Published online May 15, 2020. doi: 10.4251/wjgo.v12.i5.526
Peer-review started: January 20, 2020
First decision: March 24, 2020
Revised: March 29, 2020
Accepted: April 15, 2020
Article in press: April 15, 2020
Published online: May 15, 2020
Processing time: 115 Days and 0.7 Hours
The integrin β6 gene, which is expressed in epithelial cancer, plays a pivotal role in various aspects of cancer progression. The present research for integrin β6 regulation mainly focuses on the post-transcription and translation related regulation mechanism and its role in tumorigenesis. The mechanisms of how the integrin β6 gene is regulated transcriptionally, and the promoter and transcription factors responsible for basic transcription of integrin β6 gene remain unknown.
To clone and characterize the integrin β6 promoter.
Software analysis was used to predict the region of integrin β6 promoter. Luciferase reporter plasmids, which contained the integrin β6 promoter, were constructed. Element deletion analysis was performed to identify the location of core promoter and binding sites for transcription factors.
The regulatory elements for the transcription of the integrin β6 gene were located between -286 and -85 and contained binding sites for transcription factors such as STAT3 and Ets-1.
For the first time, we found the region of β6 core promoter and demonstrated the binding sites for transcription factors such as Ets-1 and STAT3, which are important for integrin β6 promoter transcription activity. These findings are important for investigating the mechanism of integrin β6 activation in cancer progression.
Core tip: In this paper, the transcription regulation of integrin β6 is described and the region of core promoter region is identified. Meanwhile, the mechanism of transcription regulation of integrin β6 is explored.
- Citation: Niu W, Bo QY, Niu J, Niu ZC, Peng C, Zou XQ, Zhang ZY. Identification of integrin β6 gene promoter and analysis of its transcription regulation in colon cancer cells. World J Gastrointest Oncol 2020; 12(5): 526-534
- URL: https://www.wjgnet.com/1948-5204/full/v12/i5/526.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i5.526
As a member of the cell adhesion molecular family, integrin αvβ6 is a subtype of integrin, expressed strictly in epithelia, up-regulated in parallel with embryo formation, oncogenesis, and epithelial repair, and rarely expressed in normal tissue[1,2]. Previous studies revealed that integrin αvβ6 played an important role in invasion, proliferation, apoptosis, tumor immunity, and epithelial-mesenchymal transition of malignant tumor cells[3-6]. It is worth noting that the expression of integrin αvβ6 was closely related with clinicopathological features of many carcinomas, as well as patient prognosis[7,8]. Integrin αvβ6 expression depends on integrin β6 expression, because integrin β6 only partners with subunit αv, forming a single heterodimer. Therefore, it is important to explore the mechanisms underlying the regulation of integrin β6 expression.
The expression of integrin β6 is regulated at the level of transcription initiation[9,10]. Although it has been reported that transcription factors, such as Smad3 and AP-1[11,12], regulate the expression of integrin β6, the mechanism and core regulated factors that mediate the transcription of integrin β6 are still unclear. Furthermore, little is known about the regulation of integrin β6 in colon cancer cell lines, in which the expression of integrin β6 is up-regulated abnormally. In the present study, the transcription regulation of integrin β6 was investigated and the region of core promoter region was identified. Meanwhile, the mechanism of transcription regulation of integrin β6 was explored.
Human colon cancer cell lines HT-29 and WiDr were purchased from the American Type Culture Collection. HT-29 and WiDr cells express integrin β6 constantly. The cells were maintained as monolayers in Dulbecco's Modified Eagle Medium containing 10% fetal bovine serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin, at 37°C in a 5% CO2 incubator. Dulbecco's Modified Eagle Medium, fetal bovine serum, penicillin, and streptomycin were all obtained from Hyclone.
Total RNA was isolated from cells by using an RNA Extraction Kit (Invitrogen), according to the manufacturer’s protocol. Total RNA was reverse transcribed using a Fast Quant RT Kit (Boshang Biotech, Shanghai, China) and reverse transcription-polymerase chain reaction (RT-PCR) was performed by using a SuperReal PreMix (Boshang Biotech, Shanghai, China).
Prediction of human integrin β6 promoter region: The 5’-flanking region spanning -3000 to +500 of the human integrin β6 gene was analyzed with FirstEF, MatInspector, and Proscan Prediction software.
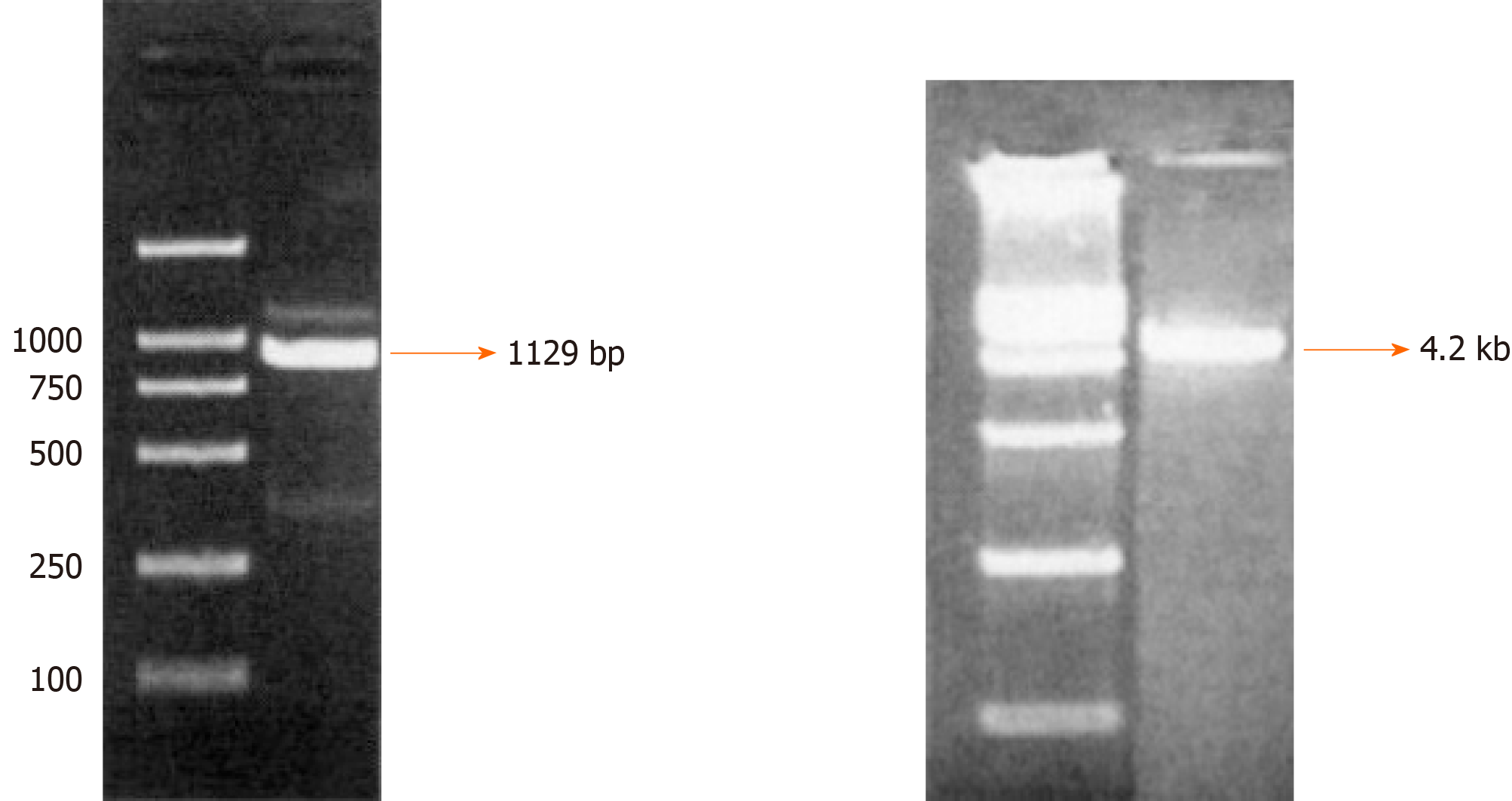
Cloning of integrin β6 promoter and construction and confirmation of pGL4-β6-B1: A 1129-bp DNA fragment, corresponding to the region -921/+208 of integrin β6 (the transcription start was designated as +1), was amplified from DNA obtained from HT-29 and WiDr cells by PCR using the primers sense -921 and antisense +208 listed in Table 1. A series of 5’ deletion elements were divided into -921/+208, -755/+208, -513/+208, -350/+208, and -85/+208, then using PCR products generated the fragments. The primers are listed in Table 1. To obtain the fragment of the integrin β6 promoter, the suitable primers which contain Kpn I and Hind III were used for PCR. Then, β6-B1 was cloned into the multiple cloning site of a pGL4-Basic plasmid to construct pGL4-β6-B1 with the correct sequence. Alkaline denaturation was performed to obtain the correct plasmid from engineering bacteria named JM109 containing pGL4-Basic.All products were digested with Kpn I and Hind III(TAKARA, Dalian, China), and primers were synthesized by Boshang Bio Company, and subcloned into the pGL4-Basic luciferase vector (Tianwei Tec, Beijing, China).
| Construct | Primer | Sequence (5’-3’) |
| pGL4-B1 (-921/+208) | Sense -921 | GGGGTACCCACAGGCATTGACCTG |
| pGL4-B2 (-755/+208) | Sense -755 | GGGGTACTGACACCGATTGTCGAT |
| pGL4-B3 (-513/+208) | Sense -513 | GGGGTAGACTGACTGCCATTGAGGTGAT |
| pGL4-B4 (-350/+208) | Sense -350 | GGGGTCCCCACGGATTCCGTAGACCTC |
| pGL4-B5 (-85/+208) | Sense -85 | GGGGTAGGTACAGGAACCGACCTA |
| Antisense | GGGGTCCCGACATTCGATTGTCGCCTT |
Transfection and dual-luciferase reporter assay: For the analysis of promoter activity, HT-29 and WiDr cells were seeded in 96-well plates. Approximately 100 ng of each plasmid combined with 0.25 ng of pRL-TK was transfected into cells. Then, cells were harvested after 24 h. Firefly luciferase activity was determined using a dual-luciferase reporter assay system (Promega, China) and analyzed using a Lumat luminometer.
The plasmids constructed above and pGL4-Basic (negative control) were co-transfected with pRL-TK into colon cancer cell lines HT-29 and WiDr. The firefly luciferase activity (M1) was normalized with the Renilla luciferase activity of pRL-TK (M2) and the ratio of M1/M2 stands for the relative activity of plasmids.
Fragments of -350/-85, -286/-85, -210/-85, and -156/-85 named pGL4-B6, B7, B8, and B9 were selected to construct an array of truncated promoter plasmids, using pGL4-β6-B1 as a template (Table 2). The plasmids constructed above andpGL4-Basic were transiently co-transfected into HT-29 and WiDr cells. Luciferase reporter gene assay was performed 24 h after transfection.
| Construct | Primer | Sequence (5’-3’) |
| pGL4-B6 (-350/-85) | Sense -350 | GGGGTACCCAAAGGCACTGATTTGC |
| pGL4-B7 (-286/-85) | Sense -286 | GGGGTACTGACAGGCATTGTCGATTGAG |
| pGL4-B8 (-210/-85) | Sense -210 | GGGGTAGACTGACAATTATTGAGGTGAT |
| pGL4-B9 (-156/-85) | Sense -156 | GGGGTCCTTGCGGATTCCGTAGACGTC |
| Antisense | GGGGTCGGGACAACGAATGTCGGCTTC |
To identify regulatory elements involved in basal transcriptional activity and positive regulation of integrin β6, sequence analysis was performed with TRANSFAC-TESS or AliBaba2 analysis software.
Prediction of integrin β6 promoter: Three kinds of software were used to predict the integrin β6 promoter, including FirstEF, Mat Inspector, and Proscan Prediction software. Mat Inspector predicted that the region between -756 and +79 was the main sequence where integrin β6 promoter was located. FirstEF predicted that the region between -560 and +325 was the main sequence, and Proscan Prediction software predicted that the region between -316 and +198 was the main sequence that positively regulates integrin β6 transcription. Since the prediction was based on mathematical statistics and the region of transcription was generally wide, we finally identified that the region between -921 and +198 was the main fragment.
Cloning of integrin β6 promoter and construction of the reporter plasmid pGL4-β6-B1: The successful construction of the reporter plasmid pGL4-β6-B1 ensured the fidelity of amplification and correct luciferase reporter plasmids. The results of electrophoresis showed a fragment of DNA, which were consistent with that of designed target sequences (Figure 1). Alkaline denaturation was performed to obtain the correct plasmid from engineering bacteria named JM109 containing pGL4-Basic (Figure 1).
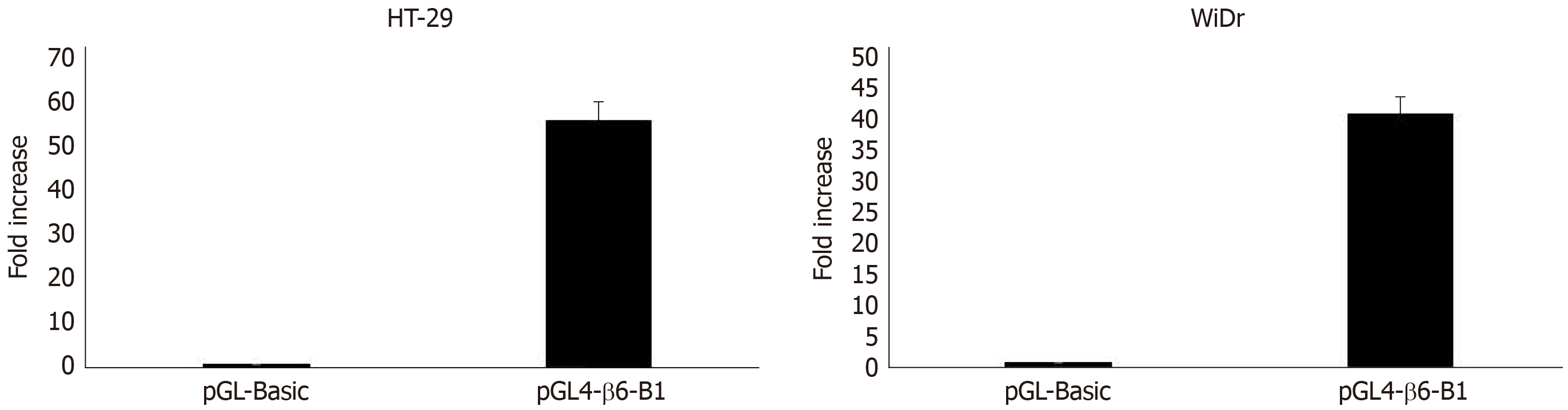
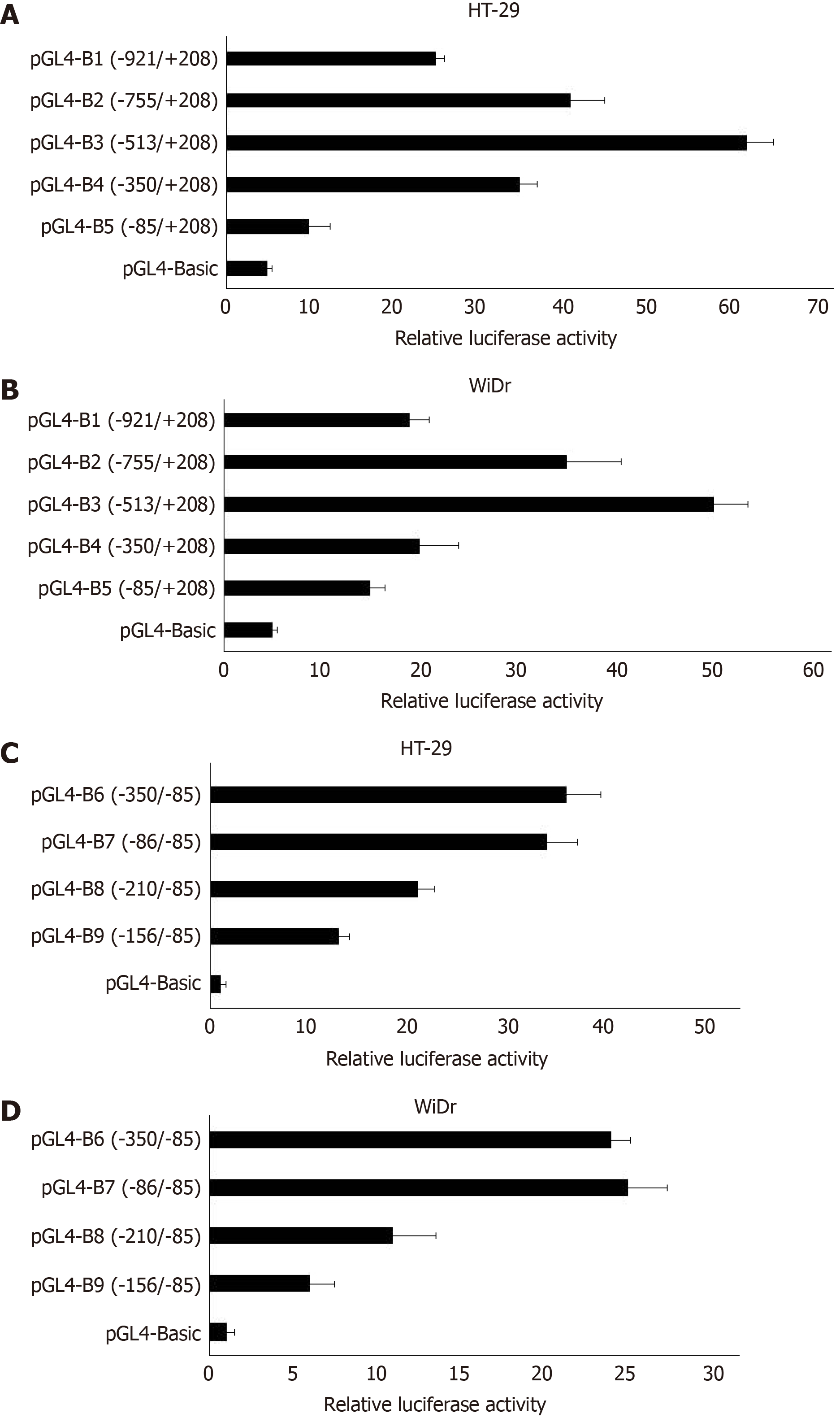
Transfection and dual-luciferase reporter assay: The results of dual-luciferase reporter assays showed that pGL4-β6-B1 could highly express about 56 times of pGL4-Basic in HT-29 cells (Figure 2). In WiDr cells, the luciferase activity of pGL4-β6-B1 was about 41 times of pGL4-Basic (Figure 2). The results showed that pGL4-β6-B1 contains the maximum promoter activity.
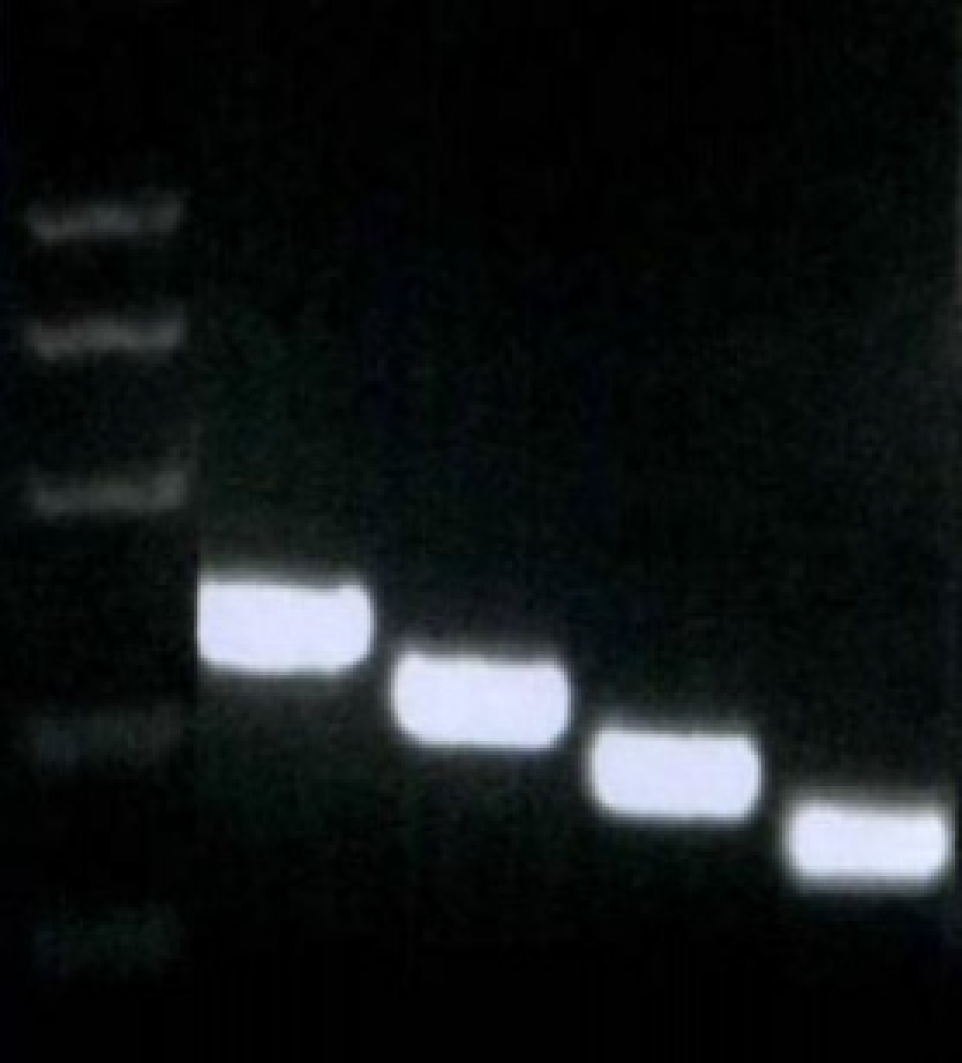
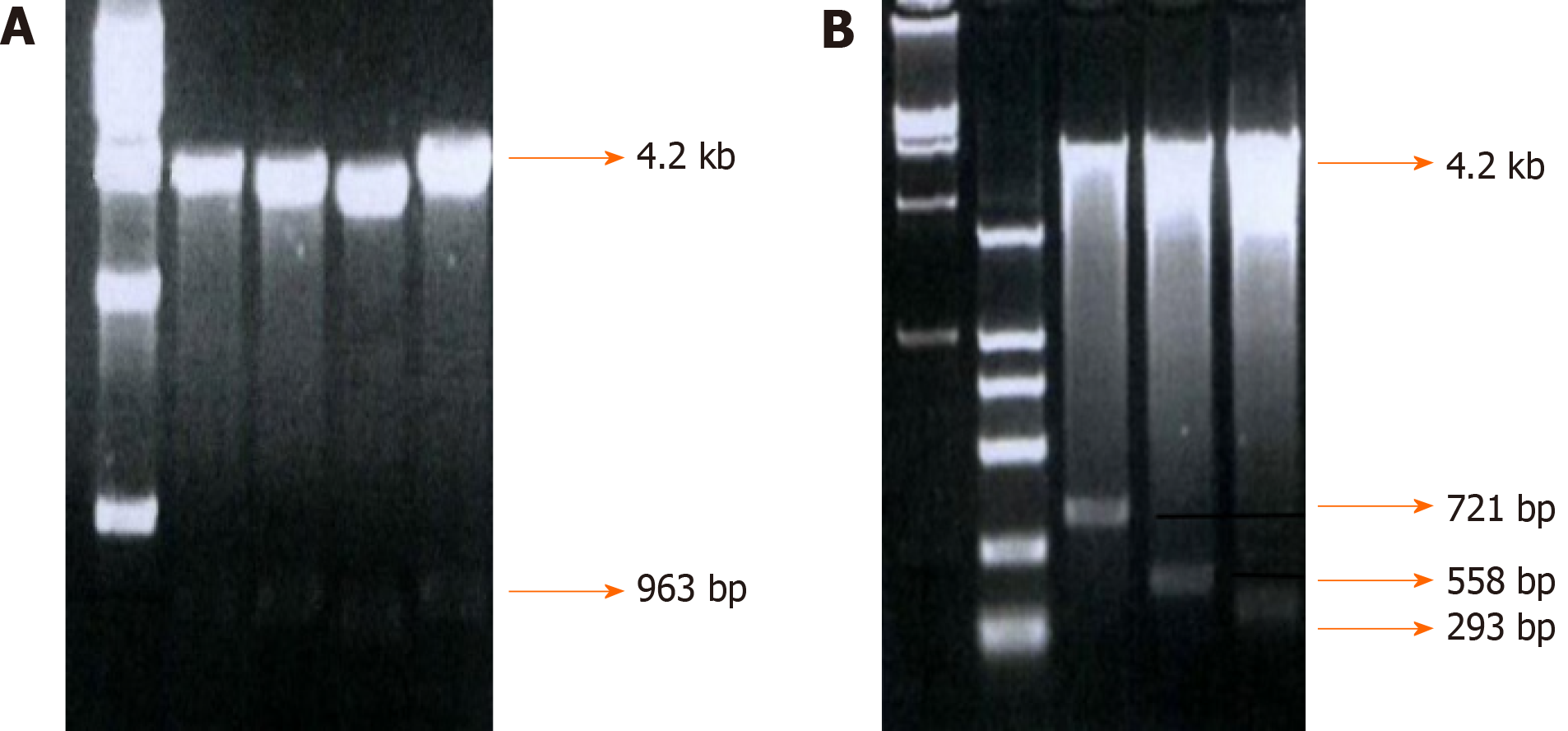
Fragments of -755-+198, -513-+198, -350-+198, and -85-+198, which were named integrin β6-B2, B3, B4, and B5, respectively, were selected to construct an array of truncated promoter plasmids, using pGL4-β6-B1 as the template (Figure 3). Then, identification of pGL4-B3, B4, and B5 was performed by enzyme analysis (Figure 4). A similar level of expression was observed using the -755-+198 region in both cell lines. However, when the region between -921 and -513 was deleted, the -513-+198 region of integrin β6 revealed the maximum luciferase activity (Figure 5). The data displayed that the region between -350 and -85 is the main sequence that positively regulates integrin β6 transcription.
To further describe the regulatory fragments that positively regulate integrin β6 transcription, a series of 5’ deletion of the region between -350 and -85 were divided into -350/-85,-286/-85, -210/-85, and -156/-85. Then, the expression of luciferase activity was analyzed. Truncation of the sequence from -286 to -85 resulted in a decrease in luciferase expression by 1.7 times and 2.1 times in the HT-29 cells and WiDr cells, respectively (Figure 5). These results displayed that the regulatory elements for integrin β6 positive transcription are located between the -286 to -85 region of integrin β6 gene.
Sequence analysis revealed the existence of many transcription regulatory elements spanning -300 to +10, for example, the binding sites for Ets, STAT3, and AP-1. To investigate the transcription regulation of integrin β6 by Ets-1 and STAT3, sequence analysis was performed with AliBaba2 software, which revealed the existence of three putative STAT3 binding sites and one Ets-1 binding site in the core promoter region of integrin β6 (Figure 6). To study the influence of STAT3 on the transcription of integrin β6, dual-luciferase reporter assay was performed. Our previous results in HT-29 cells showed that pGL-B7 can highly express about 36 times of pGL-Basic (negative control). With the truncation of 5’-flanking region, the luciferase activity ofpGL-B8, which lacked two STAT3 binding sites, decreased to about 13 times of pGL-Basic. When it was truncated to -156, allSTAT3 binding sites were cut off, and the activity decreased obviously. The results indicated that STAT3 may be responsible for the basic transcription level. The same result was obtained in the WiDr group.
To explore the effect of the potential Ets-1 binding site on integrin β6 transcription activity, a binding site mutant pGL4-B5-M-Ets in the vector pGL4-B5 was generated (Forward: 5’-ATAAGGAGAACAAGGAAGTAAATCATG-3’; reverse: 5’-ATAACGAGTTTACGGATGTGGATCATCG). After transient transfection, the relative luciferase activities of these constructs were assayed. The promoter activity was significantly reduced. The results suggested that the putative Ets-1 binding site was important for integrin β6 promoter transcription activity.
To detect the activity of integrin β6-B1 promoter, we constructed luciferase reporter plasmids. The reporter gene was used to detect the effect of gene expression and interaction of trans-acting factors. pGL4-Basic is a kind of reporter plasmid and usually used to detect the promoter activity of elements based on the luciferase reporter.
STAT3 mediates the biological behavior of cells in the presence of extracellular signals and plays an important role in chronic inflammatory and carcinogenesis[13]. Karin et al[14] reported that the deficiency of STAT3 was sufficient to inhibit tumorigenesis and progression in a colitis associated cancer model[15]. Conversely, the activation of STAT3 could promote the development of colitis associated cancer[16]. Meanwhile, STAT3 up-regulated the expression of Cyclin-D, CDC25A, c-Myc, and Pim1 proteins, which significantly promoted cell proliferation and reduced apoptosis[17]. Clinical research reported that in 60% of liver cancer cases, the expression of STAT3 increased abnormally[18]. On the one hand, STAT3 promoted cell proliferation and reduced apoptosis. On the other hand, it induced tumor invasion and metastasis. Besides, STAT3 enhanced angiogenesis via up-regulating the activity of vascular endothelial growth factor (VEGF), epidermal growth factor, platelet-derived growth factor, and hepatocyte growth factor[19,20].The expression of VEGF was mediated via STAT3, which increased the invasion of tumor associated macrophages and promoted tumor related disease[21,22]. We found that integrin β6 had a potential binding site for STAT3. Deletion of this site decreased the activity of integrin β6 expression. This result suggested that STAT3 could up-regulate the expression of integrin β6. However, the underlying mechanisms need further study, as the activity of STAT3 in cell lines is still unclear.
As an oncogenic protein of the Ets family, Ets-1 can regulate cell proliferation, angiogenesis, and apoptosis[23-25]. All Ets family members contain an 85 amino acid DNA binding domain, and regulate a number of cellular genes. So, we can predict that Ets-1 plays an important role in regulating integrin β6 expression and promoting the development of cancer.
The main limitation of this study is that the effect of transcriptional regulatory factors on the expression of integrin β6 was only verified at the molecular level, and its role has not been further verified in cell tests and animal experiments. In the future, we will carefully resolve this issue.
Integrin β6 is a critical subunit of the integrin αvβ6 and plays an important role in the development of cancer and wound healing. To investigate the transcriptional regulation of integrin β6 gene in cancer cells, we have identified its promoter. Furthermore, our results indicated that the transcription factors STAT3 and Ets-1 are both involved in the regulation of integrin β6 transcription in colon cancer cell lines.
The integrin β6 gene, which is expressed in epithelial cancer, plays a pivotal role in various aspects of cancer progression. The present research for integrin β6 regulation mainly focuses on the post-transcription and translation related regulation mechanism and its role in tumorigenesis.
The mechanism of how the integrin β6 gene is regulated, and the promoter and transcription factors responsible for basic transcription of integrin β6 gene remain unknown.
This study aimed to clone and characterize the integrin β6 promoter.
The region containing the integrin β6 promoter was predicted and luciferase reporter plasmids were constructed. The location of core promoter and binding site for transcription factors were identified by element deletion analysis.
We found that the regulatory elements for transcription of integrin β6 gene were located between -286 and-85 and contained binding sites for transcription factors such as STAT3 and Ets-1.
We have identified the region of β6 core promoter and the binding sites for transcription factors such as Ets-1 and STAT3, which are important for integrin β6 promoter transcription activity.
The findings of this study are important for investigating the mechanism of integrin β6 activation in cancer progression.
| 1. | Olof Olsson P, Gustafsson R, Salnikov AV, Göthe M, Zeller KS, Friman T, Baldetorp B, Koopman LA, Weinreb PH, Violette SM, Kalamajski S, Heldin NE, Rubin K. Inhibition of integrin αVβ6 changes fibril thickness of stromal collagen in experimental carcinomas. Cell Commun Signal. 2018;16:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1292] [Cited by in RCA: 1297] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 3. | Li Z, Lin P, Gao C, Peng C, Liu S, Gao H, Wang B, Wang J, Niu J, Niu W. Integrin β6 acts as an unfavorable prognostic indicator and promotes cellular malignant behaviors via ERK-ETS1 pathway in pancreatic ductal adenocarcinoma (PDAC). Tumour Biol. 2016;37:5117-5131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Liang B, Li L, Miao R, Wang J, Chen Y, Li Z, Zou X, Zhou M. Expression of Interleukin-6 and Integrin ανβ6 in Colon Cancer: Association with Clinical Outcomes and Prognostic Implications. Cancer Invest. 2019;37:174-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 5. | Peng C, Zou X, Xia W, Gao H, Li Z, Liu N, Xu Z, Gao C, He Z, Niu W, Fang R, Biswas S, Agrez M, Zhi X, Niu J. Integrin αvβ6 plays a bi-directional regulation role between colon cancer cells and cancer-associated fibroblasts. Biosci Rep. 2018;38:BSR20180243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 6. | Bates RC, Bellovin DI, Brown C, Maynard E, Wu B, Kawakatsu H, Sheppard D, Oettgen P, Mercurio AM. Transcriptional activation of integrin beta6 during the epithelial-mesenchymal transition defines a novel prognostic indicator of aggressive colon carcinoma. J Clin Invest. 2005;115:339-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 155] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 7. | Koivisto L, Bi J, Häkkinen L, Larjava H. Integrin αvβ6: Structure, function and role in health and disease. Int J Biochem Cell Biol. 2018;99:186-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 8. | Zhang ZY, Xu KS, Wang JS, Yang GY, Wang W, Wang JY, Niu WB, Liu EY, Mi YT, Niu J. Integrin alphanvbeta6 acts as a prognostic indicator in gastric carcinoma. Clin Oncol (R Coll Radiol). 2008;20:61-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 91] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Singh C, Shyanti RK, Singh V, Kale RK, Mishra JPN, Singh RP. Integrin expression and glycosylation patterns regulate cell-matrix adhesion and alter with breast cancer progression. Biochem Biophys Res Commun. 2018;499:374-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 10. | Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 414] [Cited by in RCA: 364] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 11. | Xu M, Yin L, Cai Y, Hu Q, Huang J, Ji Q, Hu Y, Huang W, Liu F, Shi S, Deng X. Epigenetic regulation of integrin β6 transcription induced by TGF-β1 in human oral squamous cell carcinoma cells. J Cell Biochem. 2018;119:4193-4204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Sullivan BP, Kassel KM, Manley S, Baker AK, Luyendyk JP. Regulation of transforming growth factor-β1-dependent integrin β6 expression by p38 mitogen-activated protein kinase in bile duct epithelial cells. J Pharmacol Exp Ther. 2011;337:471-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 13. | Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2935] [Cited by in RCA: 3500] [Article Influence: 205.9] [Reference Citation Analysis (20)] |
| 14. | Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103-113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1747] [Cited by in RCA: 1808] [Article Influence: 106.4] [Reference Citation Analysis (0)] |
| 15. | Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T, Canli O, Schwitalla S, Matthews V, Schmid RM, Kirchner T, Arkan MC, Ernst M, Greten FR. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 805] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 16. | Rigby RJ, Simmons JG, Greenhalgh CJ, Alexander WS, Lund PK. Suppressor of cytokine signaling 3 (SOCS3) limits damage-induced crypt hyper-proliferation and inflammation-associated tumorigenesis in the colon. Oncogene. 2007;26:4833-4841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 150] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 17. | Wang Y, Shen Y, Wang S, Shen Q, Zhou X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 2018;415:117-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 287] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 18. | He G, Yu GY, Temkin V, Ogata H, Kuntzen C, Sakurai T, Sieghart W, Peck-Radosavljevic M, Leffert HL, Karin M. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286-297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 375] [Cited by in RCA: 380] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 19. | Aggarwal BB, Gehlot P. Inflammation and cancer: how friendly is the relationship for cancer patients? Curr Opin Pharmacol. 2009;9:351-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 281] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 20. | Waldner MJ, Wirtz S, Jefremow A, Warntjen M, Neufert C, Atreya R, Becker C, Weigmann B, Vieth M, Rose-John S, Neurath MF. VEGF receptor signaling links inflammation and tumorigenesis in colitis-associated cancer. J Exp Med. 2010;207:2855-2868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 163] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 21. | Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, Peng Z, Huang S, Xiong HQ, Abbruzzese JL, Xie K. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 450] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 22. | Kujawski M, Kortylewski M, Lee H, Herrmann A, Kay H, Yu H. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J Clin Invest. 2008;118:3367-3377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 464] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 23. | Tanaka H, Terada Y, Kobayashi T, Okado T, Inoshita S, Kuwahara M, Seth A, Sato Y, Sasaki S. Expression and function of Ets-1 during experimental acute renal failure in rats. J Am Soc Nephrol. 2004;15:3083-3092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Ghosh S, Basu M, Roy SS. ETS-1 protein regulates vascular endothelial growth factor-induced matrix metalloproteinase-9 and matrix metalloproteinase-13 expression in human ovarian carcinoma cell line SKOV-3. J Biol Chem. 2012;287:15001-15015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 25. | Hashiya N, Jo N, Aoki M, Matsumoto K, Nakamura T, Sato Y, Ogata N, Ogihara T, Kaneda Y, Morishita R. In vivo evidence of angiogenesis induced by transcription factor Ets-1: Ets-1 is located upstream of angiogenesis cascade. Circulation. 2004;109:3035-3041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 83] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chandrakesan P, Kahrilas PJ, Nusrat S S-Editor: Wang JL L-Editor: Wang TQ E-Editor: Qi LL