©The Author(s) 2025.
World J Gastrointest Oncol. Oct 15, 2025; 17(10): 109398
Published online Oct 15, 2025. doi: 10.4251/wjgo.v17.i10.109398
Published online Oct 15, 2025. doi: 10.4251/wjgo.v17.i10.109398
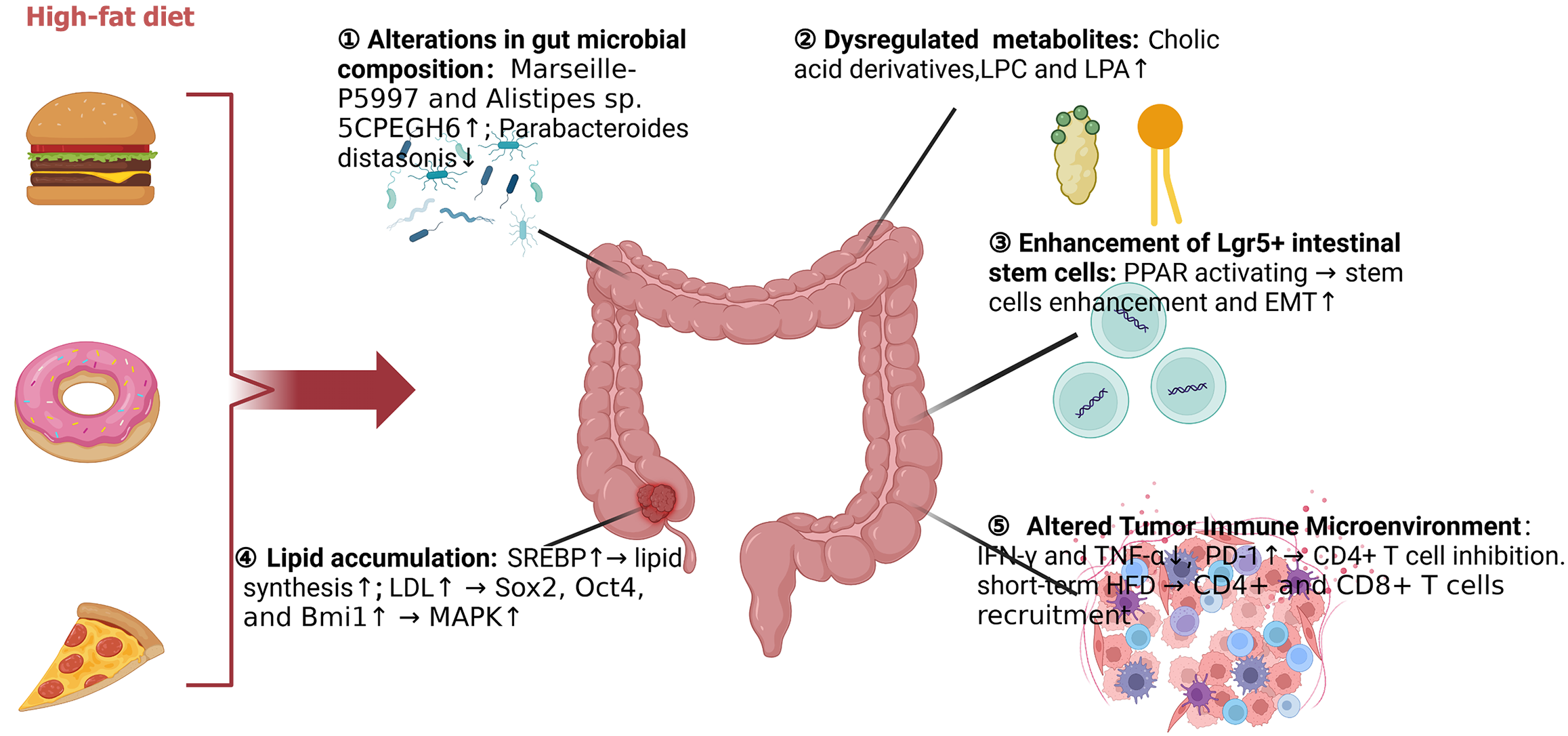
Figure 1 Schematic overview of high-fat diet-induced pathological remodeling in the intestinal microenvironment.
High-fat foods promote microbial dysbiosis and metabolic reprogramming, leading to stem cell expansion, lipid accumulation, and immunosuppression. LPC: Lysophosphatidylcholine; LPA: Lysophosphatidic acid; PPAR: Peroxisome proliferator-activated receptor; EMT: Epithelial-mesenchymal transition; IFN: Interferon; TNF: Tumor necrosis factor; PD-1: Programmed cell death protein 1; CD: Cluster of differentiation; SREBP: Sterol regulatory element-binding protein; LDL: Low-density lipoprotein; MAPK: Mitogen-activated protein kinase.
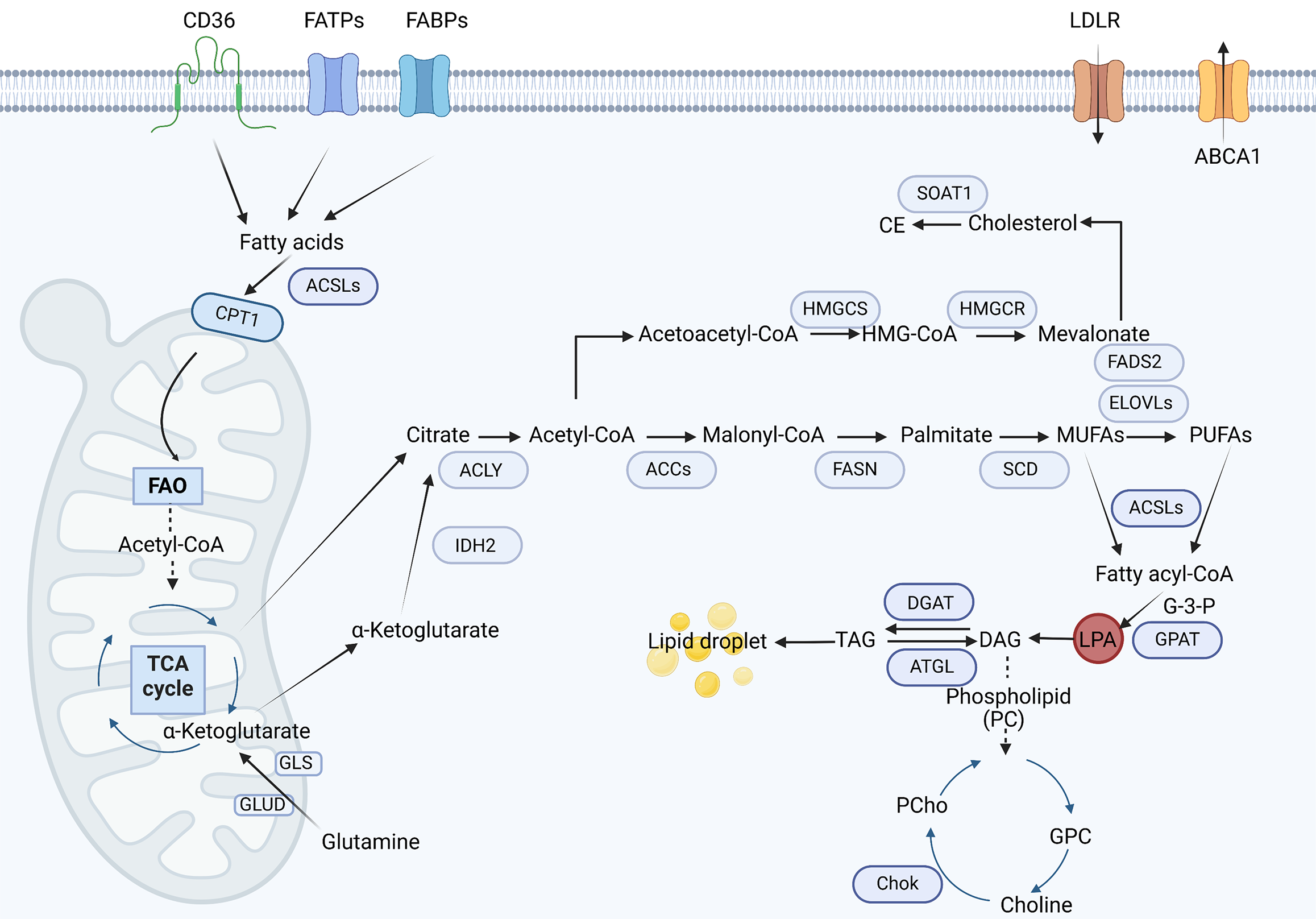
Figure 2 The process of lipid metabolism: Lipid metabolic processes include de novo lipogenesis, fatty acid uptake, fatty acid oxidation, cholesterol synthesis and lipid storage.
Tumor cells enhance lipid metabolism by augmenting exogenous lipid uptake and de novo lipogenesis, thereby elevating intracellular lipid content. Upregulation of lipid transport proteins-including cluster of differentiation 36, fatty acid transport proteins, fatty acid-binding proteins, and low-density lipoprotein receptor promotes the cellular internalization of saturated fatty acids (SFAs), polyunsaturated fatty acids, and cholesterol. Meanwhile, endogenous lipid synthesis originates from mitochondrial citrate exported to the cytosol via the tricarboxylic acid cycle citrate carrier (SLC25A1). This citrate is converted to acetyl-CoA by adenosine triphosphate (ATP)-citrate lyase, serving as a substrate for sterol regulatory element-binding protein-driven synthesis of SFAs and cholesterol. The process is catalyzed by key enzymes such as fatty acid synthase and stearoyl-CoA desaturase, which introduce double bonds into SFAs to generate monounsaturated fatty acids. Subsequently, fatty acids are activated by acyl-CoA synthetases to form acyl-CoA, which fuels either phospholipid synthesis (e.g., phosphatidylcholine via glycerol-3-phosphate acyltransferase/acylglycerol-3-phosphate acyltransferase enzymes) or fatty acid oxidation in mitochondria to generate ATP and support tumor growth. CD: Cluster of differentiation; FATPs: Fatty acid transport proteins; FABPs: Fatty acid-binding protein; LDLR: Low-density lipoprotein receptor; ACSLs: Acyl-CoA synthetase long chain; CPT: Carnitine palmitoyl transferase; FAO: Fatty acid oxidation; TCA: Tricarboxylic acid cycle; GLS: Glutaminase; GLUD: Glutamate dehydrogenase; IDH: Isocitrate dehydrogenase; ACLY: Adenosine triphosphate-citrate lyase; CE: Cholesterol ester; HMGCS: Hydroxy methyl glutaryl-CoA synthase; HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase; HMG: Hydroxymethylglutarate; FADS2: Fatty acid desaturase 2; ELOVLs: Elongation of very long-chain fatty acid protein; ACC: Acetyl-CoA carboxylase; FASN: Fatty acid synthase; SCD: Stearoyl-CoA desaturase; MUFAs: Monounsaturated fatty acids; PUFAs: Polyunsaturated fatty acids; GPAT: Glycerol-3-phosphate acyltransferase; LPA: Lysophosphatidic acid; DAG: Diacylglycerol; DGAT: Diacylglycerol acyltransferase; ATGL: Adipose triglyceride lipase; TAG: Triacylglycerol; PC: Phosphatidylcholine; PCho: Phosphorylcholine; ChoK: Choline kinase; GPC: Choline glycerophosphate.
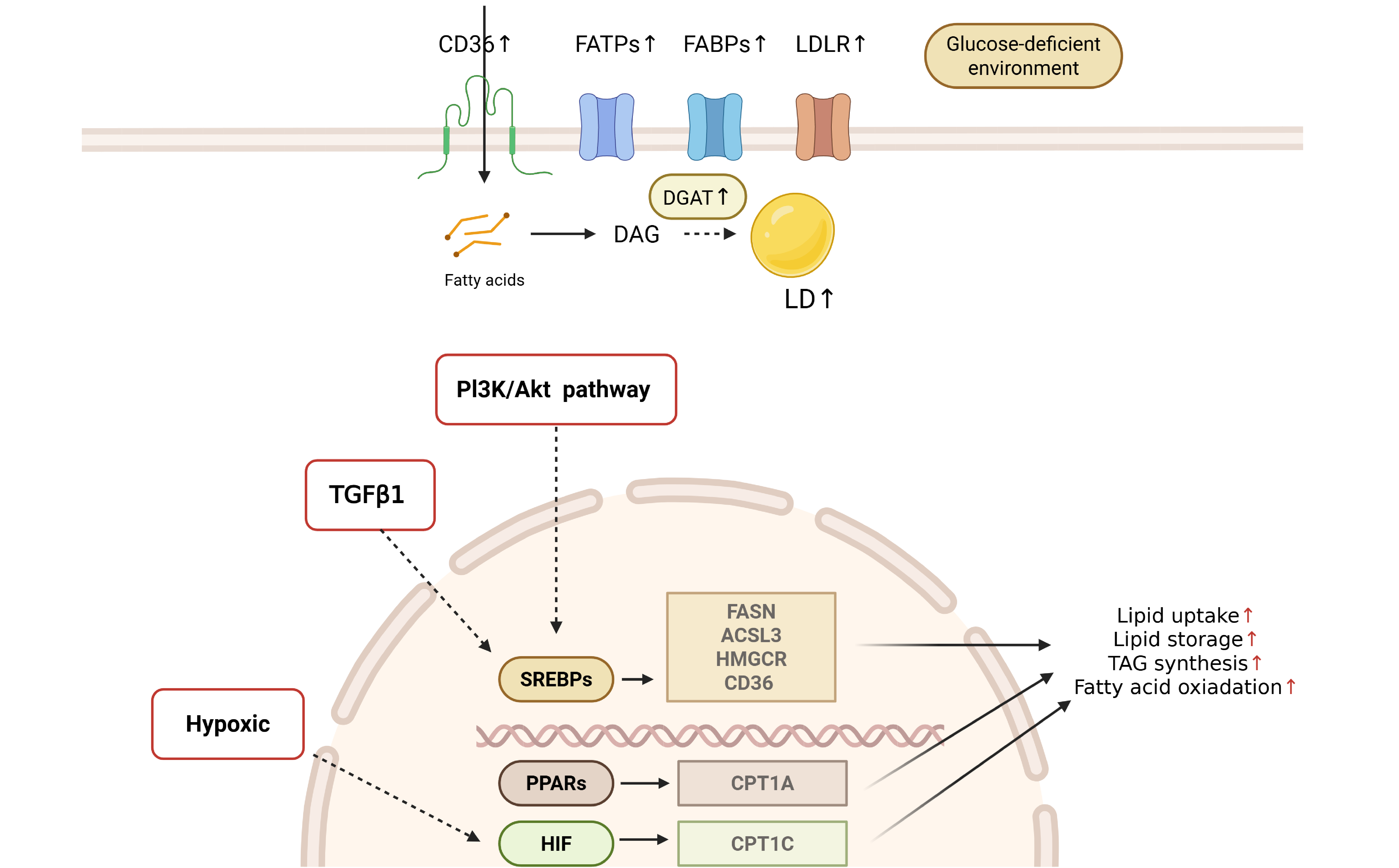
Figure 3 Lipid metabolic alterations in colorectal cancer cells.
Hypoxia and signal pathway including transforming growth factor-β1 and phosphatidylinositol 3-kinase/protein kinase B attributes to the reprogramming lipid metabolism in colorectal cancer cells. CD: Cluster of differentiation; FATPs: Fatty acid transport proteins; FABPs: Fatty acid-binding protein; LDLR: Low-density lipoprotein receptor; DAG: Diacylglycerol; DGAT: Diacylglycerol acyltransferase; LD: Lipid droplet; PI3K: Phosphatidylinositol 3-kinase; AKT: Protein kinase B; TGF: Transforming growth factor; TAG: Triacylglycerol; SREBP: Sterol regulatory element-binding protein; PPAR: Peroxisome proliferator-activated receptor; HIF: Hypoxia inducible factor; FASN: Fatty acid synthase; ACSL: Acyl-CoA synthetase long chain; HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase.
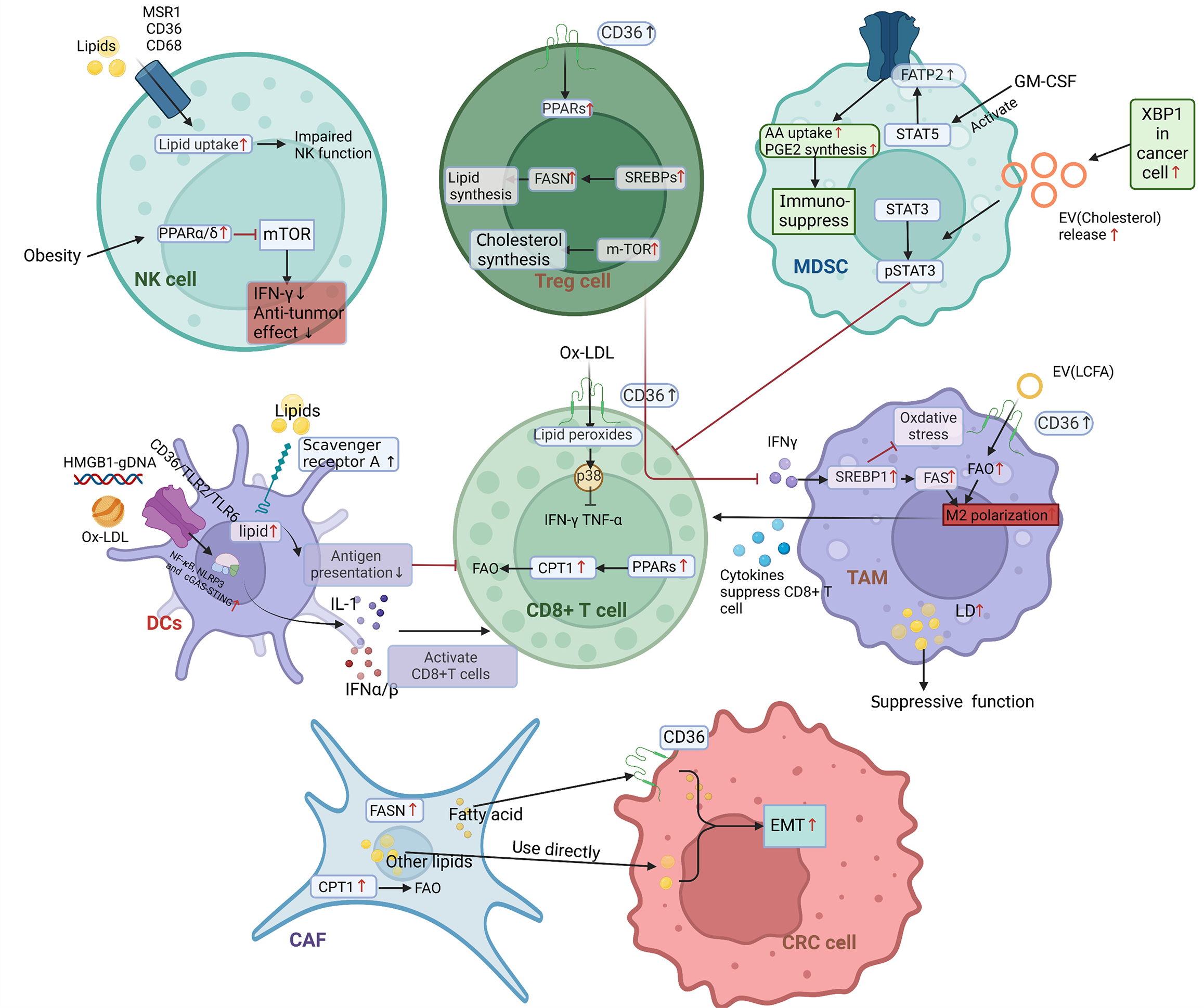
Figure 4 Mechanism of lipid metabolic reprogramming in immune cells.
Overexpression of cluster of differentiation (CD) 36 on CD8 + tumor-infiltrating lymphocytes (TILs) promotes metabolic transition by activating peroxisome proliferator-activated receptor (PPAR), which results in enhanced fatty acid oxidation (FAO). Lipid metabolism in regulatory T cells is enhanced by CD36-PPAR-β signaling, mammalian target of rapamycin-cholesterol signaling, and sterol regulatory element-binding protein (SREBP)-mediated overexpression of fatty acid synthase (FASN). Tumor-associated macrophages can uptake exosomes containing long-chain fatty acids, SREBPs-mediated overexpression of FASN. These effects together promote M2 polarization. Natural killer (NK) cells uptake lipids and activate PPAR signaling, which impairs NK cells’ function. In dendritic cells, CD36/toll-like receptor (TLR) 2/TLR6 complex internalizes oxidized low-density lipoprotein, activates nuclear factor kappa-B/NLRP3/cyclic guanosine monophosphate-adenosine monophosphate synthase-stimulator of interferon genes signaling and CD8 + TILs. Overexpression of fatty acid transport protein 2 in myeloid-derived suppressor cells mediates arachidonic acid uptake and upregulate prostaglandin E2 synthesis. Cancer-associated fibroblasts upregulate FASN and enhance FAO through increasing carnitine palmitoyl transferase 1, which finally promotes epithelial-mesenchymal transition of colorectal cancer cells. CD: Cluster of differentiation; NK: Natural killer; mTOR: Mammalian target of rapamycin; IFN: Interferon; PPAR: Peroxisome proliferator-activated receptor; FASN: Fatty acid synthase; SREBP: Sterol regulatory element-binding protein; Treg: Regulatory T cell; AA: Arachidonic acid; PGE2: Prostaglandin E2; MDSCs: Myeloid-derived suppressor cells; FATP2: Fatty acid transport protein 2; STAT: Signal transducer and activator of transcription; p-STAT: Phospho-signal transducer and activator of transcription; GM-CSF: Granulocyte-macrophage colony-stimulating factor; XBP1: X-box binding protein 1; EV: Extracellular vesicle; HMGB1: High mobility group box-1 protein; OxLDL: Oxidized low-density lipoproteins; TLR: Toll-like receptor; DCs: Dendritic cells; NF-κB: Nuclear factor kappa-B; cGAS/STING: Cyclic guanosine monophosphate-adenosine monophosphate synthase-stimulator of interferon genes signaling; IL: Interleukin; IFN: Interferon; TNF: Tumor necrosis factor; FAO: Fatty acid oxidation; CPT: Carnitine palmitoyl transferase; LCFA: Long-chain fatty acids; LD: Lipid droplet; TAM: Tumor-associated macrophages; CAF: Cancer-associated fibroblast; EMT: Epithelial-mesenchymal transition; CRC: Colorectal cancer.
- Citation: Wang CD, Zhang BX, Song J. Lipid metabolic reprogramming in colorectal cancer: Insights to mechanisms and therapeutics. World J Gastrointest Oncol 2025; 17(10): 109398
- URL: https://www.wjgnet.com/1948-5204/full/v17/i10/109398.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i10.109398