©The Author(s) 2025.
World J Gastrointest Oncol. Dec 15, 2025; 17(12): 112873
Published online Dec 15, 2025. doi: 10.4251/wjgo.v17.i12.112873
Published online Dec 15, 2025. doi: 10.4251/wjgo.v17.i12.112873
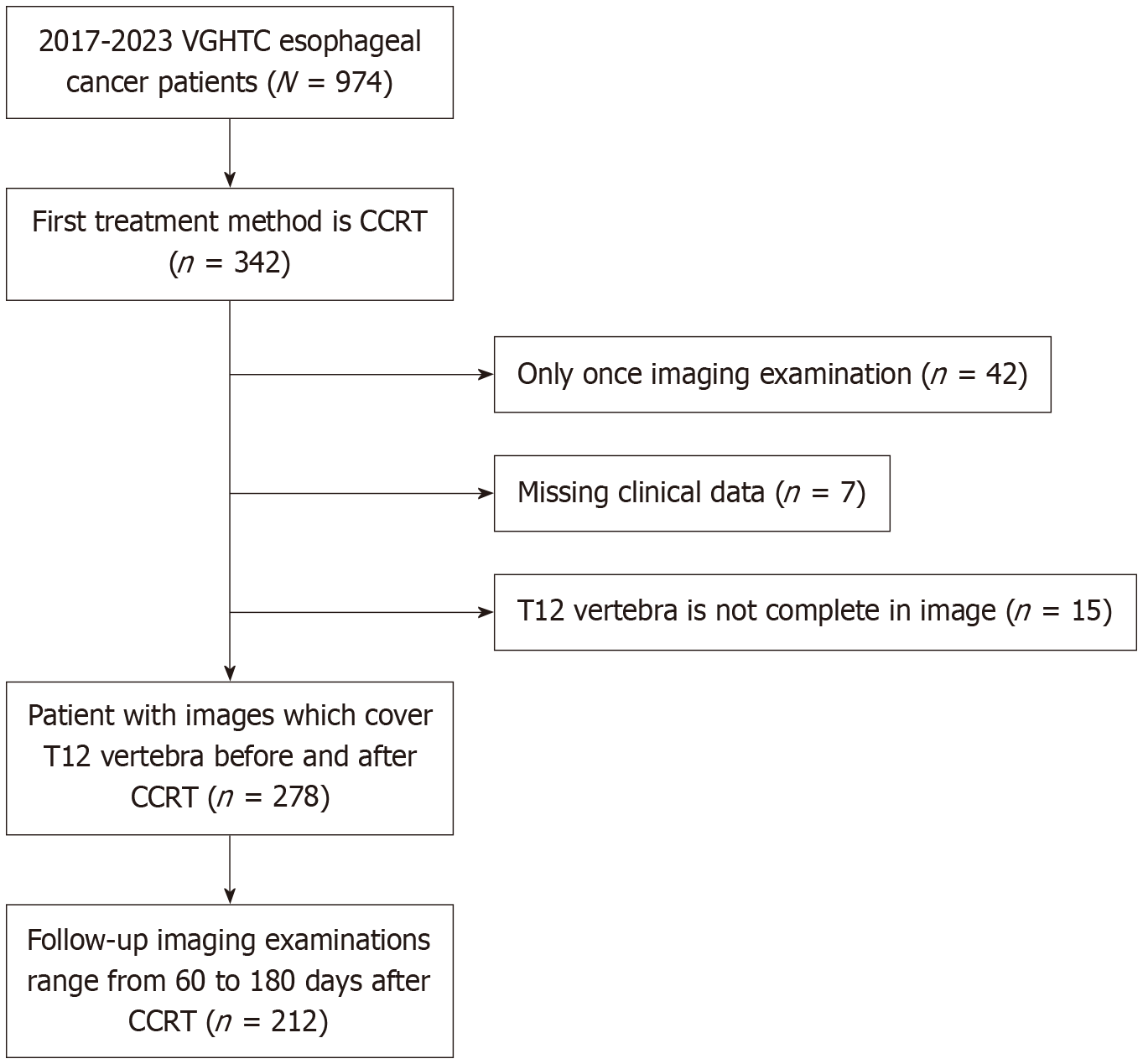
Figure 1 Patient selection flowchart.
A total of 974 patients with esophageal cancer were initially identified at Taichung Veterans General Hospital between 2017 and 2023. Among them, 342 patients received concurrent chemoradiotherapy (CCRT) as the initial treatment. Patients were excluded for the following reasons: Only one imaging examination available (n = 42), missing clinical data (n = 7), or incomplete imaging of the T12 vertebra (n = 15). As a result, 278 patients had both pre- and post-CCRT computed tomography images covering the T12 vertebra. From these, 212 patients underwent follow-up computed tomography scans performed 60-180 days after CCRT, forming the final cohort for analysis. VGHTC: Taichung Veterans General Hospital; CCRT: Concurrent chemoradiotherapy.
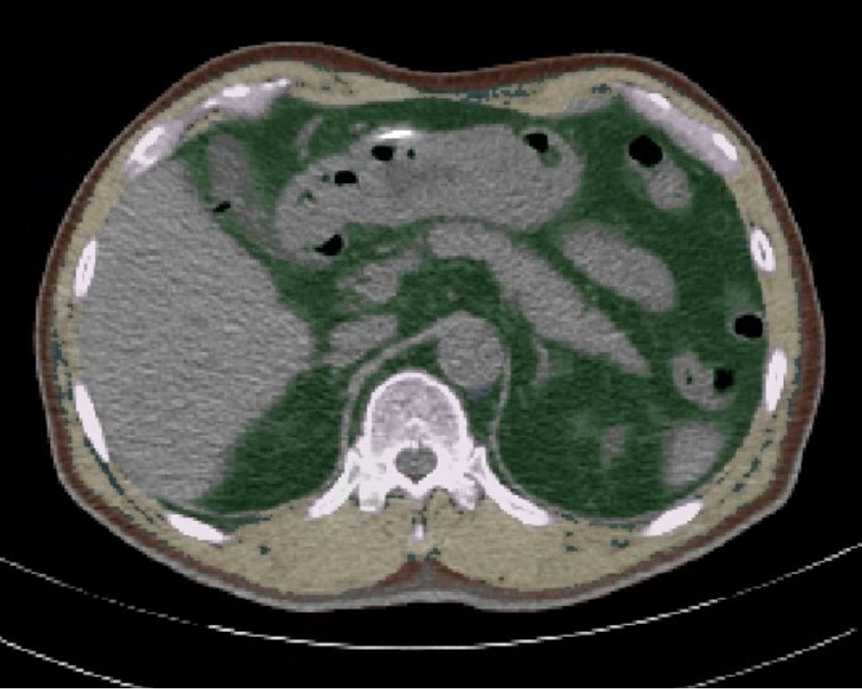
Figure 2
Exemplary cross-sectional images of automated segmentations of subcutaneous adipose tissue (brown), visceral adipose tissue (green), and total skeletal muscle area (yellow) at the T12 level.
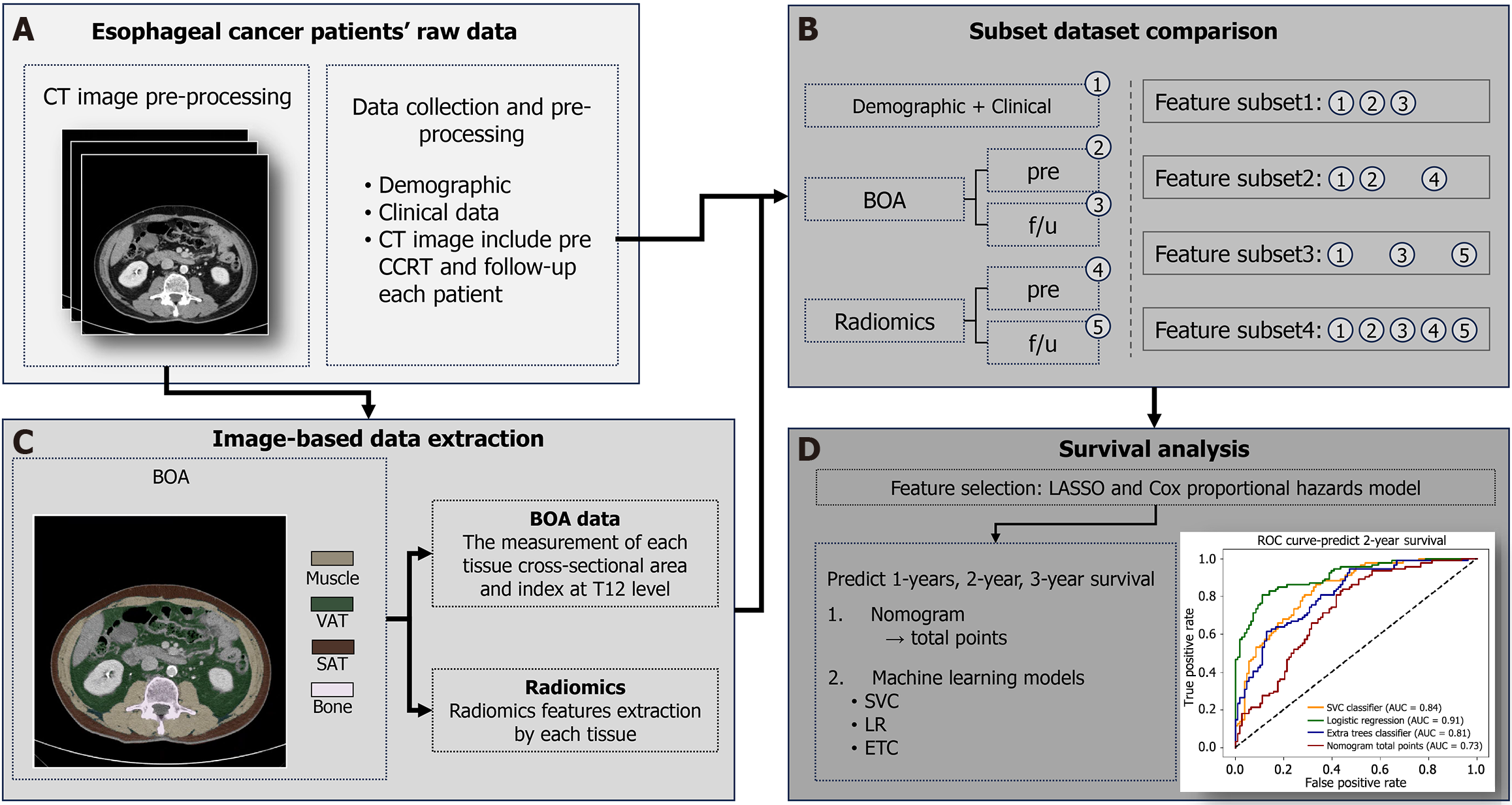
Figure 3 Study workflow and methodology.
The diagram illustrates the four-step process for predicting survival in esophageal cancer patients treated with concurrent chemoradiotherapy. A: This section shows the initial data collection, including demographic, clinical, and computed tomography (CT) imaging data. CT scans were acquired both before and after concurrent chemoradiotherapy, as well as during follow-up. An example of a transverse CT image is shown; B: This involves measuring the cross-sectional area and indices for muscle, visceral adipose tissue, and subcutaneous adipose tissue, along with radiomics features are extracted from each of the identified tissues; C: Four different combinations of features were used to create four distinct feature subsets for analysis. The features include demographic, clinical, body composition analysis, and radiomics data from both pretreatment (pre) and follow-up scans; D: This final step shows the methods used to predict patient survival. The least absolute shrinkage and selection operator and Cox proportional hazards models were used to select the most relevant features. Nomogram and three different machine learning models (support vector classification classifier, logistic regression, and the extra trees classifier) were used for predicting 1-, 2-, and 3-year survival. CT: Computed tomography; CCRT: Concurrent chemoradiotherapy; BOA: Body organ analysis; pre: Pretreatment; f/u: Follow-up; SAT: Subcutaneous adipose tissue; VAT: Visceral adipose tissue; LASSO: Least absolute shrinkage and selection operator; SVC: Support vector classification; LR: Logistic regression; ETC: Extra trees classifier; ROC: Receiver operating characteristic; AUC: Area under the time-dependent receiver operating characteristic curve.
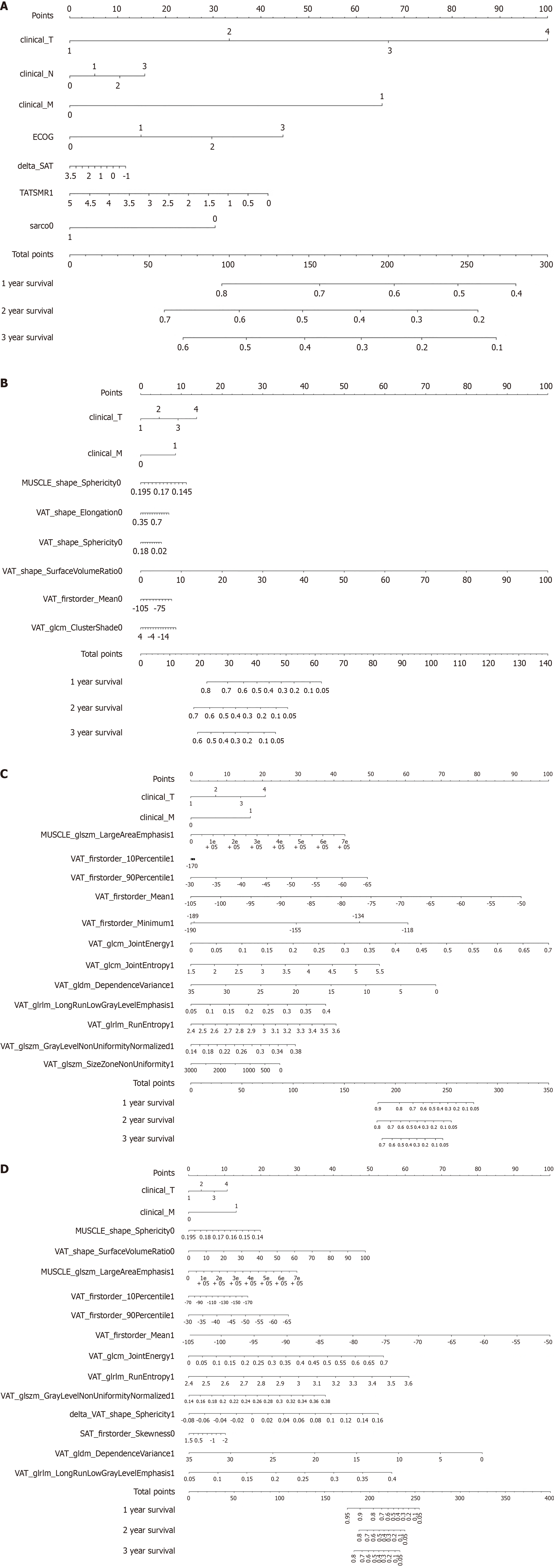
Figure 4 Nomogram: Estimating overall survival for 1-year to 3-year by using feature subset 1, subset 2, subset 3, subset 4 as input data.
A: Nomogram: Estimating overall survival (OS) for 1-year to 3-year by using feature subset 1 as input data; B: Nomogram: Estimating OS for 1-year to 3-year by using feature subset 2 as input data; C: Nomogram: Estimating OS for 1-year to 3-year by using feature subset 3 as input data; D: Nomogram: Estimating OS for 1-year to 3-year by using feature subset 4 as input data. ECOG: Eastern Cooperative Oncology Group; TATSMR1: Total adipose tissue index/skeletal muscle index ratio; sarco: Sarcopenia; 0: Pretreatment; VAT: Visceral adipose tissue; 1: Follow-up; glcm: Gray-level co-occurrence matrix; SAT: Subcutaneous adipose tissue; glszm: Gray-level size zone matrix; glrlm: Gray-level run length matrix; gldm: Gray-level dependence matrix.
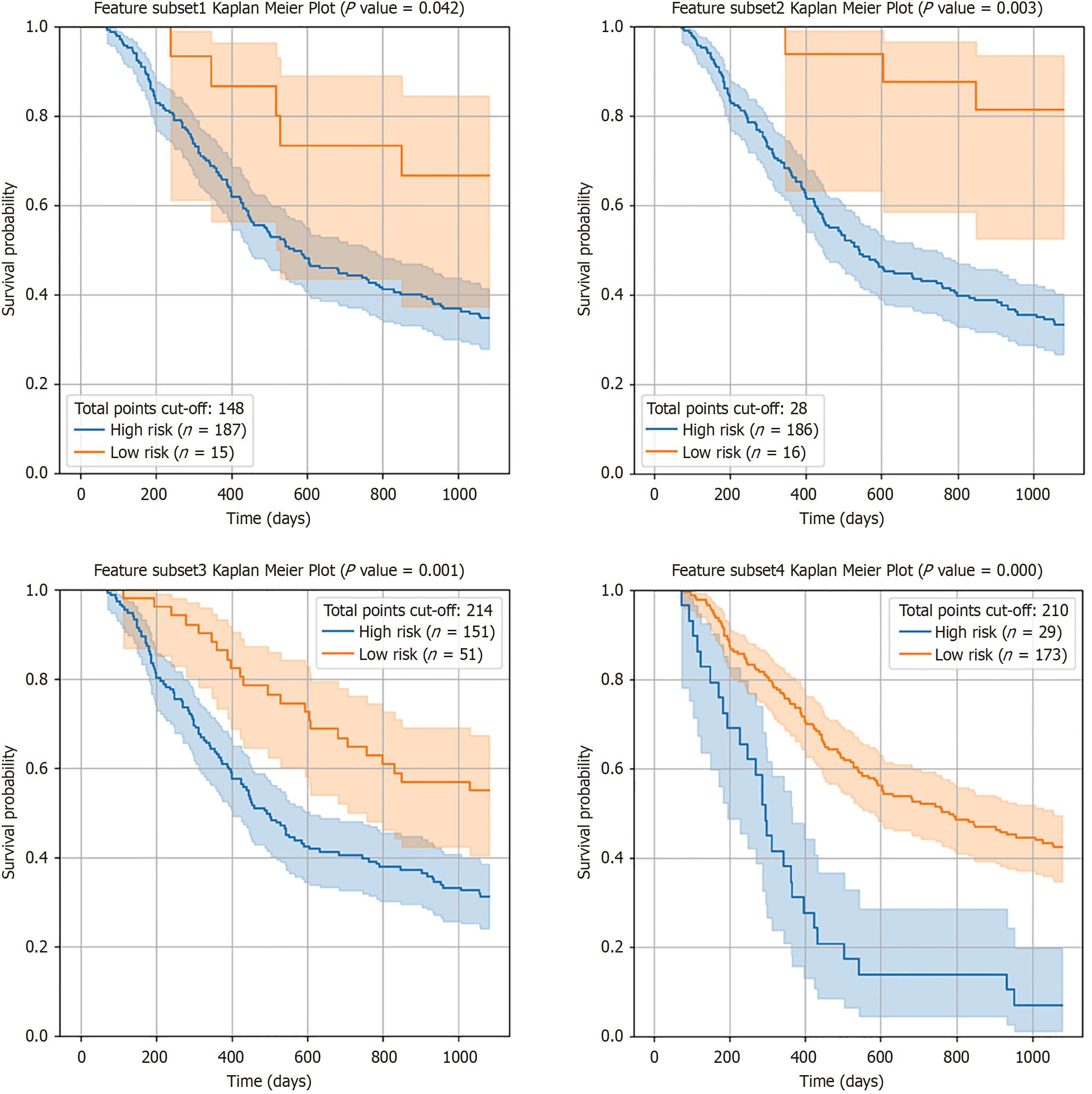
Figure 5 Results of the Kaplan-Meier survival analyses of 2-year survival rates according to the risk in feature subset 1 (upper left), feature subset 2 (lower left), feature subset 3 (upper right) and feature subset 4 (lower right) as input data.
The blue and orange lines represent patients classified as high-risk and low-risk for 2-year survival, respectively, based on the total points from the nomogram.
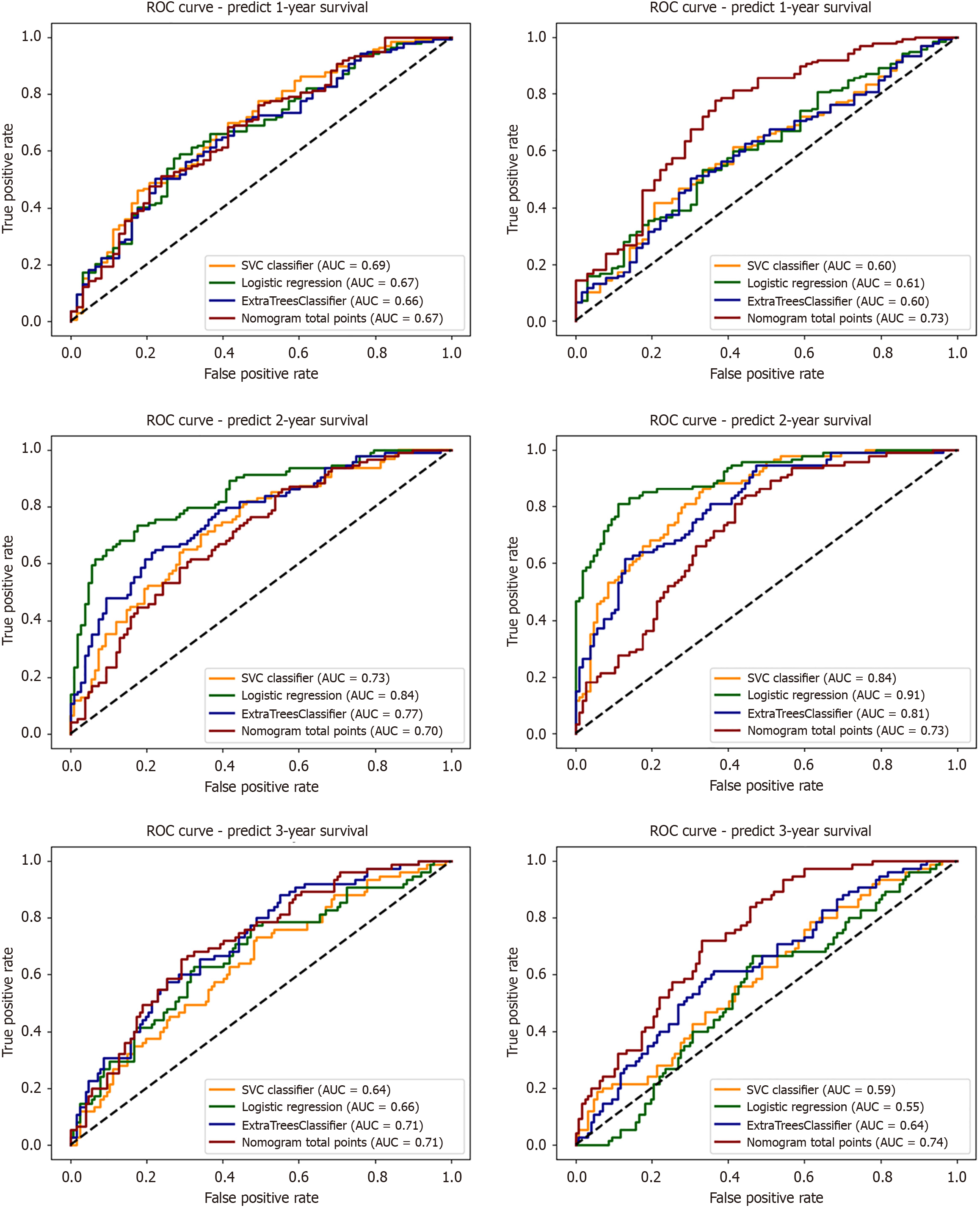
Figure 6 Area under the curves for 1-, 2-, and 3-year overall survival prediction using support vector classifier, logistic regression, extra trees classifier, and nomogram models for feature subsets 1 (no-radiomics) and 4 (combined input).
ROC: Receiver operating characteristic; AUC: Area under the time-dependent receiver operating characteristic curve.
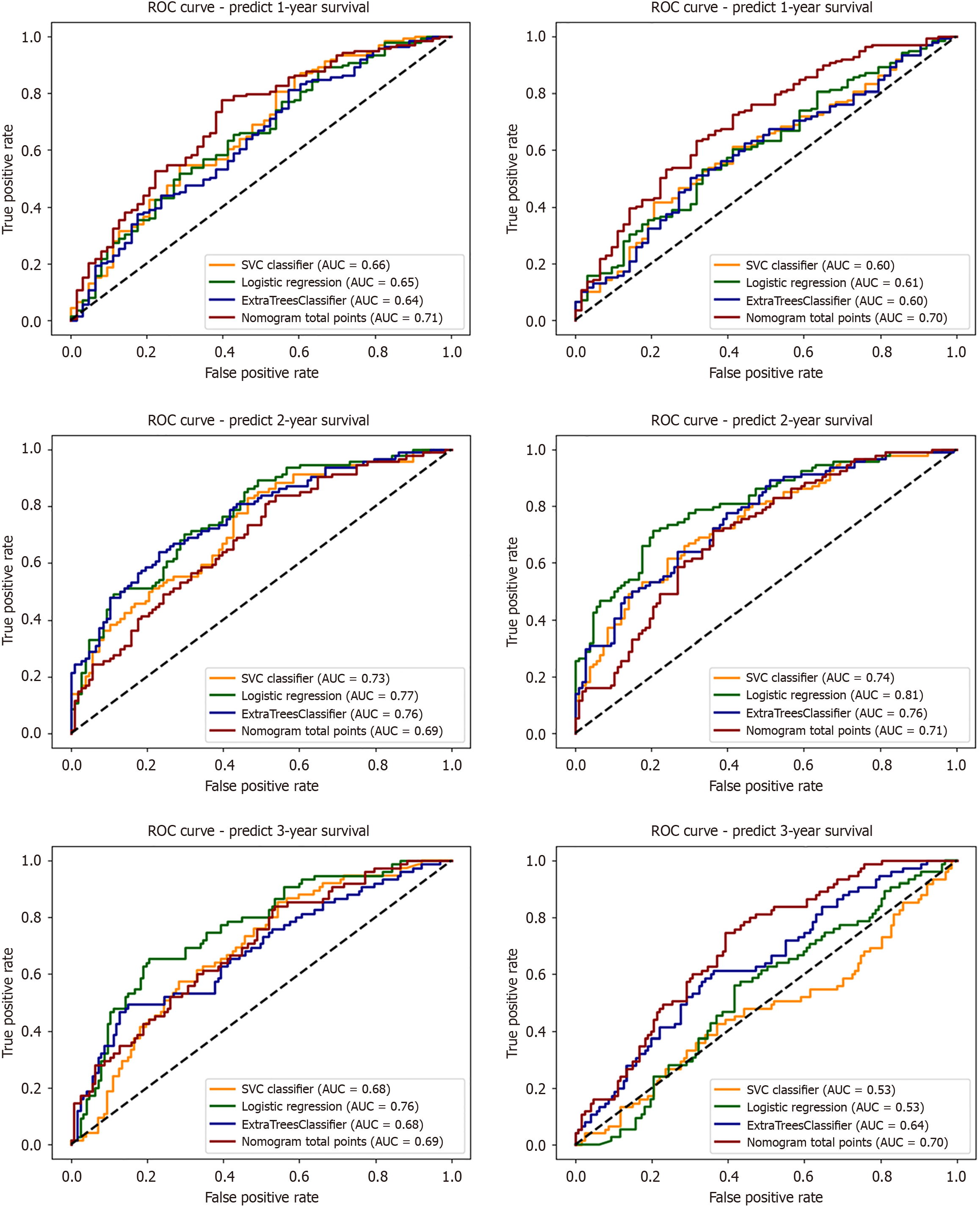
Figure 7 Area under the curves for 1-, 2-, and 3-year overall survival prediction using support vector classifier, logistic regression, extra trees classifier, and nomogram models for feature subsets 2 (pretreatment) and 3 (follow-up).
ROC: Receiver operating characteristic; AUC: Area under the time-dependent receiver operating characteristic curve.
- Citation: Liu MC, Cheng YY, Lin SC, Lin CH, Chuang CY, Chen WH, Liao CH, Hsieh CH, Hsieh MF, Liu YJ. Machine learning survival prediction in esophageal cancer using radiomics and body composition from pretreatment and follow-up T12-level computed tomography. World J Gastrointest Oncol 2025; 17(12): 112873
- URL: https://www.wjgnet.com/1948-5204/full/v17/i12/112873.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i12.112873