©The Author(s) 2025.
World J Gastrointest Oncol. Dec 15, 2025; 17(12): 112936
Published online Dec 15, 2025. doi: 10.4251/wjgo.v17.i12.112936
Published online Dec 15, 2025. doi: 10.4251/wjgo.v17.i12.112936
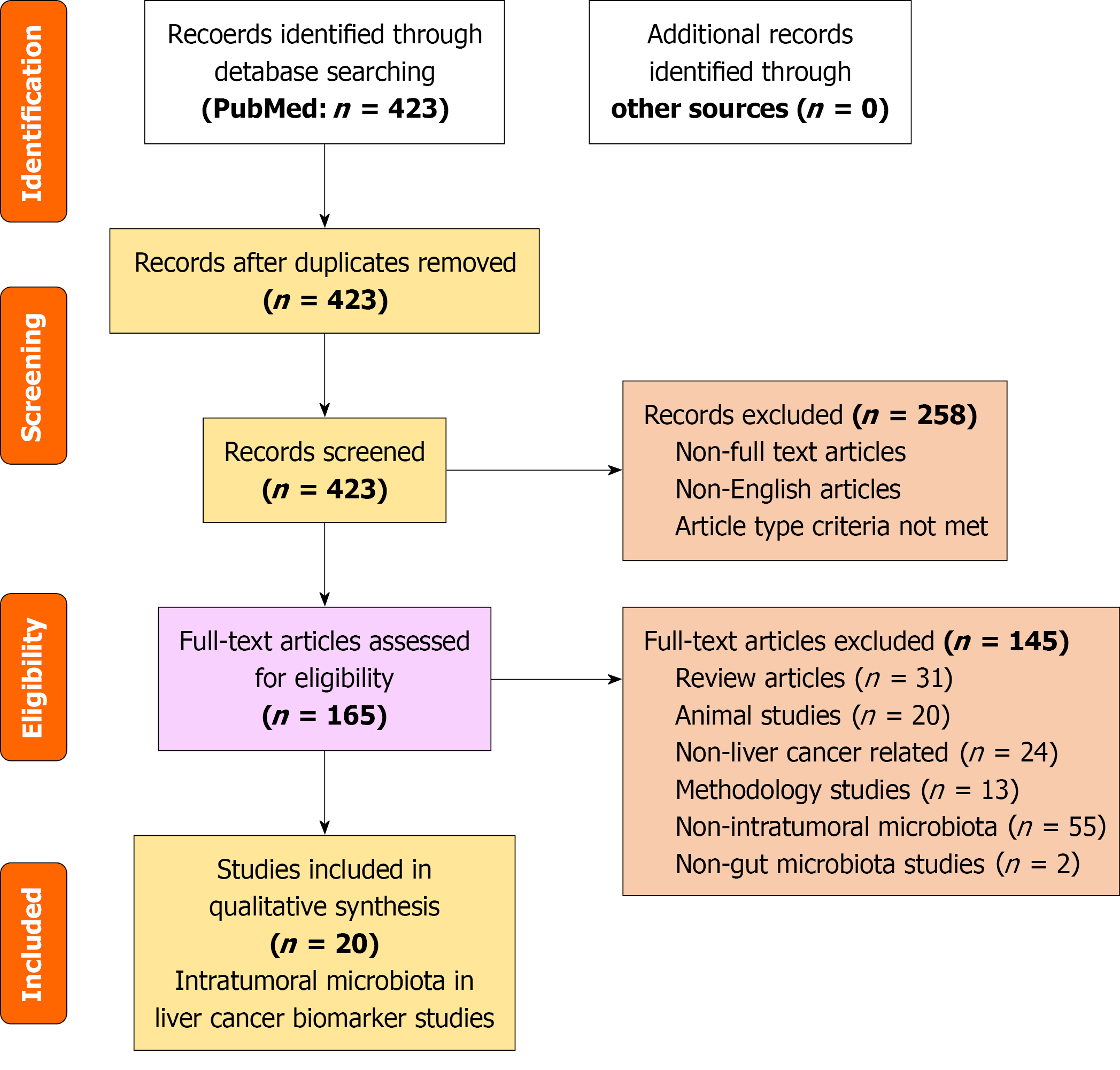
Figure 1 PRISMA flow diagram.
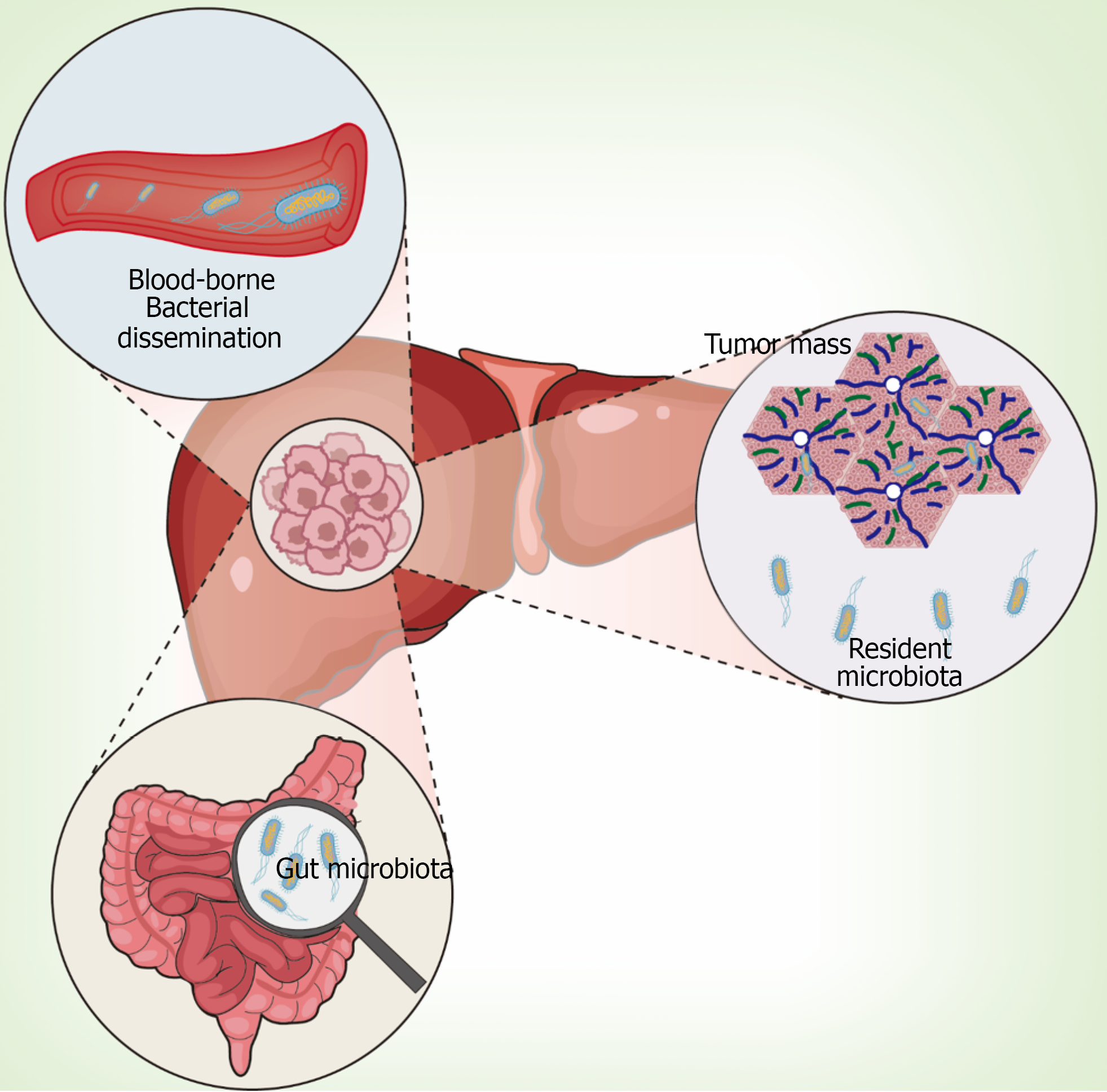
Figure 2 Anatomical pathways of microbial colonization in hepatic malignancies.
The figure shows the human digestive system with liver tumor and three distinct routes of microbial entry: Upper panel depicts blood-borne bacterial dissemination from systemic circulation; Central liver shows tumor mass (pink nodule) with resident microbial communities; Lower panel illustrates gut microbiota (bacterial symbols) with directional arrows indicating translocation pathways. Right panel shows magnified view of diverse bacterial morphologies within tumor microenvironment. The anatomical representation demonstrates spatial relationships between source organs and target liver tissue.
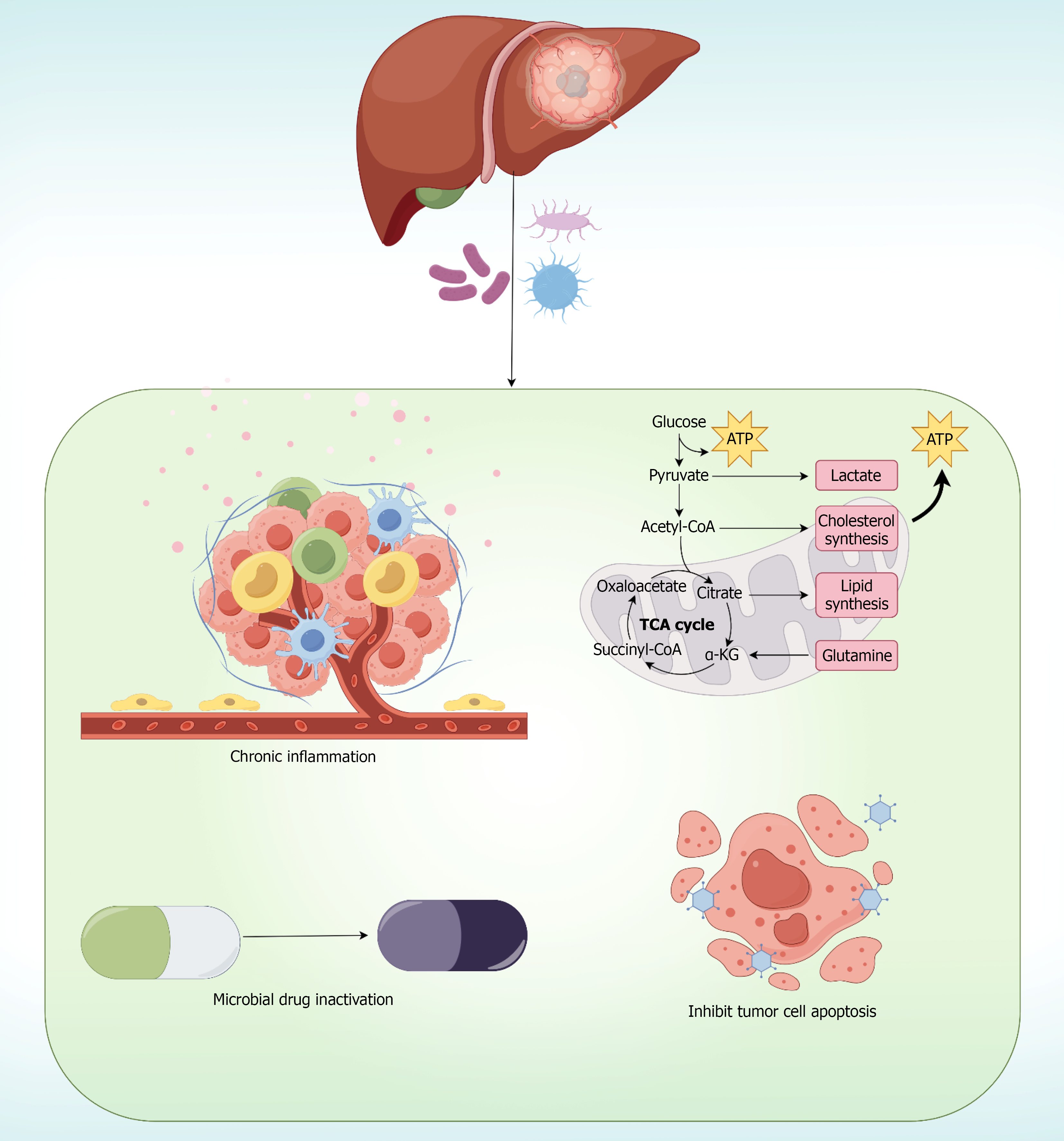
Figure 3 Possible mechanisms by which intratumoral microbes affect tumor development and drug sensitivity in hepatic malignancies.
The figure illustrates four key mechanisms: Chronic inflammation through immune cell infiltration and inflammatory mediator release; metabolic network modulation affecting glucose metabolism, TCA cycle, and biosynthetic pathways; microbial drug inactivation through enzymatic degradation; and inhibition of tumor cell apoptosis promoting tumor survival. These interconnected pathways demonstrate the multifaceted role of tumor-resident microbiota in hepatocarcinogenesis and therapeutic resistance.
- Citation: Song S, Xu LS, Wang LQ, Zhou X, Jiang X, Li CP. Tumor-resident microorganisms as clinical biomarkers in primary liver cancer: A systematic review of current evidence. World J Gastrointest Oncol 2025; 17(12): 112936
- URL: https://www.wjgnet.com/1948-5204/full/v17/i12/112936.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i12.112936