Published online Feb 27, 2026. doi: 10.4254/wjh.v18.i2.113348
Revised: October 23, 2025
Accepted: December 23, 2025
Published online: February 27, 2026
Processing time: 174 Days and 9.1 Hours
Chronic hepatitis B (CHB) is a leading cause of liver-related mortality, progres
To develop and validate a predictive model for EHLI in CHB patients using a cohort from Hunan Province, China, to facilitate early risk identification and opti
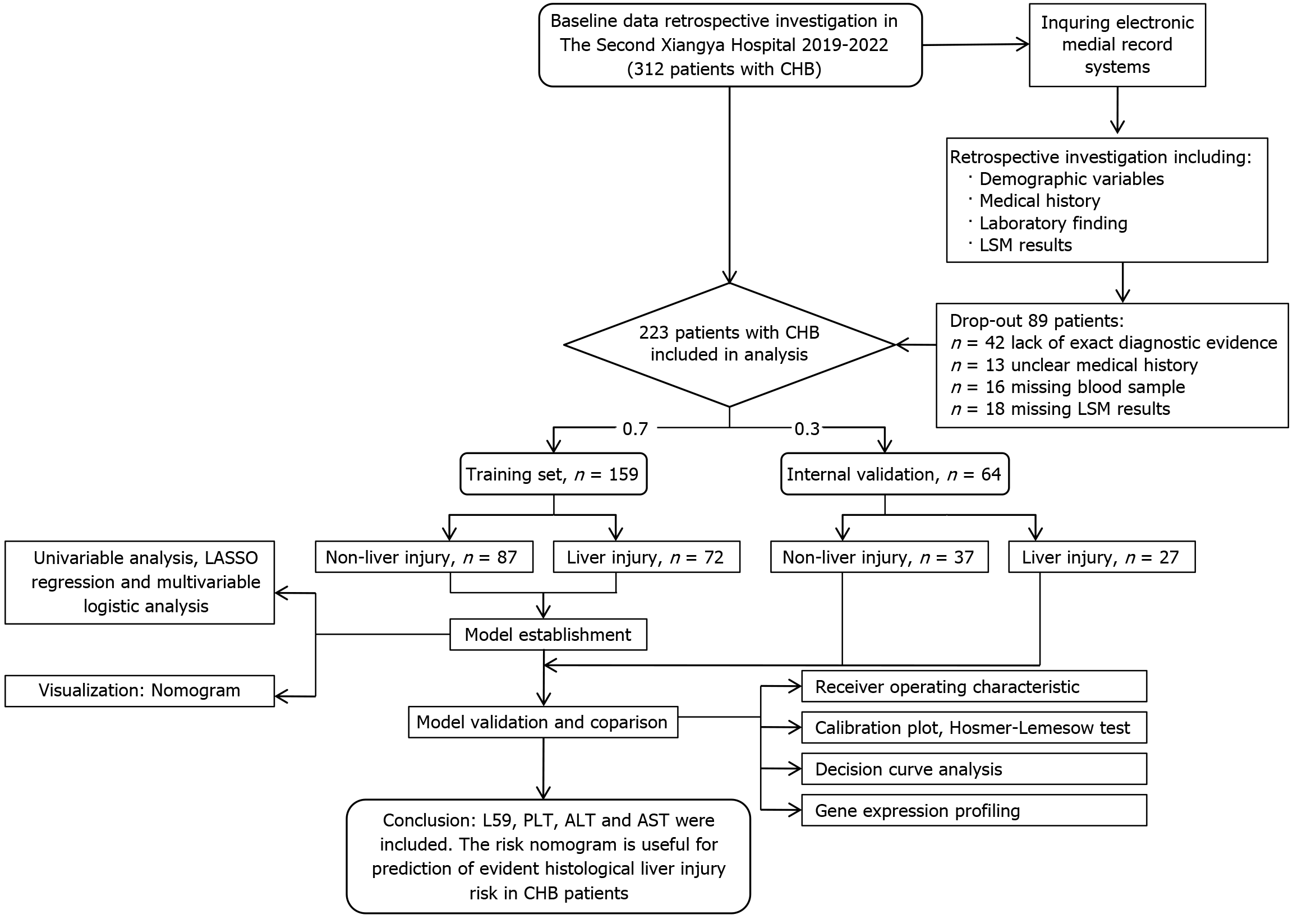
This observational real-world study enrolled 223 CHB patients (August 2020 to March 2022) from the Second Xiangya Hospital, divided into development (n = 159) and validation (n = 64) cohorts (7:3 ratio). EHLI was defined as Ishak fibrosis stage ≥ 3 and/or histologic activity index ≥ 9. Variables were screened via uni
L59, platelet count (PLT), alanine transaminase (ALT), and aspartate transaminase (AST) were identified as independent predictors of EHLI. The model showed high discriminative ability, with AUC of 0.921 [95% con
A novel nomogram incorporating L59, PLT, ALT, and AST robustly predicts EHLI in CHB patients. This model, using routinely measured variables, aids clinical decision-making and optimizes resource allocation.
Core Tip: This study developed and validated a novel nomogram incorporating L59, platelet count, alanine aminotransferase, and aspartate aminotransferase to predict evident histological liver injury (Ishak ≥ 3 or histologic activity index ≥ 9) in chronic hepatitis B patients. The model demonstrated high accuracy (area under the curve 0.921-0.959) and clinical utility, supported by transcriptomic insights implicating COL1A2 and transforming growth factor-β/Smad pathways. This tool enables noninvasive risk stratification, optimizing resource allocation and guiding early intervention.
- Citation: Dai ZS, Cao X, Jiang YF, He B. Predictive tool for evident histological liver injury in chronic hepatitis B patients: Development and validation. World J Hepatol 2026; 18(2): 113348
- URL: https://www.wjgnet.com/1948-5182/full/v18/i2/113348.htm
- DOI: https://dx.doi.org/10.4254/wjh.v18.i2.113348
Chronic hepatitis B (CHB) is a leading cause of liver-related mortality and is already the leading etiology of liver disease requiring liver transplantation[1,2]. Persistent hepatitis B virus (HBV) infection inexorably progresses to liver fibrosis, cirrhosis and end-stage liver disease, and potentially leads to hepatocellular carcinoma (HCC)[3]. The burden of end-stage liver disease is expected to increase over the coming decade given the high prevalence of HBV infection[4].
Noninvasive screening techniques for liver histological liver injury mainly include serological markers and liver stiffness measurement (LSM). Serological markers are mainly evaluated based on the aspartate aminotransferase to platelet ratio index (APRI) and fibrosis-4 index (FIB-4) scores; both of which have the advantages of being simple, practical, and easy to obtain in the clinic. APRI exhibits low accuracy in assessing HBV-related fibrosis[5], while dynamic changes in FIB-4 fail to reflect fibrosis reversal after antiviral therapy[6]. These limitations highlight the need for a more robust noninvasive tool for evident histological liver injury (EHLI) prediction. LSM is widely used in clinical practice and is able to accurately identify progressive hepatic fibrosis and early cirrhosis, but it has been found to be unstable and is affected by a variety of factors such as hepatic inflammation and necrosis, cholestasis, and severe steatosis[7,8]. Liver biopsy has limited clinical use due to its invasiveness and poor acceptability, and its popularity is further limited by sampling, subject variability, and postoperative complications[9]. Traditional noninvasive screening techniques and liver biopsy still have limitations in the assessment of probability of EHLI in patients with HBV infection, so new models based on CHB prognostic-related risk factors are constantly being explored.
In seven cohorts of secondary/tertiary hepatology clinics, Sanyal et al[10] reported that risk factors associated with development of advanced fibrosis and cirrhosis included age, LSM, aspartate transaminase (AST)/alanine transaminase (ALT) ratio, platelets, sex and diabetes status. They concluded that their two novel noninvasive scores (Agile 4 and Agile 3+) improved identification of cirrhosis or advanced fibrosis among individuals attending liver clinics and reduced the need for liver biopsy in this population. Chang et al[11] established an observational study of patients with hepatitis B e antigen (HBeAg)-positive chronic HBV infection patients from 14 centers in China and constructed a predictive risk score. Age, LSM, ALT, alkaline phosphatase, and albumin were identified as independent predictors for EHLI and used to develop a nomogram that demonstrated good performance in predicting EHLI with area under the receiver operating characteristic curve (AUROC) of 0.92 in the training cohort.
The early identification of individuals at high risk for EHLI is imperative, as it facilitates the provision of appropriate care and optimizes the allocation of limited resources. To assist in identifying patients likely to develop EHLI upon hospital admission, we sought to develop and validate a predictive model derived from a cohort of patients with CHB in Hunan Province, China.
This study was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University (No. 2020-047). The study was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from participants or their families for prospectively collected data.
This real-world and observational study recruited patients with CHB from the Second Xiangya Hospital of Central South University, from August 2020 to March 2022. All participants had a clear diagnosis of CHB according to WHO interim guidance, based on the positive high-throughput sequencing or real-time reverse transcription polymerase chain reaction for blood specimens[12]. Demographic variables, laboratory findings, and LSM results were extracted from electronic medical records and through interviews with attending physicians. A team of trained physicians and medical students was responsible for data collection.
Demographic variables collected for the study included sex and age. Laboratory findings included L (59) LAP-DPs, CHI3 L1, platelet count (PLT), ALT, AST, total protein, albumin, total bilirubin, direct bilirubin, HBV-DNA level, hepatitis B surface antigen, HBeAg, and hepatitis B core antibody. The FIB-4 was calculated by the formula: Age (years) × AST (U/L)/[PLT (109/L) × ALT (U/L)1/2][13]. APRI was calculated by the formula: AST level (ULN)/PLT (109/L) × 100[14]. The LSM by vibration-controlled transient elastography (FibroScan, Echosens, France) result ranged from 1.0 kPa to 80 kPa.
Patients were divided into the EHLI group (n = 99) and non-EHLI group (n = 124). We defined CHB EHLI as Ishak fibrosis staging ≥ 3 and/or histologic activity index (HAI) ≥ 9[15]. We chose this composite end point since CHB EHLI outcomes such as Ishak fibrosis staging and HAI have been used in prior studies to gauge the seriousness of liver histological damage[16,17].
Baseline characteristics and laboratory indicators of patients were presented as categorical variables, expressed in terms of frequencies and percentages. Stratified randomization and split-sample techniques were used to divide the study population into development (n = 159) and validation (n = 64) cohorts with a 7:3 ratio. This maintained categorical variable proportions consistent with original data distribution. Initial variable selection was conducted using univariable logistic regression. Variables for the univariable logistic regression were drawn from existing clinical practice, previous literature, and statistically significant variables. Review of existing literature guided the inclusion of variables related to EHLI in CHB, supplemented by indicators derived from clinical experience[11,17]. Thirty-two variables were included in the selection process as described here. Predictive features were selected from the original dataset using least absolute shrinkage and selection operator (LASSO) regression, renowned for high-dimensional predictor selection.
The predictive model was constructed using variables that demonstrated prognostic significance in the multivariable logistic regression analysis. Ultimately, a nomogram was developed to visualize the model. The predictive accuracy of the multivariable logistic regression model in the development and validation cohorts was evaluated using AUROC, receiver operating characteristic (ROC) curve, Hosmer-Lemeshow test, calibration plot, and decision curve analysis (DCA). To internally validate accuracy estimates and minimize overfitting, we used 1000 bootstrap resamples, considering P < 0.05 as statistically significant.
The analytical pipeline was executed as follows. Raw data underwent pre-processing, including encoding of categorical variables and standardization of continuous features. The LASSO regression analysis was performed using the “glmnet” package in R. A sequence of potential regularization parameters (λ) was generated, and the optimal λmin was identified through 10-fold cross-validation. These predictors were used to fit the final multivariable logistic regression model, whose coefficients were estimated via maximum likelihood. The probability of EHLI is subsequently derived using the logistic function: P (EHLI) = 1/(1 + e-LP). This probability was used to generate the receiver operating characteristic curves and to perform the DCA presented in the results.
The gene expression profiles used for bioinformatic analysis were sourced from the Gene Expression Omnibus database. The specific dataset utilized was under the accession number GSE84044, which was generated on the GPL570 platform. This dataset comprised a total of 124 samples, including 81 liver tissue samples from CHB patients and 43 samples from healthy controls. Gene expression profiles of CHB patients were analyzed to identify immune-related differentially expressed genes (DEGs). The methodology used included weighted gene co-expression network analysis for constructing networks, machine learning algorithms for biomarker identification, formation of subclusters associated with immune reactions, and single-sample gene set enrichment analysis to investigate correlation of hub genes with immune cell infiltration and immune pathway activation.
Descriptive statistics are articulated as interquartile ranges for continuous variables and as n (%) for categorical variables. Differences in the distribution of patient characteristics between the EHLI and non-EHLI cohorts are expressed with 95% confidence intervals (CI). Continuous variables underwent analysis via the Mann-Whitney U test, while categorical variables were compared between subgroups using the χ2 test or Fisher’s exact test. Statistical analyses were executed using R software (version 4.1.3, R Foundation), with significance defined at P < 0.05.
Figure 1 illustrates the study workflow. Of the 312 patients, 89 were excluded based on the exclusion criteria, resulting in the enrollment of 223 patients. As demonstrated in Table 1, detailed baseline characteristics and laboratory data were obtained from the electronic medical records of these patients. Data for the development cohort were gathered from 159 patients at the Second Xiangya Hospital of Central South University, Hunan Province, between August 2020 and March 2022. At hospital admission, 99 of the 223 patients (44.3%) exhibited EHLI, while the remaining 124 patients (55.7%) displayed non-EHLI as assessed by the Ishak fibrosis staging and HAI. The mean age of patients in the cohort was 46.28 ± 10.84 years, with 115 (72.3%) being men. Comprehensive laboratory findings for the development cohort are detailed in Table 1.
| Characteristics | Total (n = 159) | Evident histological liver injury | P value | |
| No (n = 87) | Yes (n = 72) | |||
| Demographic variables | ||||
| Male | 115/159 (72.3) | 55/87 (63.2) | 60/72 (83.3) | 0.005 |
| Age, year | 46.28 (10.84) | 42.98 (10.08) | 50.26 (10.44) | < 0.001 |
| Laboratory findings | ||||
| PLT, × 109/L | 167.87 (71.18) | 199.38 (59.86) | 129.81 (65.17) | < 0.001 |
| ALT, U/L | 41.20 (59.35) | 27.69 (23.82) | 57.52 (81.59) | 0.001 |
| AST, U/L | 35.45 (36.51) | 24.56 (10.74) | 48.61 (50.06) | < 0.001 |
| TP, g/L | 75.48 (5.01) | 75.56 (4.81) | 75.38 (5.27) | 0.83 |
| ALB, g/L | 46.46 (4.49) | 47.85 (3.49) | 44.79 (5.00) | < 0.001 |
| TBIL, μmol/L | 16.91 (15.96) | 15.27 (18.97) | 18.90 (11.14) | 0.15 |
| DBIL, μmol/L | 6.19 (8.66) | 4.90 (8.96) | 7.76 (8.06) | 0.04 |
| HBV-DNA, × 105 IU/mL | 17.68 (135.71) | 31.04 (182.56) | 1.55 (11.71) | 0.17 |
| HBsAg, IU/mL | 1439.43 (6674.76) | 1938.84 (8631.85) | 835.98 (2867.01) | 0.30 |
| HBeAg, IU/mL | 93.11 (571.93) | 166.89 (767.11) | 3.95 (20.55) | 0.05 |
| HBcAb, IU/mL | 187.65 (313.01) | 222.54 (324.45) | 145.48 (295.34) | 0.12 |
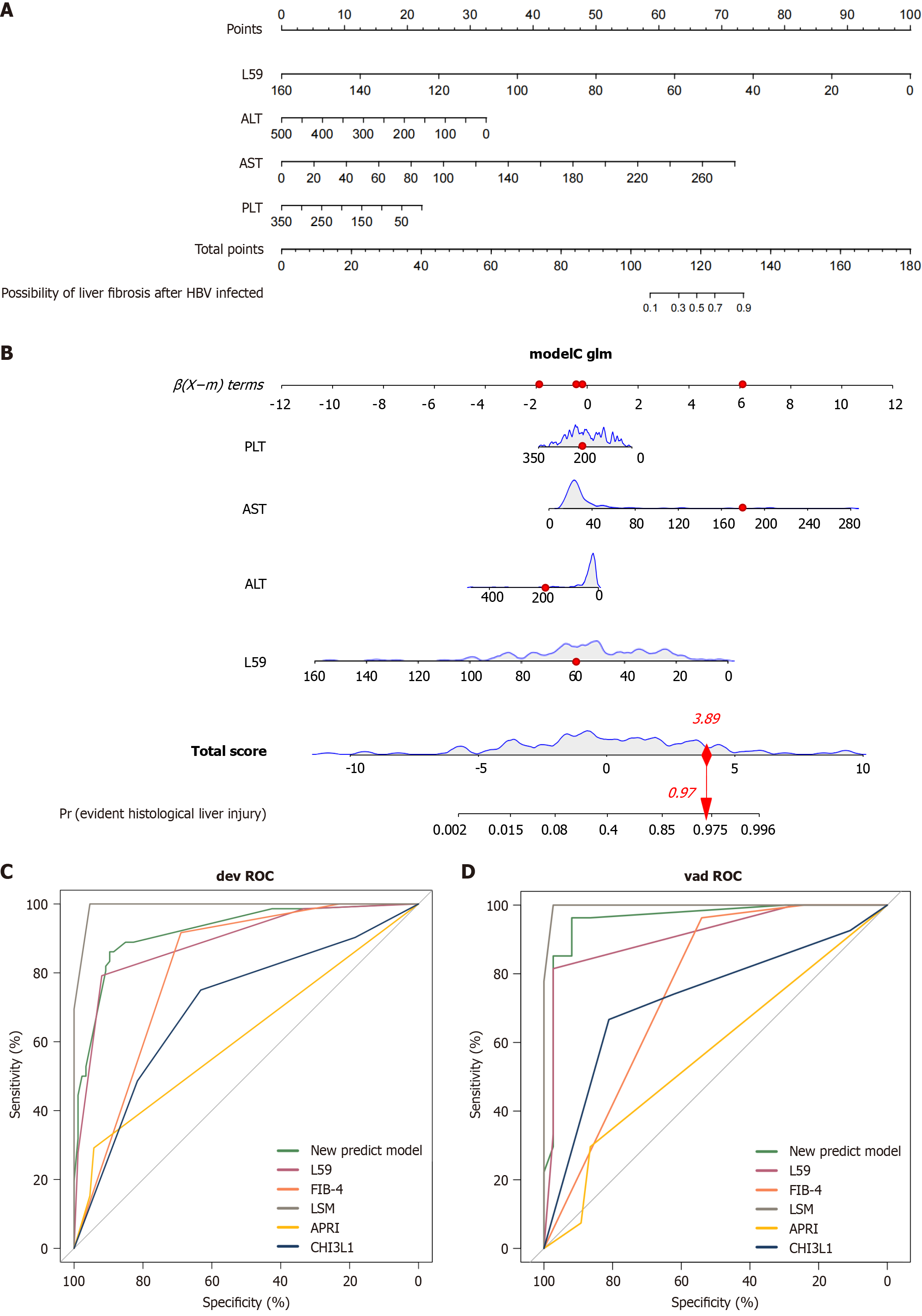
Initially, these parameters were analyzed through univariable logistic regression. Significant variables processed via LASSO logistic regression were distilled to the most robust indicators, enhancing our predictive modeling framework. Table 2 presents the variables identified in the univariable logistic regression analysis. LASSO logistic regression was used to select features (Supplementary Figure 1). The minimum value (λ_min) served as the screening criterion. When λ_min reached 0.032, the indicators in the development cohort were narrowed down to five potential predictors by reducing the total absolute value of the regression coefficients below a specified threshold. The four predictive factors identified by the LASSO logistic regression analysis included LSM, L59, PLT, ALT, and AST, with corresponding LASSO coefficients of 3.235320, -0.105827, -0.475601, -0.321742, and -0.284133, respectively. The intercept of the equation was calculated to be 1.022.
| Factor | Univariable analysis | Multivariable analysis | ||||
| OR | 95%CI | P value | OR | 95%CI | P value | |
| Male | 0.34 | 0.16-0.73 | 0.006 | |||
| Age, year | 2.04 | 1.43-2.92 | < 0.001 | |||
| L59 | 0.07 | 0.03-0.15 | < 0.001 | 0.15 | 0.06-0.38 | < 0.001 |
| PLT, × 109/L | 2.24 | 1.37-3.66 | 0.001 | 2.71 | 1.09-6.75 | 0.03 |
| ALT, U/L | 1.79 | 1.02-3.18 | 0.048 | 7.20 | 1.35-28.8 | 0.02 |
| AST, U/L | 5.56 | 2.14-14.47 | < 0.001 | 8.40 | 1.10-63.8 | 0.04 |
| ALB, g/L | 5.69 | 1.23-26.33 | 0.026 | |||
| TBIL, μmol/L | 2.31 | 1.33-4.02 | 0.003 | |||
| DBIL, μmol/L | 3.54 | 1.79-6.99 | < 0.001 | |||
Incorporating these nine variables into a multivariable logistic regression model yielded four variables as independent predictors statistically significant for EHLI, which were subsequently integrated into the risk score. These variables comprise L59 (OR = 0.15; 95%CI: 0.06-0.38; P < 0.001), PLT (OR = 2.71; 95%CI: 1.09-6.75; P = 0.03), ALT (OR = 7.20; 95%CI: 1.35-28.8; P = 0.02), and AST (OR = 8.40; 95%CI: 1.10-63.8; P = 0.04) (Table 2). Figure 2 illustrates the nomogram developed to estimate the probability of EHLI in CHB patients, building upon the predictive model including L59, PLT, ALT, and AST.
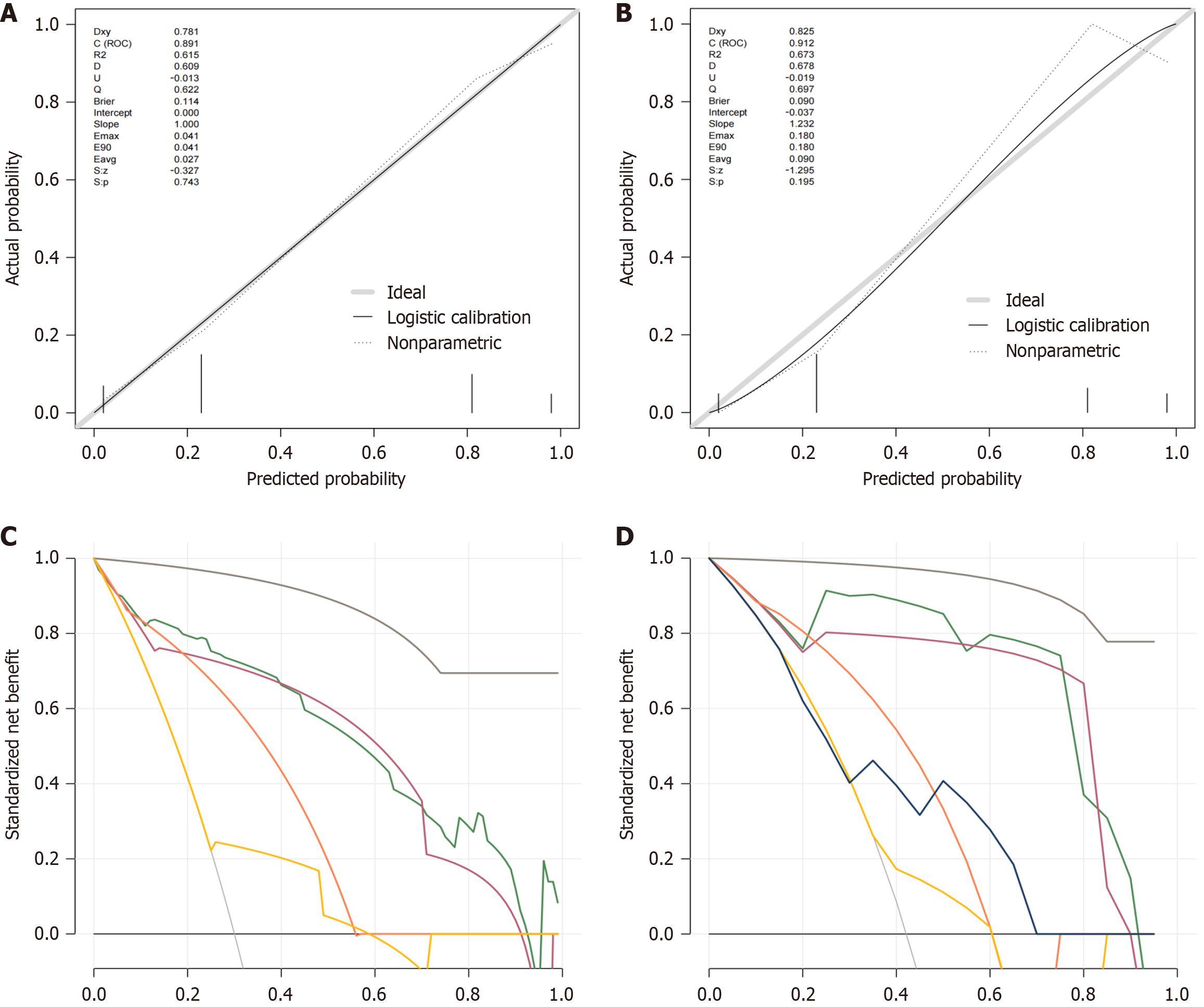
The performance of the multivariable logistic regression model and nomogram was evaluated using area under the curve (AUC), Hosmer-Lemeshow test, calibration plots, and DCA. ROC curves for the likelihood of EHLI were constructed for the development and validation cohorts. The mean AUC for predicting the likelihood of EHLI from the development cohort data was 0.921 (95%CI: 0.880-0.963) (Figure 2). To further assess the generalizability of the model, we performed 1000 bootstrap validations. The accuracy of the CHB risk score in the validation cohort closely mirrored that of the development cohort, with an AUC of 0.959 (95%CI: 0.910-1.0), signifying the strong discriminatory capability of the model. Supplementary Table 1 provides the complete set of performance metrics (AUC, accuracy, sensitivity, specificity, precision, and F1-score) for our nomogram and all comparison models in the training and validation sets. To evaluate the importance of each variable, shown in Supplementary Figure 2, we performed an ablation study. The results de
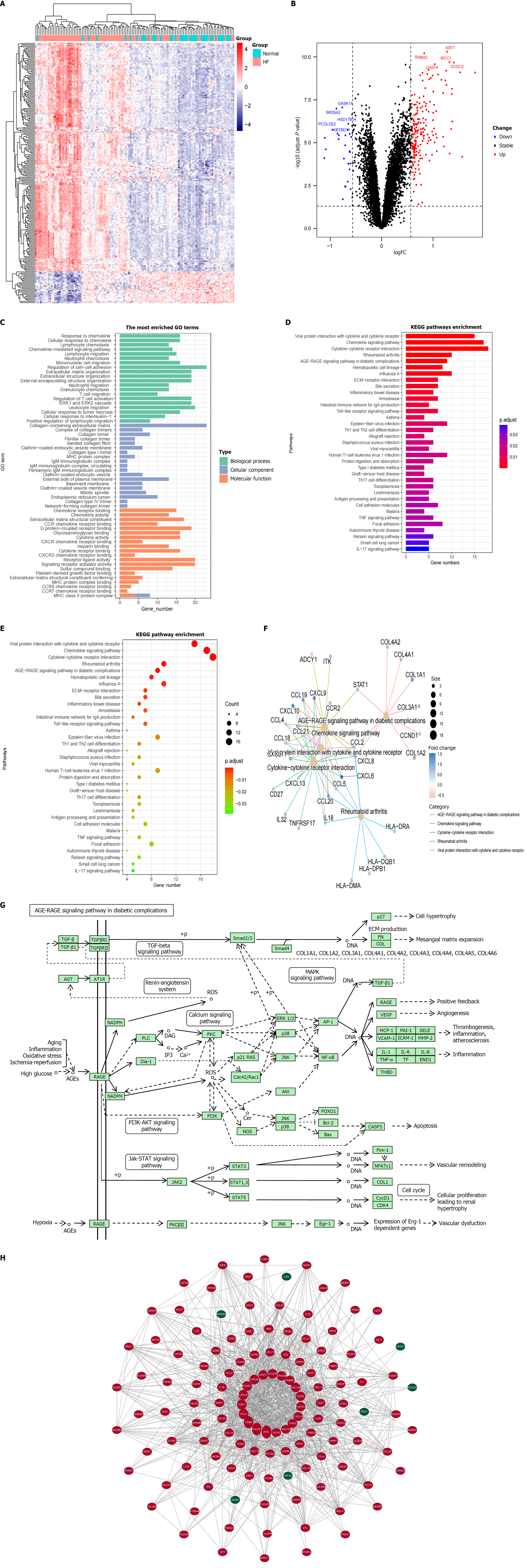
The study identified 210 DEGs that distinguish healthy individuals from CHB patients, underscoring the critical role of the immune response in disease progression (Figure 4). Five hub genes, COL1A2, COL3A1, COL1A1, COL4A1, and COL4A2, validated as potential diagnostic biomarkers for high-risk CHB patients. Kyoto Encyclopedia of Genes and Genomes pathway analysis enriched the transforming growth factor (TGF)-β/Smad pathway, with L59 reflecting activation of TGF-β, suggesting the potential influence of L59 on liver histological fibrosis injury.
In this study, we developed and validated a clinical risk score and a novel nomogram to predict the onset of EHLI in patients infected with HBV. The risk score and nomogram demonstrated satisfactory performance, achieving AUC-based accuracies of 0.92 and 0.95 in the development and validation cohorts, respectively. Calibration and clinical applicability were robustly demonstrated across both cohorts. The four variables necessary to calculate the risk of EHLI are typically gathered during routine patient data collection, namely, L59, PLT, ALT, and AST. The risk-factor-based nomogram allows clinicians to easily and efficiently estimate the likelihood of EHLI in patients with HBV infection.
The therapeutic goals of CHB are to maximize long-term inhibition of HBV replication, reduce hepatocellular inflammation and necrosis and hepatic fibrous tissue proliferation, delay and reduce the onset of cirrhosis loss, improve quality of life, and prolong survival time[18]. To assess the severity of the histological liver injury at an early stage and optimize the allocation of resources, several studies to predict the risk factors of EHLI with CHB have been carried out. APRI and FIB-4 are two classic scales used to assess the degree of liver fibrosis from CHB. However, the risk assessment of EHLI remains controversial. Innes et al[19] used a two-stage study design to assess the performance of existing risk scores for predicting cirrhosis complications in the community. They found that the cumulative incidence of cirrhosis complications at 10 years was 0.58% from 197509 United Kingdom biobank participants and the top performing risk scores were FIB-4 (C-index 0.78; 95%CI: 0.76-0.79) and APRI (C-index 0.80; 95%CI: 0.78-0.82). APRI and FIB-4 accessible risk scores, in particular, can be repurposed to assess 10-year risk of cirrhosis morbidity in the community. Li et al[20] found that among patients who had liver-related events at 5 years, using the high cut-off, SAFE score could predict 84.9% of patients accurately, compared to 40.9% for FIB-4 and 27.2% for APRI. Zhang et al[21] aimed to validate the performance of the Baveno VII algorithm in patients with HBV-related cirrhosis and they found that missed high-risk varices rates were > 5% for the measure: 11.3% for LSM-longitudinal spleen diameter to platelet ratio score. As shown by AUC, calibration plots and DCA threshold probabilities, compared with APRI, FIB-4 and LSM score, our new predictive model demonstrated good discrimination and calibration in predicting the probability of EHLI with CHB, indicating good performance and predictive value for clinical use. In this study, we confirmed that the four indicators were significant predictive factors associated with progression of HBV infection to EHLI.
L59, PLT, ALT, and AST were included in the CHB risk score. Previous studies have found several of these variables to be risk factors for EHLI related to CHB. Chun et al[22] found that older age, male, lower PLT, comorbidity with diabetes, and moderate HBV DNA levels were associated with a higher risk of developing HCC in patients infected with HBV. In addition, their model validation showed a time-dependent AUROC of 0.81 for the prediction of HCC development at 5 years. When stratified by risk score, HCC risk was significantly higher in high-risk than in low-risk patients (sub-distribution hazard ratio = 8.43, P < 0.001). Ndow et al[23] found that the high-risk patients with HBV-related cirrhosis had lower PLT, partial or complete portal thrombosis, and above-moderate ascites. However, compared to the PAGE-B score and our predictive model, the cohort was small and confined to Africa, and lacked external validation, affecting its generalizability. Kim et al[24] discovered that in HBV-infected patients, higher ALT, older age, higher α-fetoprotein, and moderate or higher levels of HBV DNA were linked to a higher chance of developing HCC. RWS-HCC and AASL-HCC models were valuable and practical tools for screening CHB patients who were at risk of progression to HCC and required additional early intervention.
The transcriptomic analysis was conducted to provide biological plausibility for the clinical model, particularly for its novel component, L59. The identification of the TGF-β/Smad pathway as significantly enriched directly supports the role of L59, which reflects TGF-β activation, in CHB-related liver injury. Upregulation of key hub genes involved in collagen deposition (e.g., COL1A2, COL3A1, and COL1A1) provides a molecular counterpart to the histological fibrosis that the model predicts. L59 reflects TGF-β activation, and while the precise mechanisms in CHB remain uncertain, our bioinformatic analysis and prior evidence linking L59 to fibrosis stages[25] support its inclusion in the model. This integration of clinical and genomic data significantly strengthens the biological rationale for our predictor.
Our study had several limitations. Firstly, our cohort was not population-based, with participants recruited from a tertiary hospital in China, potentially limiting the generalizability of our findings to the wider Chinese CHB patient population. This limited sample size also manifested in two specific ways: It contributed to wide CIs for estimates of biomarkers with high biological variability (e.g., ALT and AST). For this statistical issue, we used LASSO and bootstrap resampling to enhance stability, and the robust validation performance (AUC 0.959) of the model mitigates concern regarding overfitting. Secondly, the absence of complete laboratory testing for all patients resulted in a reduced sample size for nomogram construction. Thirdly, future research should aim to include a larger patient cohort to enhance the application of this predictive model and verify its clinical practicality.
We developed a risk score and an innovative nomogram designed to estimate the likelihood of EHLI in CHB patients, utilizing four variables routinely assessed upon hospital admission. The model exhibited strong performance and could aid clinicians in decision-making processes, thereby optimizing the allocation of medical resources. Future work will focus on the external validation of this nomogram in multicenter, observational cohorts and the development of a user-friendly web-based calculator to facilitate its adoption in routine clinical practice.
We would like to thank all participants for collecting the data of this study.
| 1. | Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1260] [Cited by in RCA: 1261] [Article Influence: 157.6] [Reference Citation Analysis (6)] |
| 2. | Jiang P, Jia H, Qian X, Tang T, Han Y, Zhang Z, Jiang L, Yu Z, Zheng L, Yu G, Cai H, Zhang S, Zhang X, Gu J, Ye C, Yang L, Lu Y, Liu H, Lu X, Jin C, Ren Y, Lu M, Xu L, Yu J, Jin X, Yang Y, Qian P. Single-cell RNA sequencing reveals the immunoregulatory roles of PegIFN-α in patients with chronic hepatitis B. Hepatology. 2024;79:167-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 34] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 3. | Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM, Brown RS Jr, Bzowej NH, Wong JB. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67:1560-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2290] [Cited by in RCA: 3100] [Article Influence: 387.5] [Reference Citation Analysis (1)] |
| 4. | Lin C, Luo L, Xun Z, Zhu C, Huang Y, Ye Y, Zhang J, Chen T, Wu S, Zhan F, Yang B, Liu C, Ran N, Ou Q. Novel function of MOTS-c in mitochondrial remodelling contributes to its antiviral role during HBV infection. Gut. 2024;73:338-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 5. | Duah A, Nartey YA. Clinical Profile and Limitations in the Management of HBV Patients Attending Clinic at a District Hospital in Ghana. Int J Hepatol. 2023;2023:4424718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Tsai MY, Yen YH, Huang PY, Sou FM, Lin CC, Cho WR, Wang HM, Chen DW, Chang KC, Wu CK, Hu TH, Tsai MC. The Pre- and Postoperative FIB-4 Indexes Are Good Predictors to the Outcomes of HBV-Related HCC Patients after Resection. Gastroenterol Res Pract. 2019;2019:8945798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 7. | de Franchis R, Bosch J, Garcia-Tsao G, Reiberger T, Ripoll C; Baveno VII Faculty. Baveno VII - Renewing consensus in portal hypertension. J Hepatol. 2022;76:959-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1537] [Cited by in RCA: 1864] [Article Influence: 466.0] [Reference Citation Analysis (3)] |
| 8. | Wang H, Wen B, Chang X, Wu Q, Wen W, Zhou F, Guo Y, Ji Y, Gu Y, Lai Q, He Q, Li J, Chen J, Hou J. Baveno VI criteria and spleen stiffness measurement rule out high-risk varices in virally suppressed HBV-related cirrhosis. J Hepatol. 2021;74:584-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 9. | Rockey DC, Caldwell SH, Goodman ZD, Nelson RC, Smith AD; American Association for the Study of Liver Diseases. Liver biopsy. Hepatology. 2009;49:1017-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1449] [Cited by in RCA: 1636] [Article Influence: 96.2] [Reference Citation Analysis (2)] |
| 10. | Sanyal AJ, Foucquier J, Younossi ZM, Harrison SA, Newsome PN, Chan WK, Yilmaz Y, De Ledinghen V, Costentin C, Zheng MH, Wai-Sun Wong V, Elkhashab M, Huss RS, Myers RP, Roux M, Labourdette A, Destro M, Fournier-Poizat C, Miette V, Sandrin L, Boursier J. Enhanced diagnosis of advanced fibrosis and cirrhosis in individuals with NAFLD using FibroScan-based Agile scores. J Hepatol. 2023;78:247-259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 153] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 11. | Chang X, Wang J, Chen Y, Long Q, Song L, Li Q, Liu H, Shang Q, Yu Z, Jiang L, Xiao G, Li L, Chen L, Wang X, Li Z, Chen D, Dong Z, An L, Tan L, Chen Y, Yang Y. A novel nomogram to predict evident histological liver injury in patients with HBeAg-positive chronic hepatitis B virus infection. EBioMedicine. 2021;67:103389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 12. | Aberra H, Desalegn H, Berhe N, Mekasha B, Medhin G, Gundersen SG, Johannessen A. The WHO guidelines for chronic hepatitis B fail to detect half of the patients in need of treatment in Ethiopia. J Hepatol. 2019;70:1065-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 13. | Vallet-Pichard A, Mallet V, Nalpas B, Verkarre V, Nalpas A, Dhalluin-Venier V, Fontaine H, Pol S. FIB-4: an inexpensive and accurate marker of fibrosis in HCV infection. comparison with liver biopsy and fibrotest. Hepatology. 2007;46:32-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1288] [Cited by in RCA: 1669] [Article Influence: 87.8] [Reference Citation Analysis (1)] |
| 14. | Lok AS, Ghany MG, Goodman ZD, Wright EC, Everson GT, Sterling RK, Everhart JE, Lindsay KL, Bonkovsky HL, Di Bisceglie AM, Lee WM, Morgan TR, Dienstag JL, Morishima C. Predicting cirrhosis in patients with hepatitis C based on standard laboratory tests: results of the HALT-C cohort. Hepatology. 2005;42:282-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 248] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 15. | Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H, Desmet V, Korb G, MacSween RN. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3521] [Cited by in RCA: 3839] [Article Influence: 123.8] [Reference Citation Analysis (2)] |
| 16. | Rozario R, Ramakrishna B. Histopathological study of chronic hepatitis B and C: a comparison of two scoring systems. J Hepatol. 2003;38:223-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 94] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 17. | Sonneveld MJ, Brouwer WP, Hansen BE, Chan HL, Piratvisuth T, Jia JD, Zeuzem S, Chien RN, Choi H, de Knegt RJ, Wat C, Pavlovic V, Gaggar A, Xie Q, Buti M, de Man RA, Janssen HLA; SONIC-B Study Group. Very low probability of significant liver inflammation in chronic hepatitis B patients with low ALT levels in the absence of liver fibrosis. Aliment Pharmacol Ther. 2020;52:1399-1406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 18. | Lim SG, Baumert TF, Boni C, Gane E, Levrero M, Lok AS, Maini MK, Terrault NA, Zoulim F. The scientific basis of combination therapy for chronic hepatitis B functional cure. Nat Rev Gastroenterol Hepatol. 2023;20:238-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 110] [Reference Citation Analysis (0)] |
| 19. | Innes H, Morling JR, Buch S, Hamill V, Stickel F, Guha IN. Performance of routine risk scores for predicting cirrhosis-related morbidity in the community. J Hepatol. 2022;77:365-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 20. | Li G, Lin H, Sripongpun P, Liang LY, Zhang X, Wong VWS, Wong GLH, Kim WR, Yip TCF. Diagnostic and prognostic performance of the SAFE score in non-alcoholic fatty liver disease. Liver Int. 2024;44:15-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 21. | Zhang X, Song J, Zhang Y, Wen B, Dai L, Xi R, Wu Q, Li Y, Luo X, Lan X, He Q, Luo W, Lai Q, Ji Y, Zhou L, Qi T, Liu M, Zhou F, Wen W, Li H, Liu Z, Chen Y, Zhu Y, Li J, Huang J, Cheng X, Tu M, Hou J, Wang H, Chen J. Baveno VII algorithm outperformed other models in ruling out high-risk varices in individuals with HBV-related cirrhosis. J Hepatol. 2023;78:574-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 22. | Chun HS, Papatheodoridis GV, Lee M, Lee HA, Kim YH, Kim SH, Oh YS, Park SJ, Kim J, Lee HA, Kim HY, Kim TH, Yoon EL, Jun DW, Ahn SH, Sypsa V, Yurdaydin C, Lampertico P, Calleja JL, Janssen H, Dalekos GN, Goulis J, Berg T, Buti M, Kim SU, Kim YJ. PAGE-B incorporating moderate HBV DNA levels predicts risk of HCC among patients entering into HBeAg-positive chronic hepatitis B. J Hepatol. 2024;80:20-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 23. | Ndow G, Vo-Quang E, Shimakawa Y, Ceesay A, Tamba S, Njai HF, Bojang L, Hateley C, Takao Y, Opoku E, Warsop Z, Ingiliz P, D'Alessandro U, Chemin I, Mendy M, Thursz M, Njie R, Lemoine M. Clinical characteristics and outcomes of patients with cirrhosis and hepatocellular carcinoma in The Gambia, west Africa: a prospective cohort study. Lancet Glob Health. 2023;11:e1383-e1392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 24. | Kim HS, Yu X, Kramer J, Thrift AP, Richardson P, Hsu YC, Flores A, El-Serag HB, Kanwal F. Comparative performance of risk prediction models for hepatitis B-related hepatocellular carcinoma in the United States. J Hepatol. 2022;76:294-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 25. | Yokoyama H, Masaki T, Inoue I, Nakamura M, Mezaki Y, Saeki C, Oikawa T, Saruta M, Takahashi H, Ikegami M, Hano H, Ikejima K, Kojima S, Matsuura T. Histological and biochemical evaluation of transforming growth factor-β activation and its clinical significance in patients with chronic liver disease. Heliyon. 2019;5:e01231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/