©The Author(s) 2025.
World J Gastrointest Oncol. Oct 15, 2025; 17(10): 110661
Published online Oct 15, 2025. doi: 10.4251/wjgo.v17.i10.110661
Published online Oct 15, 2025. doi: 10.4251/wjgo.v17.i10.110661
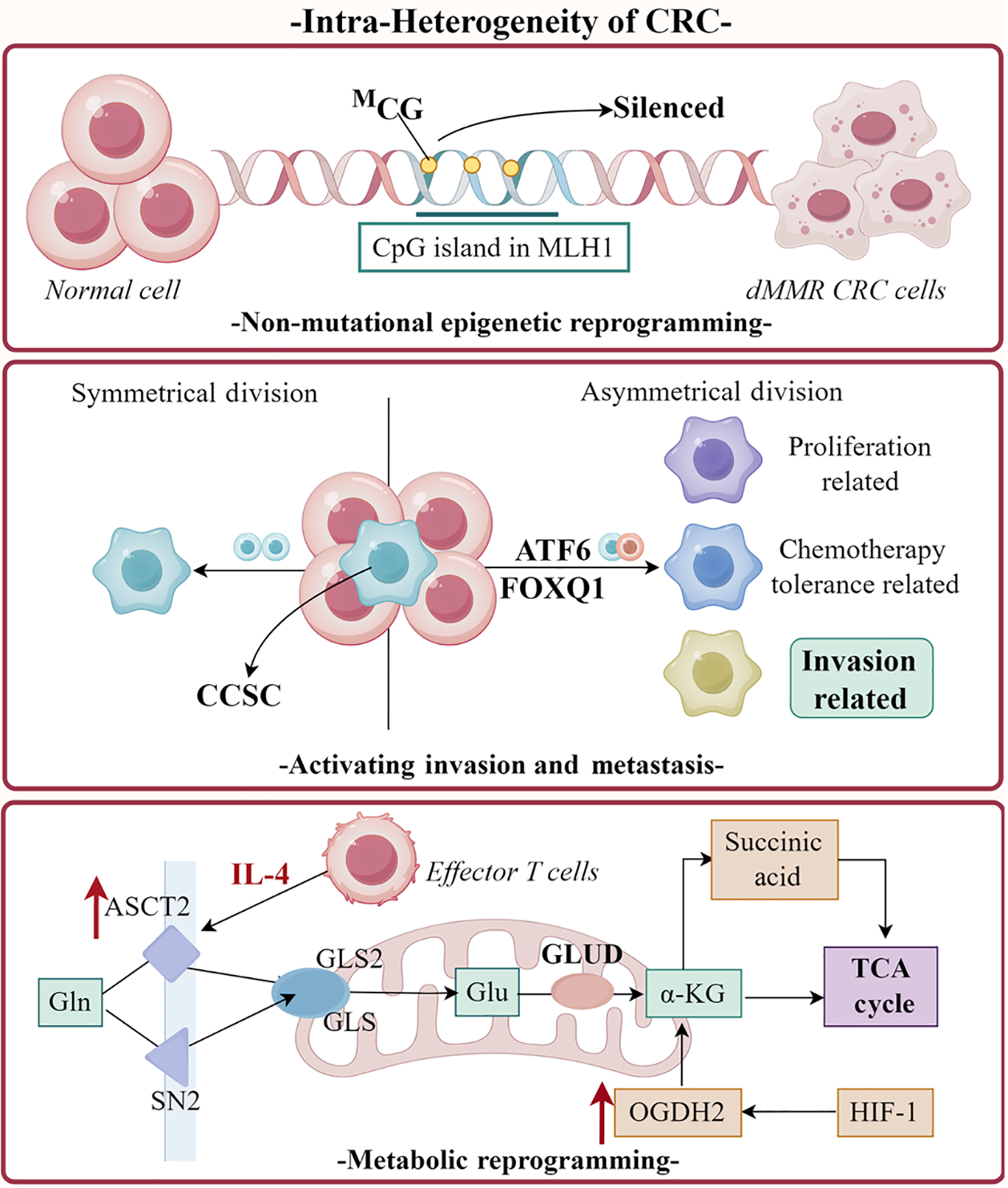
Figure 1 Overview of the integration of colorectal cancer heterogeneity with single-cell RNA sequencing and spatial transcriptomics.
This schematic illustrates key drivers of cellular diversity within defective mismatch repair (dMMR) colorectal cancer (CRC) tumors. Epigenetic origin: Hypermethylation of the MLH1 gene promoter CpG island drives the transition from normal colonic epithelium to dMMR CRC. Cellular heterogeneity: Non-mutational epigenetic reprogramming enables both symmetric division (expanding the cancer cell pool) and asymmetric division, generating distinct subpopulations with divergent properties: Proliferation-related, chemotherapy tolerance-related, invasion-related. Key regulators in this process include ATF6 and FOXQ1. Activating invasion and metastasis: Invasion-prone subclusters exhibit metabolic reprogramming centered on glutamine/amino acid metabolism, generating α-ketoglutaric acid to fuel the tricarboxylic acid cycle. Activation of hypoxia-inducible factor 1-alpha and oxoglutarate dehydrogenase, ultimately promoting invasive and metastatic behavior. dMMR: Defective mismatch repair; CRC: Colorectal cancer; CCSC: Cancer stem cell-like cell; ASCT2: Alanine-serine-cysteine transporter 2; SN2: Sodium-coupled neutral amino acid transporter 2; GLS: Glutaminase; GLS2: Glutaminase 2; IL-4: Interleukin-4; Gln: Glutamine; Glu: Glucose; GLUD: Glutamate dehydrogenase; OGDH2: Oxoglutarate dehydrogenase; α-KG: Alpha-ketoglutarate; TCA cycle: Tricarboxylic acid cycle; HIF-1: Hypoxia-inducible factor 1-alpha.
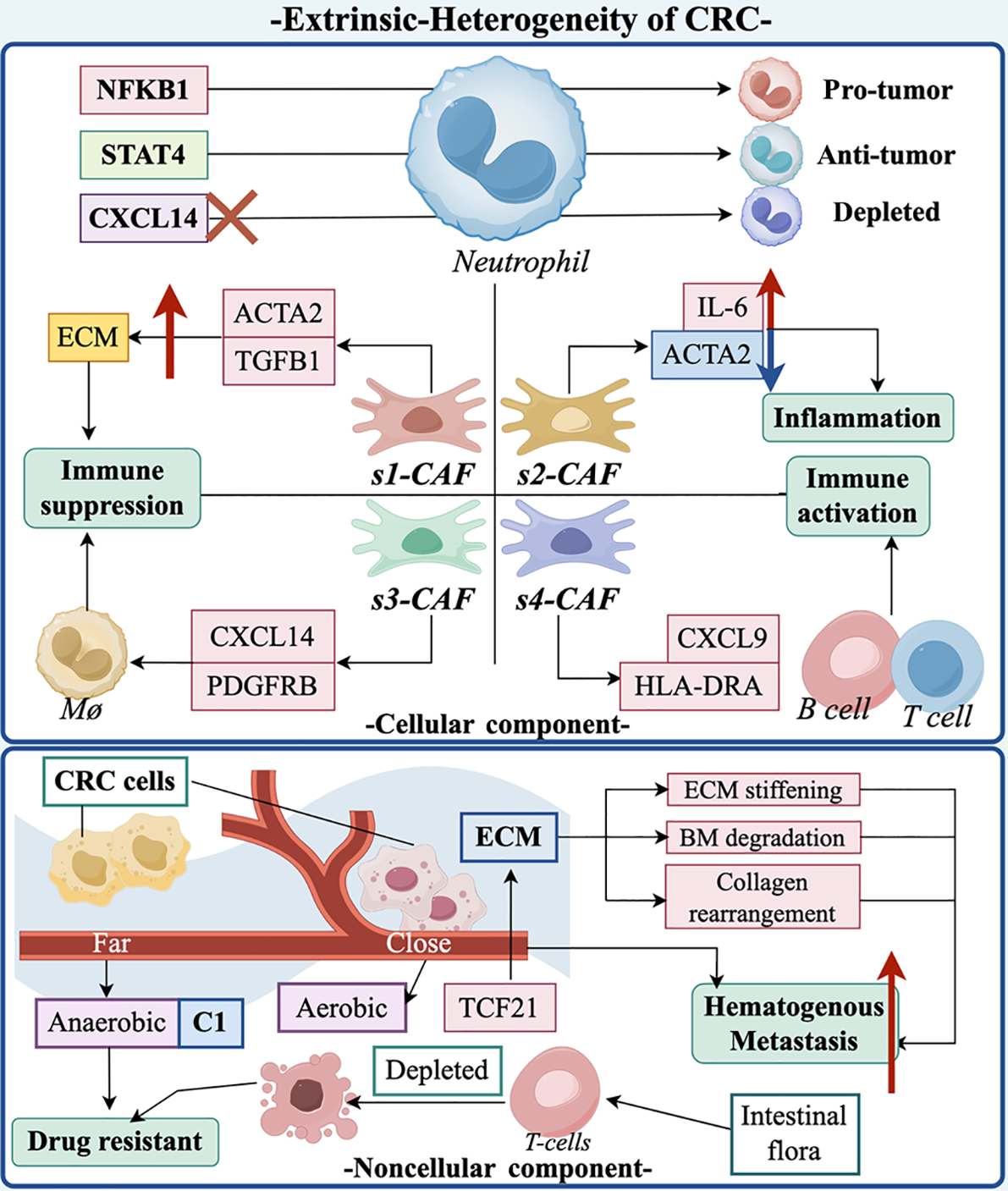
Figure 2 Overview of the extrinsic of colorectal cancer heterogeneity with single-cell RNA sequencing and spatial transcriptomics.
This figure shows representative regulatory mechanisms in the cellular vs non-cellular components of colorectal cancer (CRC) extrinsic heterogeneity. First, different genes induce different types of neutrophil formation such as tumor-promoting, tumor-suppressing, and depleted types. Second, different subtypes of CRC-associated fibroblasts have different roles. S1-cancer-associated fibroblast (CAF) causes immunosuppression by affecting the extracellular matrix (ECM) under the regulation of actin alpha 2 (ACTA2) and transforming growth factor-β-1, and also causes immunosuppression by the subtype S3-CAF, which is achieved through the modulation of macrophages by CXCL14 and PDGFRB. S2-CAF reduces the number of macrophages through the up-regulation of interleukin-6, the ACTA2 expression to elicit an inflammatory response. S4-CAF, on the other hand, is associated with a favorable prognosis for inflammatory bowel disease in precancerous lesions of CRC. In the noncellular component, pro-vascular CRC cells and CRC cells distant from the vasculature produce different metabolites due to different metabolic patterns resulting in different CRC subtypes. CRC cells far from the vasculature tend to produce drug-resistant subtypes, and some metabolites of intestinal flora are also involved in the formation of this resistance. In addition, regulation of the ECM, such as transcription factor 21, shapes CRC subtypes that are susceptible to hematogenous metastasis by increasing the stiffness of the ECM, degrading the basement membrane, and collagen rearrangement. CRC: Colorectal cancer; ECM: Extracellular matrix; ACTA2: Actin alpha 2; TGFB1: Transforming growth factor-β-1; CAF: Cancer-associated fibroblasts; IL-6: Interleukin-6; TCF21: Transcription factor 21; BM: Basement membrane.
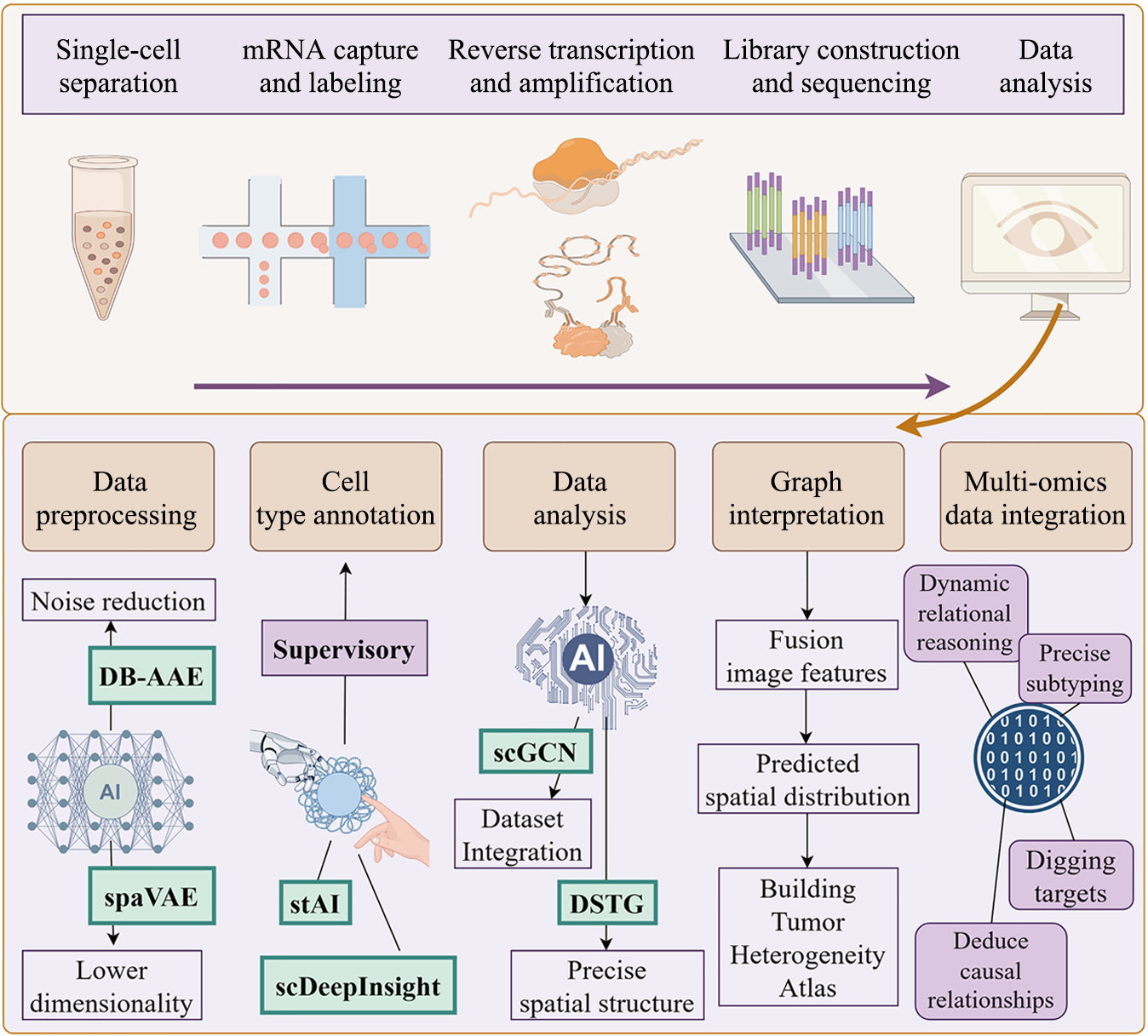
Figure 3 Artificial intelligence-powered analytical framework for single-cell RNA sequencing and spatial transcriptomics in colorectal cancer heterogeneity.
Standard transcriptomics workflow: Illustrates sequential steps from single-cell separation through messenger RNA capture and labeling. Reverse transcription and amplification, library construction and sequencing, to data analysis. Artificial intelligence (AI)-enhanced analytics: AI transforms multi-modal data interpretation through: Advanced data preprocessing (noise reduction and lower dimension via AI models such as dynamic batching adversarial autoencoder and spatial variational autoencoder); Cell type annotation (supervision, supplementation, and optimization through deep learning models such as stAI and scDeepInsight); Data analysis (dataset integration via single-cell graph convolutional network and accurate resolution of spatial information through deconvoluting spatial transcriptomics data through graph-based convolutional networks); Ensemble image features for accurate prediction; Integrating multi-omics analysis for causal analysis, target prediction, and precision medicine. mRNA: Messenger RNA; DB-AAE: Dynamic batching adversarial autoencoder; spaVAE: Spatial variational autoencoder; AI: Artificial intelligence; scGCN: Single-cell graph convolutional network; DSTG: Deconvoluting spatial transcriptomics.
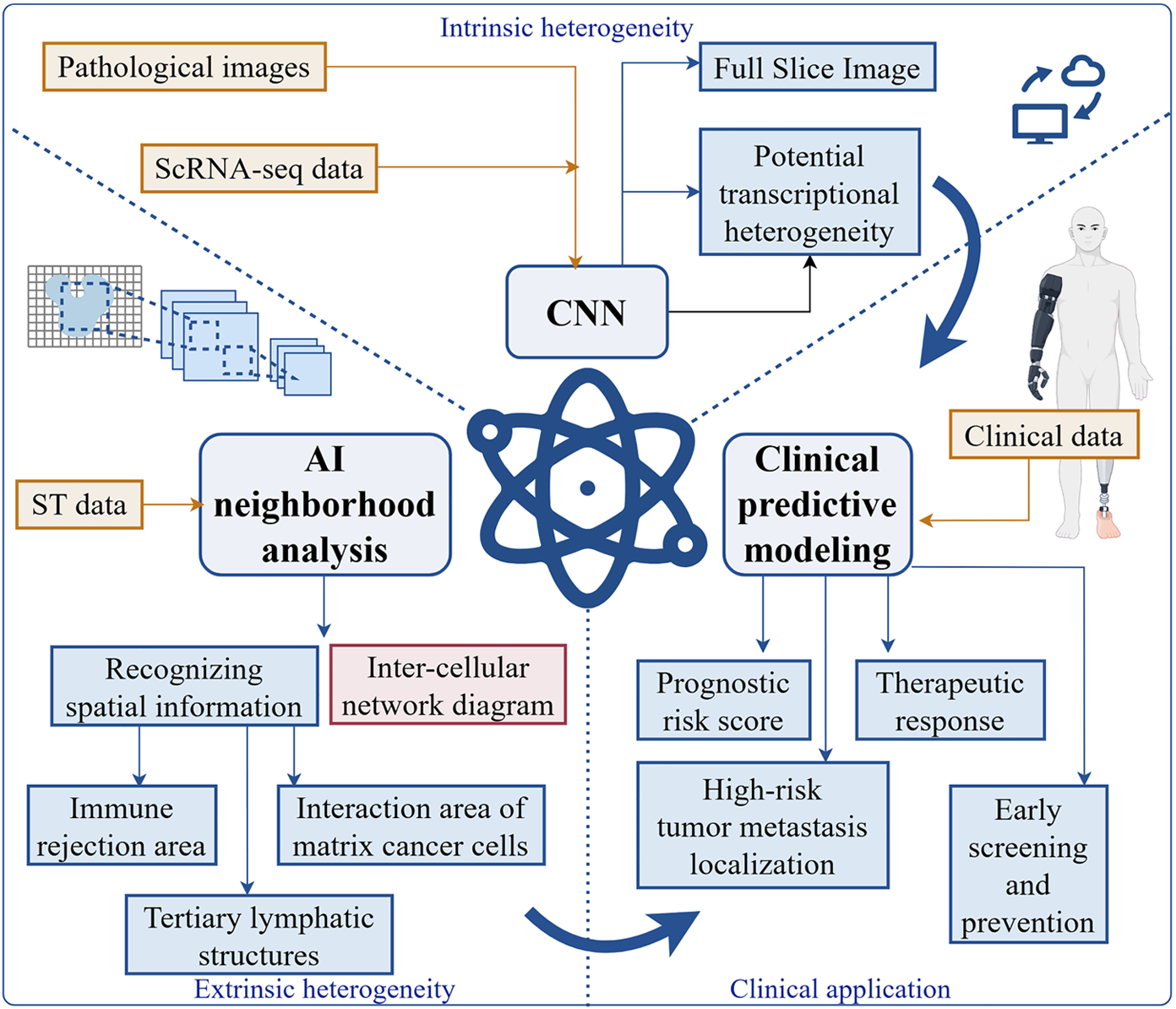
Figure 4 Artificial intelligence-enabled single-cell RNA sequencing and spatial transcriptomics-involved colorectal cancer heterogeneity analysis and its guided clinical decision-making.
This figure includes an artificial intelligence-based graphic neural convolutional network to drive clinical translation of colorectal cancer (CRC) intrinsic heterogeneity research through enhanced pathology image characterization and integrated data analysis. Clinical translation of CRC extrinsic heterogeneity research is driven by artificial intelligence-mediated neighborhood analysis revealing spatial information reorganization of intercellular networks, identification of immune-rejection regions highlighting stroma-cancer cell interactions, and characterization of tertiary lymphoid structures with extrinsic heterogeneity. These analyses support clinical predictive algorithms for prognostic risk scoring and treatment response prediction, enabling high-risk tumor metastasis localization and clinical precision therapy, as well as supporting early screening and prevention based on comprehensive clinical data. scRNA-seq: Single-cell RNA sequencing; CNN: Convolutional neural network; AI: Artificial intelligence; ST: Spatial transcriptomics.
- Citation: Luan WY, Zhao Q, Zhang Z, Xu ZX, Lin SX, Miao YD. Multidimensional decoding of colorectal cancer heterogeneity: Artificial intelligence-enabled precision exploration of single-cell and spatial transcriptomics. World J Gastrointest Oncol 2025; 17(10): 110661
- URL: https://www.wjgnet.com/1948-5204/full/v17/i10/110661.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i10.110661