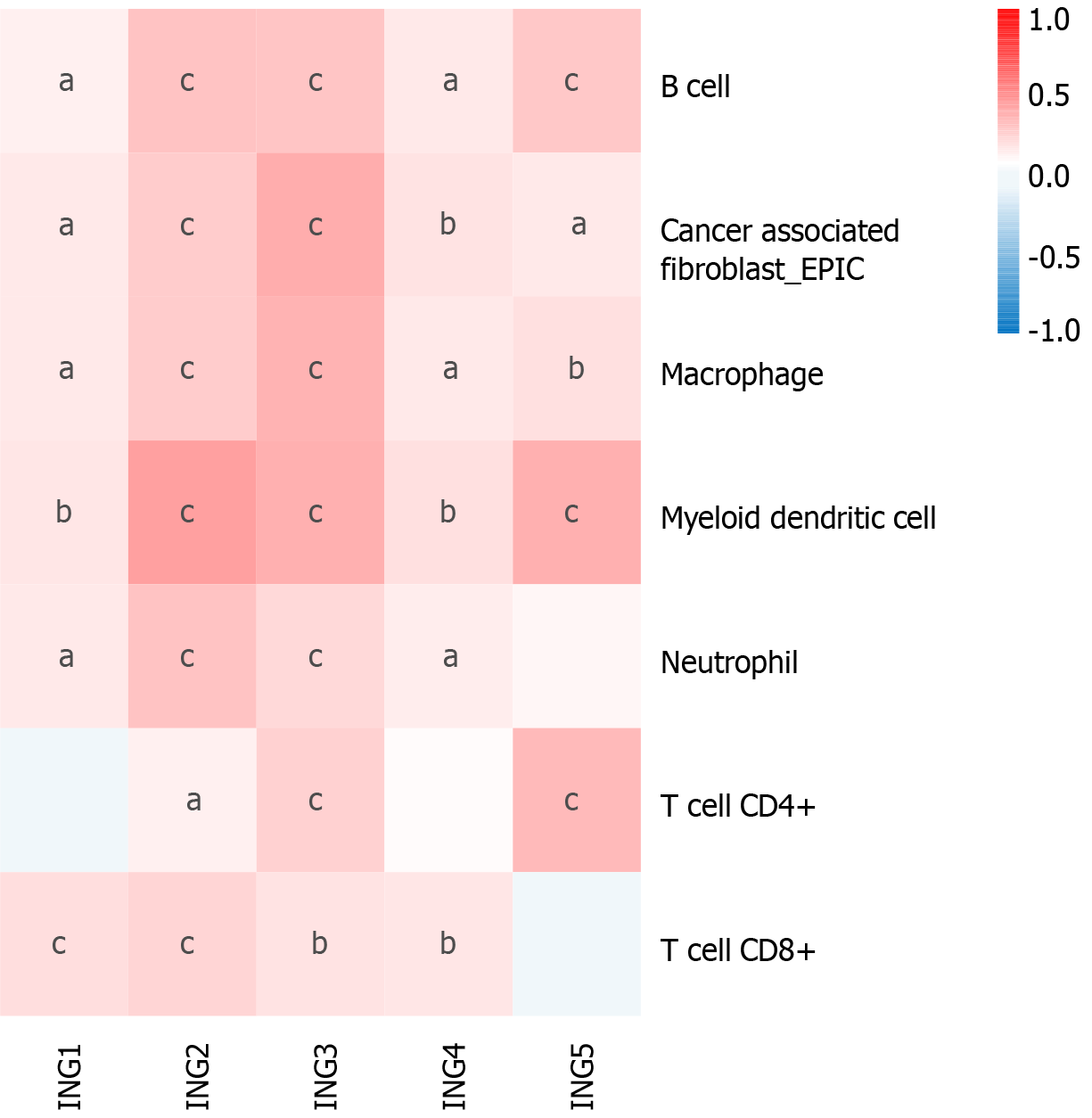
Emerging evidence supports the correlation between immune cell levels and the occurrence and progression of multiple types of tumors[19,20]. We explored the relationship between different ING members and immune cell infiltration levels in LIHC using TIMER2, which will contribute to the monitoring of immune therapy response in LIHC and the exploration of immune infiltration mechanisms. The expression of ING1-4 was significantly positively related to neutrophils and CD8+ T cells (Figure 4). Besides, the expression of ING1-5 was significantly positively related to cancer-associated fibroblasts, B cells, macrophages and myeloid dendritic cells. In addition, the expression of ING2/3/5 was significantly positively related to CD4+ T cells. Supplementary Figure 1 shows the results in detail.
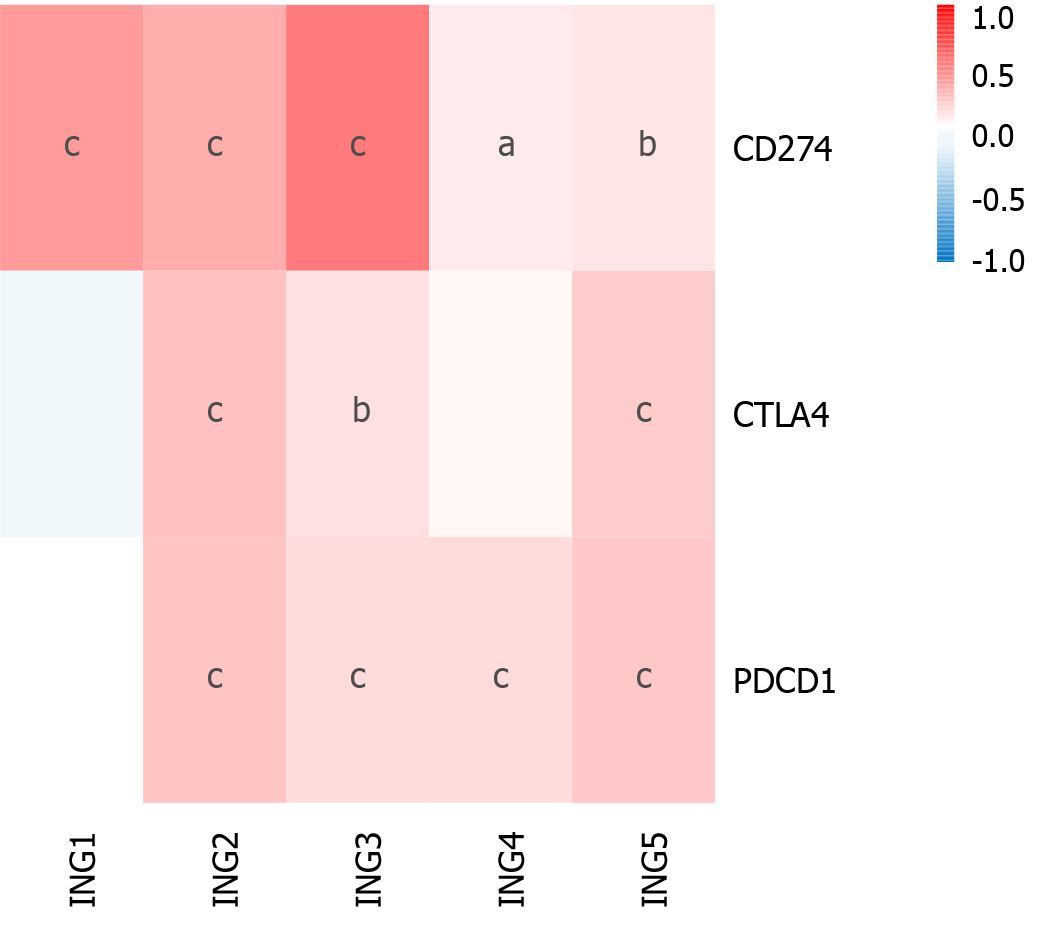
Immunotherapy based on cytotoxic T lymphocyte-associated antigen (CTLA)-4 and PD-1/PD ligand (PD-L)1 has emerged as a new pillar of LIHC treatment[21,22]. We determined the effect of ING gene family expression on LIHC immunotherapy (Figure 5). Expression of ING2/3/5 in patients with LIHC was significantly positively related to CTLA-4, PD-1, and PD-L1. Expression of ING4 in LIHC was significantly positively related to PD-L1 and PD-1. Expression of ING1 was significantly positively related to PD-L1. These results suggest that positive expression of ING family genes may be a better predictor of immunotherapy response than negative expression. Supplementary Figure 2 shows the results in detail.
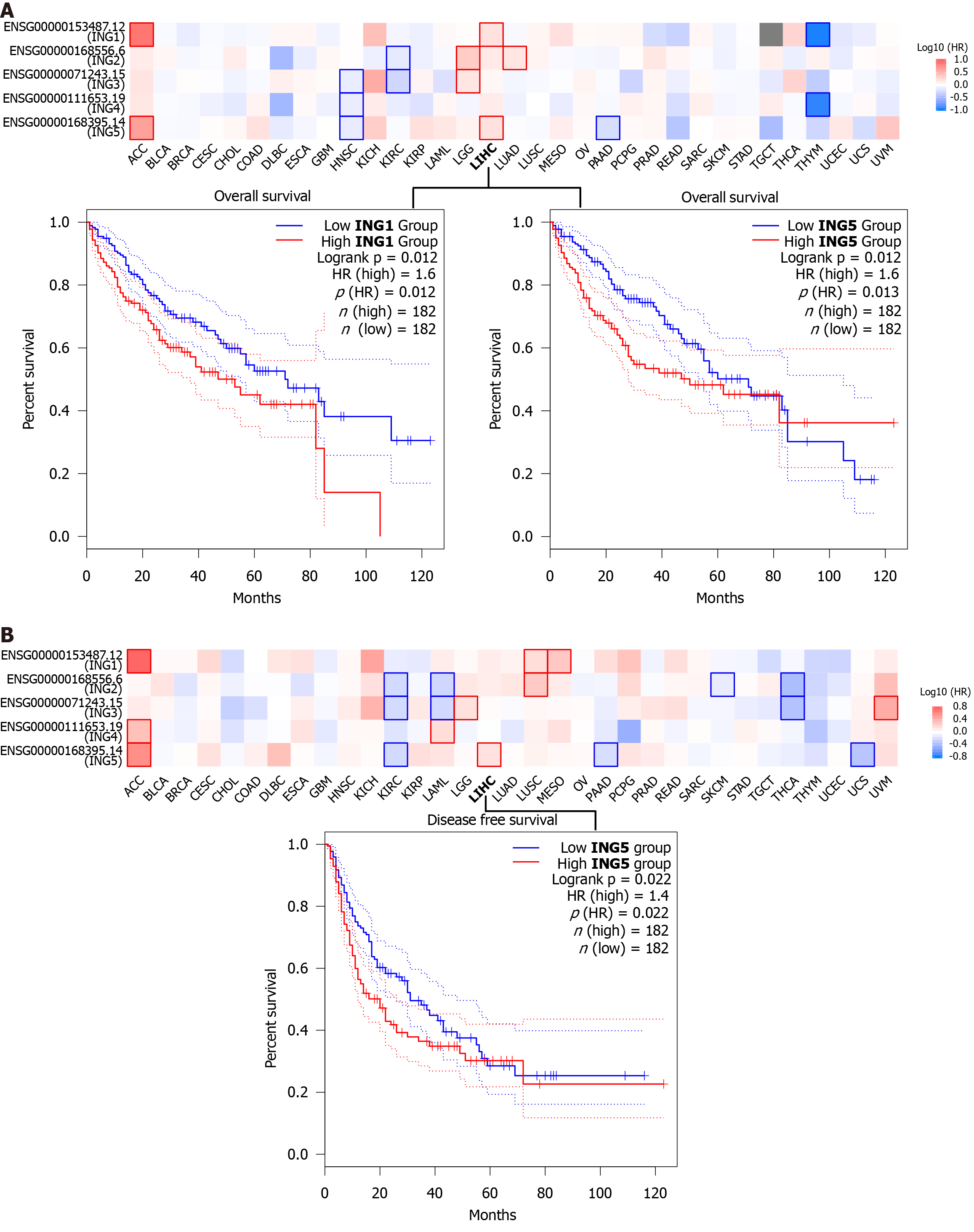
To investigate whether expression of ING family genes was related to prognosis of LIHC, we explored the prognostic potential of ING family genes in cancer using GEPIA2 tool. We were using two prognostic indicators, OS and DFS, to evaluate the prognostic potential of ING family genes. In OS analysis, high expression of ING1/5 indicated poorer survival (Figure 6A). DFS analysis showed that high expression of ING5 was related to poor prognosis (Figure 6B). The remaining ING family members were unrelated to survival time. Therefore, mRNA expression of ING1/5 was significantly related to prognosis of LIHC and could serve as a useful predictive biomarker. Finally, based on comprehensive analysis of the expression, clinicopathological parameters, and prognostic potential of ING family genes, we identified that ING1/5 may play important roles in the occurrence and progression of LIHC.
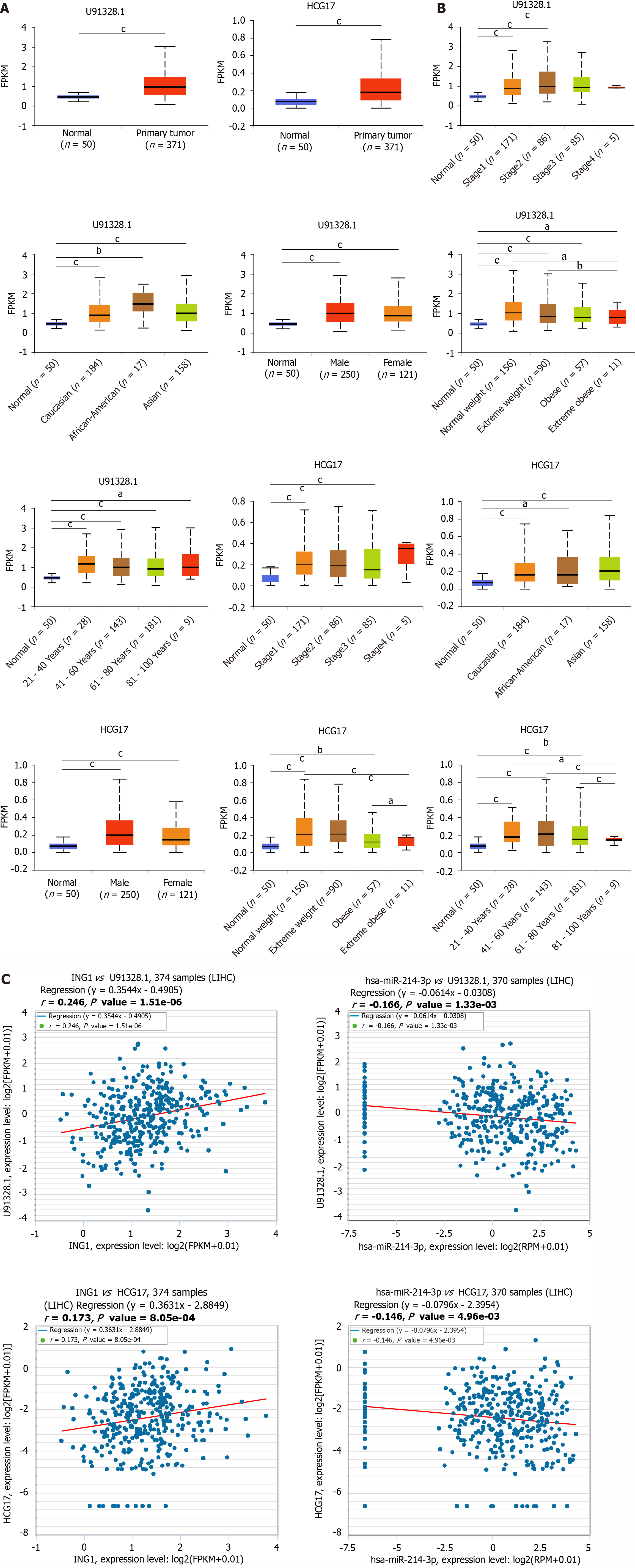
ncRNAs refer to RNA that do not translate to proteins in the transcriptome but regulate gene expression at diversified levels, such as transcription, chromatin structure editing and RNA splicing[23]. To study whether ING1 is regulated by ncRNAs, we used the ENCORI database to predict upstream miRNAs that may bind to ING1. Finally, three miRNAs were identified (hsa-miR-193a-3p, hsa-miR-193b-3p, and hsa-miR-214-3p). The results showed that hsa-miR-214-3p negatively regulated ING1 expression with r and P value of -0.143 and 5.80 × 10-3, respectively. The overall idea of miRNA and mRNA binding analysis was to find target genes and target miRNAs according to the negative correlation between miRNA expression and target gene expression in view of the targeting relationship between miRNA and mRNA[24]. hsa-miR-214-3p was identified as an upstream miRNA that might bind to ING1. We further explored whether hsa-miR-214-3p was involved in the regulation of ING1 expression in LIHC. hsa-miR-214-3p was significantly downregulated in LIHC with P = 2.11 × 10-15 (Figure 7A). We further explored the correlation between hsa-miR-214-3p miRNA expression and clinical pathological characteristics of LIHC. Expression of hsa-miR-214-3p in different pathological stages of LIHC was lower than that in normal tissues, and the same results were shown for other clinicopathological parameters (Figure 7B-G). These results indicated that the regulation of ING1 expression by hsa-miR-214-3p plays a function in the tumorigenesis of LIHC.
Using the ENCORI database, the upstream lncRNAs of hsa-miR-214-3p were explored. The differential expression of these lncRNAs in cancer and normal tissues was evaluated using the TCGA database. Only two lncRNAs were finally identified, namely U91328.1 and HCG17. Compared with normal tissue samples, their expression in LIHC was significantly upregulated (Figure 8A). The analysis showed that expression of U91328.1 and HCG17 in normal tissues was lower than that in LIHC tissues at different pathological stages (Figure 8B), and the similar results were found for other clinicopathological parameters (Figure 8B). According to the theory of competing endogenous RNA (ceRNA), lncRNAs can regulate the transcription of target genes by competing with shared miRNAs as ceRNAs[24,25]. Therefore, expression of lncRNA and mRNA must be positively correlated, while expression of miRNA and lncRNA must be negatively correlated. Subsequently, the ENCORI database was used to explore pairwise correlations between mRNAs, lncRNAs and miRNAs to determine collinearity (Figure 8C). Finally, U91328.1 and HCG17 were identified as direct targets of hsa-miR-214-3p.
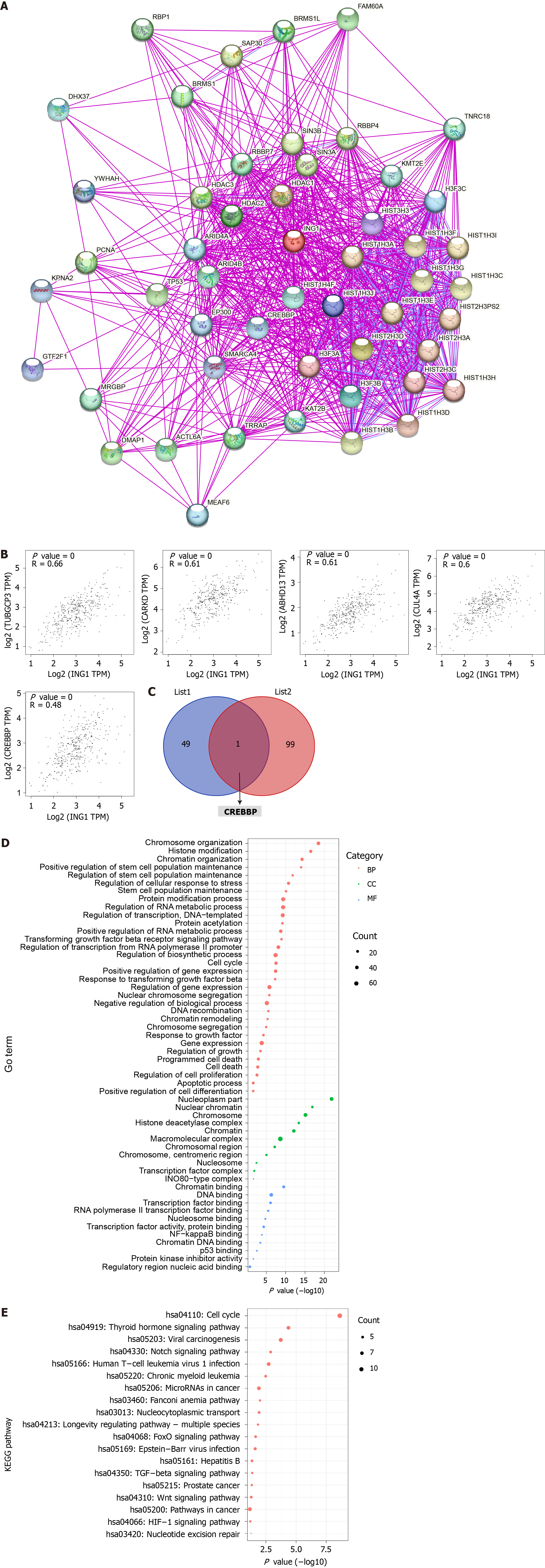
To further explore the molecular mechanism of ING1 in the occurrence of LIHC, we screened for targeted ING1 binding proteins and ING1 expression related genes, and conducted a series of functional analyses. Fifty ING1 binding proteins were obtained using the STRING database[17]. The interaction networks of these proteins were visualized in Figure 9A. Using GEPIA2[14] combined with TCGA LIHC expression data, the top 100 genes related to ING1 expression were obtained. The results indicated that expression of ING1 was positively correlated with expression of TUBGCP3 (r = 0.66), CARKD (r = 0.61), ABHD13 (r = 0.61), CUL4A (r = 0.60), and CREBBP (r = 0.48) (all P < 0.001, Figure 9B). An intersection of the two obtained gene sets was taken, and a common gene, CREBBP, was found (Figure 9C). Previous studies have confirmed that CREBBP gene is associated with various cancers (e.g., colorectal and gastric cancer)[26-28]. Therefore, we infer that CREBBP plays a functional role in LIHC.
Functional analysis was performed on all genes in these two datasets. The gene annotation results showed that these genes were mainly correlated with chromatin organization (GO:0006325), regulation of stem cell population maintenance (GO:2000036), nuclear factor-kappaB binding (GO:0051059), p53 binding (GO:0002039), chromosome organization (GO:0051276), programmed cell death (GO:0012501), and transcription factor binding (GO:0008134) (Figure 9D). Pathway annotation showed that these genes were mainly concentrated in 19 pathways (Figure 9E), including hepatitis B (hsa05161), Wnt signaling pathway (hsa04310), Notch signaling pathway (hsa04330) and pathways in cancer (hsa05200). These biological processes, molecular functions and pathways all play vital roles in cancers.
LIHC is the leading cause of death from cancer worldwide[29]. At present, the main methods to treat LIHC are radiotherapy, chemotherapy, surgery and liver transplantation[30]. Although immunotherapy has made significant progress, such as PD-1 targeted therapy, prognosis of LIHC remains poor[31]. Therefore, it is urgent to explore the pathogenesis of LIHC and find new therapeutic and prognostic biomarkers. Based on several large public databases, we investigated the expression profile, clinical relationships, prognostic significance, and immune infiltration of ING family genes in LIHC.
mRNA/protein expression of different ING family genes in LIHC was analyzed in different databases. The mRNA expression of ING family genes was significantly upregulated in LIHC tissues and in other cancers, such as gallbladder cancer, head and neck cancer. Similar results were obtained using the UALCAN database to further explore the transcription levels of ING family genes. Analysis of protein expression data showed that the ING1/3/4/5 proteins were significantly upregulated in LIHC tissue, verifying the reliability of the results. After comprehensive analysis of expression patterns of ING family genes, we further studied the correlation between mRNA expression of ING family genes and clinical pathologic characteristics in LIHC. Using the LIHC protein expression data in the CPTAC database, we further analyzed the correlation between expression of ING members and the sex and age of patients. We also evaluated the correlation between different ING family genes by analyzing their mRNA expression. There was significant positive correlation among all genes. There is increasing evidence for the important role of ING family genes in different malignancies[8,10,11]. It is reported that the ING family genes (ING1-5) were identified as tumor suppressor genes[6,7]. Our analysis combined with previous studies suggests that ING1-5 may be biomarker of LIHC.
The genetic alterations of ING family genes in LIHC were explored. The ING family genes were altered in 47 samples from 366 patients with LIHC, accounting for a 13% alteration rate. According to TCGA data, the percentage genetic chan
In recent years, immune checkpoint inhibitors (such as CTLA-4, PD-1, and PD-L1 inhibitors) have shown significant therapeutic effects in the clinical treatment of various solid tumors[22]. However, only a small percentage of cancer patients have shown significant benefits from such drugs[33]. Thus, there is a need to accurately identify patients who could benefit from immune checkpoint inhibitor therapy. To determine the influence of expression of ING family genes on LIHC immunotherapy, we studied the correlation between gene expression and CTLA-4, PD-L1 and PD-1. ING2/3/5 expression was significantly positively correlated with PD-1, PD-L1, and CTLA-4. ING4 expression was significantly positively correlated with PD-1 and PD-L1. ING1 expression was significantly positively correlated with PD-L1. Therefore, these results indicate that positive expression of ING family genes is more predictive of immune therapy response than negative expression. The potential mechanism of the correlation between ING family genes and immune checkpoint inhibitors needs to be further explored.
We conducted a survival analysis to explore the prognostic potential of ING family genes. In OS analysis, high ex
The current literature mainly focuses on the key functions and roles of the regulatory ncRNAs in the occurrence and progression of cancer. To identify the upstream regulatory miRNA of ING1, seven prediction tools were introduced to predict the possible binding miRNA of ING1. Three upstream miRNAs of ING1 were identified. By differential expression and mRNA-miRNA correlation analyses, hsa-miR-214-3p was discovered as the upstream miRNA of ING1 that affected progression of LIHC. Upstream lncRNAs of the hsa-miR-214-3p/ING1 axis were also identified. Through expression and correlation analyses, U91328.1 and HCG17, which were the two most promising upregulated lncRNAs of hsa-miR-214-3p, were identified. Compared with normal tissue samples, their expression was significantly upregulated in LIHC. The analyses results showed that expression of U91328.1 and HCG17 in LIHC at different pathological stages was higher than that in normal tissue samples, and the similar results were found for other clinicopathological parameters. According to the ceRNA theory, expression of lncRNA and mRNA must be positively correlated, while the expression of miRNA and lncRNA must be negatively correlated. Subsequently, the ENCORI database was used to explore pairwise correlations between mRNAs, lncRNAs and miRNAs to determine collinearity. Finally, U91328.1 and HCG17 were identified as direct targets of hsa-miR-214-3p. The ING1/hsa-miR-214-3p/ U91328.1 or HCG17 axis was discovered as a potential regulatory pathway.
This research studied the expression of ING family genes in LIHC, showing that ING members were highly expressed in LIHC, and ING1/5 were related to poor prognosis. An ncRNA-mediated regulatory mechanism of ING1 in occurrence and development of LIHC (ING1/hsa-miR-214-3p/U91328.1 or HCG17) was constructed. We found that the expression of ING family genes was related to immune cell infiltration and to the expression of immune checkpoint genes. Our findings provide new insights into diagnosis, treatment, and prognosis of LIHC.
| 1. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 10854] [Article Influence: 3618.0] [Reference Citation Analysis (2)] |
| 2. | Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, Laversanne M, McGlynn KA, Soerjomataram I. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77:1598-1606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 1385] [Article Influence: 346.3] [Reference Citation Analysis (1)] |
| 3. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 4414] [Article Influence: 882.8] [Reference Citation Analysis (4)] |
| 4. | Maki H, Hasegawa K. Advances in the surgical treatment of liver cancer. Biosci Trends. 2022;16:178-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 77] [Reference Citation Analysis (0)] |
| 5. | Sangro B, Sarobe P, Hervás-Stubbs S, Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18:525-543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 942] [Article Influence: 188.4] [Reference Citation Analysis (0)] |
| 6. | Guérillon C, Bigot N, Pedeux R. The ING tumor suppressor genes: status in human tumors. Cancer Lett. 2014;345:1-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 7. | Jacquet K, Binda O. ING Proteins: Tumour Suppressors or Oncoproteins. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Kumamoto K, Fujita K, Kurotani R, Saito M, Unoki M, Hagiwara N, Shiga H, Bowman ED, Yanaihara N, Okamura S, Nagashima M, Miyamoto K, Takenoshita S, Yokota J, Harris CC. ING2 is upregulated in colon cancer and increases invasion by enhanced MMP13 expression. Int J Cancer. 2009;125:1306-1315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193-3203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 175] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 10. | Unoki M, Kumamoto K, Robles AI, Shen JC, Zheng ZM, Harris CC. A novel ING2 isoform, ING2b, synergizes with ING2a to prevent cell cycle arrest and apoptosis. FEBS Lett. 2008;582:3868-3874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Doyon Y, Cayrou C, Ullah M, Landry AJ, Côté V, Selleck W, Lane WS, Tan S, Yang XJ, Côté J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 539] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 12. | Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509-W514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2451] [Cited by in RCA: 3729] [Article Influence: 621.5] [Reference Citation Analysis (0)] |
| 13. | Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 1627] [Article Influence: 406.8] [Reference Citation Analysis (0)] |
| 14. | Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1991] [Cited by in RCA: 3483] [Article Influence: 497.6] [Reference Citation Analysis (0)] |
| 15. | Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9144] [Cited by in RCA: 13454] [Article Influence: 961.0] [Reference Citation Analysis (0)] |
| 16. | Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92-D97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2607] [Cited by in RCA: 4293] [Article Influence: 330.2] [Reference Citation Analysis (0)] |
| 17. | Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, Bork P, Jensen LJ, von Mering C. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638-D646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1815] [Cited by in RCA: 5063] [Article Influence: 1687.7] [Reference Citation Analysis (0)] |
| 18. | Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216-W221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2064] [Cited by in RCA: 3943] [Article Influence: 985.8] [Reference Citation Analysis (0)] |
| 19. | Zhu H, Wang G, Zhu H, Xu A. ITGA5 is a prognostic biomarker and correlated with immune infiltration in gastrointestinal tumors. BMC Cancer. 2021;21:269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 20. | Want MY, Tsuji T, Singh PK, Thorne JL, Matsuzaki J, Karasik E, Gillard B, Cortes Gomez E, Koya RC, Lugade A, Odunsi K, Battaglia S. WHSC1/NSD2 regulates immune infiltration in prostate cancer. J Immunother Cancer. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 21. | Szeto GL, Finley SD. Integrative Approaches to Cancer Immunotherapy. Trends Cancer. 2019;5:400-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 22. | Dhasmana A, Dhasmana S, Haque S, Cobos E, Yallapu MM, Chauhan SC. Next-generation immune checkpoint inhibitors as promising functional molecules in cancer therapeutics. Cancer Metastasis Rev. 2023;42:597-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 23. | Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J, Xiao YF. Translation of noncoding RNAs and cancer. Cancer Lett. 2021;497:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 24. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5718] [Article Influence: 381.2] [Reference Citation Analysis (0)] |
| 25. | Wang P, Guo Q, Hao Y, Liu Q, Gao Y, Zhi H, Li X, Shang S, Guo S, Zhang Y, Ning S, Li X. LnCeCell: a comprehensive database of predicted lncRNA-associated ceRNA networks at single-cell resolution. Nucleic Acids Res. 2021;49:D125-D133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 26. | Schoof M, Epplen GD, Walter C, Ballast A, Holdhof D, Göbel C, Neyazi S, Varghese J, Albert TK, Kerl K, Schüller U. The tumor suppressor CREBBP and the oncogene MYCN cooperate to induce malignant brain tumors in mice. Oncogenesis. 2023;12:36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 27. | Yang J, Zhao S, Su J, Liu S, Wu Z, Ma W, Tang M, Wu J, Mao E, Han L, Liu M, Zhang J, Cao L, Shao J, Shang Y. Comprehensive genomic profiling reveals prognostic signatures and insights into the molecular landscape of colorectal cancer. Front Oncol. 2023;13:1285508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 28. | Jiang Q, Miao J, Wu F, Li F, Zhang J, Jing X, Cai S, Ma X, Wang X, Zhao L, Huang C. RNF6 promotes gastric cancer progression by regulating CCNA1/CREBBP transcription. Cell Cycle. 2023;22:2018-2037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | Donne R, Lujambio A. The liver cancer immune microenvironment: Therapeutic implications for hepatocellular carcinoma. Hepatology. 2023;77:1773-1796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 431] [Article Influence: 143.7] [Reference Citation Analysis (1)] |
| 30. | Brown ZJ, Tsilimigras DI, Ruff SM, Mohseni A, Kamel IR, Cloyd JM, Pawlik TM. Management of Hepatocellular Carcinoma: A Review. JAMA Surg. 2023;158:410-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 481] [Reference Citation Analysis (1)] |
| 31. | Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, Pikarsky E, Zhu AX, Finn RS. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 1210] [Article Influence: 302.5] [Reference Citation Analysis (3)] |
| 32. | Guo Y, Yang J, Ren K, Tian X, Gao H, Tian X, Zhang X, Kan Q. The Heterogeneity of Immune Cell Infiltration Landscape and Its Immunotherapeutic Implications in Hepatocellular Carcinoma. Front Immunol. 2022;13:861525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 33. | Banday AH, Abdalla M. Immune Checkpoint Inhibitors: Recent Clinical Advances and Future Prospects. Curr Med Chem. 2023;30:3215-3237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/