Published online Mar 26, 2015. doi: 10.4252/wjsc.v7.i2.329
Peer-review started: July 29, 2014
First decision: October 14, 2014
Revised: October 27, 2014
Accepted: November 17, 2014
Article in press: November 19, 2014
Published online: March 26, 2015
Processing time: 234 Days and 6.6 Hours
Causative mutations and variants associated with cardiac diseases have been found in genes encoding cardiac ion channels, accessory proteins, cytoskeletal components, junctional proteins, and signaling molecules. In most cases the functional evaluation of the genetic alteration has been carried out by expressing the mutated proteins in in-vitro heterologous systems. While these studies have provided a wealth of functional details that have greatly enhanced the understanding of the pathological mechanisms, it has always been clear that heterologous expression of the mutant protein bears the intrinsic limitation of the lack of a proper intracellular environment and the lack of pathological remodeling. The results obtained from the application of the next generation sequencing technique to patients suffering from cardiac diseases have identified several loci, mostly in non-coding DNA regions, which still await functional analysis. The isolation and culture of human embryonic stem cells has initially provided a constant source of cells from which cardiomyocytes (CMs) can be obtained by differentiation. Furthermore, the possibility to reprogram cellular fate to a pluripotent state, has opened this process to the study of genetic diseases. Thus induced pluripotent stem cells (iPSCs) represent a completely new cellular model that overcomes the limitations of heterologous studies. Importantly, due to the possibility to keep spontaneously beating CMs in culture for several months, during which they show a certain degree of maturation/aging, this approach will also provide a system in which to address the effect of long-term expression of the mutated proteins or any other DNA mutation, in terms of electrophysiological remodeling. Moreover, since iPSC preserve the entire patients’ genetic context, the system will help the physicians in identifying the most appropriate pharmacological intervention to correct the functional alteration. This article summarizes the current knowledge of cardiac genetic diseases modelled with iPSC.
Core tip: This paper revises the cardiac genetic diseases that have been modeled so far using the technology that starts from patient somatic cells, reprogram their fate to a pluripotent state, and then proceed to cardiomyocyte differentiation. We will describe the main steps of this procedure, from pluripotent stem cells to mature cardiomyocytes, and we will discuss the main features linked to the different cardiac pathologies that this model recapitulate in a cell culture dish.
- Citation: Dell’Era P, Benzoni P, Crescini E, Valle M, Xia E, Consiglio A, Memo M. Cardiac disease modeling using induced pluripotent stem cell-derived human cardiomyocytes. World J Stem Cells 2015; 7(2): 329-342
- URL: https://www.wjgnet.com/1948-0210/full/v7/i2/329.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v7.i2.329
In 19th century, the Italian physician Giulio Bizzozero, founder of the Italian school of experimental pathology, proposed the classification of tissues depending on their regeneration rate. He divided human tissues in three categories: labile, stable, and perennial, where the labile are physiologically subjected to a continuous renewal, e.g., keratinocytes, or bone marrow cells; stables, that are normally in a quiescent state but still retain the capacity to proliferate, e.g., hepatocytes; and perennial, the so-called post-mitotic cells, that have completely lost their regenerative potential. The most representative cells in the latter case are neurons and cardiomyocytes, highly specialized cells that, apart from their duplication inability, share also the capacity, during development, to connect to sister cells and to build an organized network capable of an electric signal transmission.
During these years we learned the methods to cultivate and to maintain labile and stable cells in culture, but neurons and cardiomyocytes are still “problematic” cells, difficult to preserve in absence of their natural support, glial cells or cardiac fibroblasts, and, moreover, almost impossible to be obtained as sample from a healthy person.
Progress in this field came from studies of developmental biologist that were able to isolate, cultivate, and differentiate adult or embryonic stem cells (ESCs). These cells basically recapitulate in vitro the developmental process and ESCs in particular are indeed a continuous source of terminally differentiated post-mitotic cells that can be currently easily studied. The isolation and the use of human ESCs, although obtained from surplus of in vitro fertilization procedures, opened a serious and profound ethical issue. The great revolution came in 2006 when Shinya Yamanaka proposed a complex protocol designed to change cellular fate by forcing transcription factor expression[1]. With this procedure, a murine fibroblast was reprogrammed to a cell that assumed the main characteristics of an ESC: high proliferation rate as well as wide differentiation potential toward the three germ layers. Nevertheless, the search for an identical physiological counterpart has lead to the conclusion that these induced pluripotent stem cells (iPSCs), although very similar to ESCs, must be considered as artificial cells created in the lab. In 2007, the same process was applied successfully to human fibroblasts by Yamanaka’s as well as Thomson’s group[2,3], thus creating cell lines that can be propagated almost indefinitely and can be used, in a close future, as a continuous source of differentiated cells for therapeutic purposes.
The application of this powerful technique to humans opened the possibility to reprogram fibroblasts isolated not only from healthy people but also from patients suffering of a genetic disease. The resulting human iPSCs (hiPSC) will keep the entire genetic information of the patient, including the mutation that has been linked to the pathology. The first examples of disease-related hiPSC involved patients with a range of human genetic diseases, whose DNA mutations, including the trisomy 21, were effectively maintained through all the reprogramming procedure[4].
Since then, many laboratories started to model in vitro human genetic disease, succeeding to get a considerable number of post-mitotic cells for detailed functional studies. Indeed, this result is particularly important and useful when studying affected human cardiomyocytes or neurons, otherwise difficult to be obtained and maintained in culture. In most cases, despite the fact that hiPSC-derived cells are not exactly equal to the physiological counterpart, the system has attested successful in vitro replication of the main cellular characteristics already known to be associated with the modeled disease.
In this review we will discuss the important discoveries of the biological mechanisms underlying some genetic cardiomyopathies, made possible by the use of the cellular reprogramming technology.
In vitro development of ESCs has been widely studied using murine ESCs (mESCs), whose differentiation procedure in culture implies the initial leukemia inhibitory factor (LIF) removal and the formation of cellular aggregates using the “hanging drop” method. These three dimensional (3D) structures, called embryoid bodies (EBs), replicate in vitro the different stages of murine embryonic development[5]. Around differentiation day (dd) 8, clusters of spontaneously beating cells appear in culture; these cells express several transcriptional and structural cardiac markers and were therefore classified generically as cardiomyocytes[6]. It should be noted that mESC differentiation starts by just removing LIF also in a 2D culture, but the resulting process does not strictly follow embryonic development (P. Dell’Era, unpublished results).
Since their isolation, human ESCs have shown different culturing needs from the murine counterpart, and their behavior revealed also minor differentiation plasticity. Cardiac differentiation is the most glaring example of this statement. The spontaneous 3D differentiation of mESC leads to the easy appearance of spontaneous beating foci in all the considered EBs, while in the case of both hESC and hiPSC, only a modest proportion of EBs contains contracting cells. This occurrence leads to the setup of different methods aimed to increase cardiac differentiation during in vitro development of pluripotent stem cells. While some of these procedures retain an initial 3D EB formation, others start from a confluent 2D monolayer.
During gastrulation, cardiomyocytes emerge from mesodermal tissue, in particular from the anterior region of the primitive streak. Bone morphogenetic protein produced by the adjacent endoderm induces the cardiomyocyte fate, whereas WNT-mediated signals from the underlying neural tube and notochord suppress cardiomyocyte specification. Therefore the timed addition of inducers and/or inhibitors of the cited pathways would guide PSC differentiation toward: mesoderm, cardiomyogenic mesoderm, cardiac precursors, and cardiomyocytes. This is conceptually the summary of several protocols. At variance the required instructive signals can be provided by a visceral endoderm-like cell line that can be used as a feeder layer during the differentiation process[7].
Within these years, several efforts have been made to identify a protocol that would give rise to high percentage of cardiomyocytes following differentiation of either hESC or hiPSC. All the attempts lead to the creation of several tools such as defined cultivation media that, avoiding animal components, will facilitate therapeutic use of pluripotent stem cells; recombinant single protein matrices such as vitronectin or laminin; aggregation plates to standardize the production of EBs; and the more recent commercial cardiomyocyte differentiation media (StemRD and Life Technologies). The main differentiation protocols that have been used over the years are outlined in Table 1.
| PSC type | Differentiation | Pre-treatment | Cardiac differentiation treatment | % Beating cells | % cTNT + cells | Ref. | ||||||||
| h-iPSC | h-ESC | 2D | 3D | Others | ROCK-I | GSK-I | BMP4 | Activin A | Wnt-I | FGF2 | Others | |||
| X | END2 | AA | < 40% | n.a. | [83] | |||||||||
| X | X | X | Specific serum | 5%-10% hiPSC | n.a. | [20] | ||||||||
| 10%-25% hESC | ||||||||||||||
| X | X | X | X | X | n.a. | > 80% hiPSC | [84] | |||||||
| 60%-80% hESC | ||||||||||||||
| X | X | X | X | AA | n.a. | 50%-70% | [85] | |||||||
| X | X | X | p38- MAPK-I | > 23% hiPSC | n.a. | [86] | ||||||||
| 50% hESC | ||||||||||||||
| X | X | Sandwich | X | X | X | X | n.a. | 98% | [87] | |||||
| X | X | X | X | X | X | X | X | AA | n.a. | 80% | [88] | |||
| X | X | X | END-2-CM | n.a. | < 10% | [89] | ||||||||
| X | X | X | X | X | X | MEF-CM | n.a. | < 60% | ||||||
| X | X | X | X | Tricho- statin A | < 50% | < 10% | [90] | |||||||
| X | X | X | X | X | X | X | X | VEGF | < 50% hiPSC | n.a. | [91] | |||
| < 40% hESC | 95% hESC | |||||||||||||
| X | X | X | X | Albumin, AA | n.a. | 90% | [8] | |||||||
| X | X | X | X | X | X | VEGF | < 40% | < 20% | [92] | |||||
| X | X | X | X | X | X | Blebbistatin | 100% | 90% | [93] | |||||
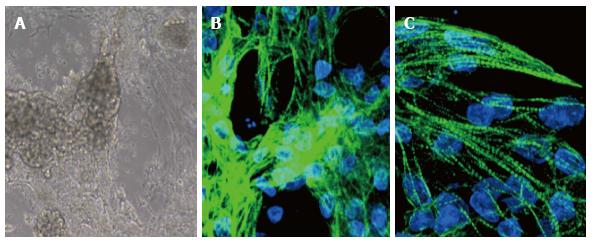
We tested several of these procedures using iPSC derived from patients’ fibroblasts and adapted to grow on Matrigel-coated dishes; in our experience, we observed beating areas only when cells were treated with modulators of the Wnt pathway, while the general TNNT2 expression was achieved in most protocols (Figure 1B). Of particular interest for therapeutic purposes the recent setup by Burridge et al[8] that employs a chemically defined medium consisting of just three components on a dish covered by synthetic biological matrices. Indeed, using this protocol we strongly increased the number of beating cells in our culture up to 50%, but the best result, around 70%, was recently obtained using the PSC Cardiomyocyte Differentiation Kit (Life Technologies).
In early development, the primitive heart tube is composed by small myocytes without distinct region of cells conducting and coordinating the stimulus. The contractile impulse begins with the primary pacemaker area in the primitive atrium that will later evolve in the sinoatrial node, where the fast beating pacemaker myocytes will reside. As development proceeds, also the conductive system will differentiate, generating CMs in anatomical different regions that will become the atrio-ventricular (AV) node, the AV bundles, and Purkinje fibers. Each of these CM subtype has its own electrical properties such as the presence and the form of the action potential (AP), the contraction rate, and the presence of specific currents that can be measured electrophysiologically.
However, CMs differentiated in vitro vary considerably from cells isolated from a mature human heart, because of the absence of humoral factors and organized mechanical and electrical stress. In general, many of the features of hPSC-CMs are reminiscent of normal fetal cells. hPSC-CMs are spontaneously beating cells co-expressing atrial-, ventricular-, and nodal- markers, with unorganized sarcomeres, immature mitochondria, and an expression profile different from adult CMs[9]. Our data indicate that, after 16 d of in vitro differentiation, iPSC-derived CMs start to segregate in the various subtypes, showing pronounced sarcomeric structures that reveal a certain degree of maturation (Figure 1C).
The CMs that arise during early hESC or hiPSC in vitro differentiation exhibit spontaneous AP, with a relatively depolarized resting membrane potential, probably due to the temporary absence of the inward rectifier potassium current (IK1)[10].
The expression of the ion channels and, consequently, the ionic currents will undergo developmental maturation over time, as assessed by modifications in current density and property[10]. hPSC-CMs immaturity is also reflected in their excitation-contraction machinery, lacking clear T-tubuli, disorganized sarcomeric striations, and immature Ca2+ handling[11-13]. Unlike primary CMs that tend to undergo apoptosis or dedifferentiate, CMs derived from hPSC develop and maintain a functional phenotype in long-term culture[14]. After surviving for 80 d, late-stage hESC-CMs show pronounced multinucleation that is accompanied by an increase in cellular perimeter, and area[15]. Ultrastructural studies demonstrated that the sarcomere of hiPSC-CMs continue to mature through a 1-year culture[16]. Young hiPSC-CMs contained a low number of unaligned myofibrils and immature high-density Z-bands. Within 6 mo the myofibrils became more tightly packed and formed parallel arrays accompanied by the appearance of mature Z-, A-, H-, and I-bands. M-bands were finally detected in 360-d-old CMs, but expression levels of M-band-specific genes remained lower in comparison with those in the adult heart[14].
Also a different gene expression accompanies these changes: late-stage CMs show increased levels of structural filaments MYH6 and MYH7, and of other specific molecules such as connexin 43, hyperpolarization activated cyclic nucleotide-gated potassium channel 4, and sarco(endo)plasmic reticulum Ca2+ ATPase[14-16]. Finally, the electrophysiological profile of late-stage hESC-CMs show a significantly enhanced AP upstroke and a hyperpolarized maximum diastolic potential, and during maturation no differences were observed for AP duration (APD) to 50% or 90% (APD50, APD90) of repolarization[15].
It must be noted that, while at early differentiation stages there are unspecified CMs that co-express different CM-subtype markers at later stages, around dd30, CMs acquire a more specific phenotype, expressing ventricular-, atrial-, and nodal-markers[8]. The relative proportion of the three CM subtypes varies among the differentiation protocols that have been used, and can be additionally modulated by supplementing chemicals, microRNAs, or biological molecules to the culture[17].
Despite all the discussed limitations, hPSC differentiation remains a powerful method to model in vitro genetic cardiac diseases, because of the capacity of these cells to give rise to terminally differentiated stable cardiac cells. The cardiac pathologies that have been modeled so far using hiPSC include some cardiomyopathies and arrhythmias, whose implicated genes are listed in Table 2.
| Cardiac disease | Gene | Protein/current | Chr | Mutation | Differentiation method | Cardiomyocyte subtype | Maturation days | Ref. |
| HCM | MYH7 | Myosin heavy chain β | 14q12 | R663H | 3D spontaneous | [19] | ||
| DCM | LMNA | Lamin A | 1q22 | R225X | END-2 co-culture | V- and A-like | 20 | [25] |
| GCCA insertion | ||||||||
| TNNT2 | Troponin T type 2 | 1q32 | R173W | 3D, activin, BMP4, DKK1, FGF2, VEGF | V-, A-, and N-like | > 30 | [27] | |
| DES | Desmin | 2q35 | A285V | END co-culture | n.d. | > 14 | [26] | |
| BTHS | TAZ | Tafazzin | Xq28 | 517delG | 2D, activin, BMP4 | n.d. | > 12 | [30] |
| c.328T>C | ||||||||
| LQT1 | KCNQ1 | Kv7.1/I(Ks) | 11p15 | R190Q | 3D spontaneous | V-, A-, and N-like | 20-30 | [36] |
| LQT2 | KCNH2 | hERG/I(Kr) | 7q36 | R176W | 3D spontaneous | V- and A-like | n.d. | [42] |
| A561T | V-, A-, and N-like | 25-30 | [38] | |||||
| A561V | V- and A-like | 21 | [45] | |||||
| A614V | V-, A-, and N-like | > 30 | [37] | |||||
| N996I | n.d. | 20-60 | [43] | |||||
| LQT3 | SCN5A | Nav1.5 /I(Na) | 3p21 | F1473C | 3D, activin, BMP4, Wnt-I, FGF2 | V- and A-like | 25-45 | [50] |
| V1763M | 3D spontaneous | V-, A-, and N-like | > 28 | [51] | ||||
| V240M, R535Q | END-2 co-culture | V-, A-, and N-like | 20-30 | [52] | ||||
| LQT8 | CACNA1C | CaV1.2/I(Ca) | 12p13 | G1216A | 3D + Wnt3a | V-, A-, and N-like | > 37 | [55] |
| CPVT | RYR2 | Ryanodine receptor 2/I(Ca) | 1q43 | F2483I | END-2 co-culture | V-, A-, and N-like | 20-30 | [61] |
| P2328S | END-2 co-culture | Mostly V-like | n.d. | [62] | ||||
| S406L | 3D spontaneous | V-, A-, and N-like | > 70 | [64] | ||||
| M4109R | 3D spontaneous | V-, A-, and N-like | > 30 | [65] | ||||
| E2311D | 3D spontaneous | V/A- and N-like | > 30 | [66] | ||||
| CASQ2 | Calsequestrin | 1p13 | D307H | 3D spontaneous | n.d. | 25-43 | [67] | |
| ARVC | PKP2 | Plakophilin-2 | 12p11 | L614P | 3D spontaneous | V-, A-, and N-like | > 28 | [72] |
| A324fs335X | 3D spontaneous | n.d. | > 30 | [73] | ||||
| T50SfsX110 | ||||||||
| Criptic splicing | 3D spontaneous | n.d. | > 60 | [74] | ||||
| c.2013delC |
Familial hypertrophic cardiomyopathy: Hypertrophic cardiomyopathy (HCM) is a heterogeneous monogenic heart disease in which a portion of the myocardium is heavily hypertrophic. It is caused by more than 1400 mutations in at least 11 genes that encode thick and thin contractile myofilaments of the sarcomere or the adjacent Z-disc[18]. HCM patients display abnormal thickening of the left ventricular myocardium in the absence of increased hemodynamic burden. Most people with familial HCM are symptom-free or have only mild symptoms, but their risk for clinical complications such as progressive heart failure, arrhythmia, and sudden cardiac death is strongly increased.
Efforts to elucidate the mechanisms underlying development of HCM have lead to the generation of patient-specific hiPSC-CMs that recapitulate in vitro a number of disease characteristics including cellular hypertrophy, and contractile arrhythmia[19]. hiPSC were generated from a family cohort carrying a hereditary HCM missense mutation (Arg663His) in the MYH7 gene, and CMs were generated using the 3D spontaneous differentiation protocol[20]. Mutant hiPSC-CMs demonstrated not only cellular enlargement and multinucleation, but also other hallmarks of HCM including expression of atrial natriuretic factor, elevation of β-myosin/α-myosin ratio, calcineurin activation, and nuclear translocation of nuclear factor of activated T cells[19]. Using this model the authors were able to show that irregular Ca2+ transients and elevation of diastolic intracellular calcium [Ca2+]i precedes the presentation of other phenotypic abnormalities, strongly implicating dysregulation of Ca2+ cycling in the pathogenesis of the disease. Pharmaceutical drug screening of mutant hiPSC-CMs further supported elevated [Ca2+]i as a central mechanism for arrhythmia development. Indeed, only pharmaceutical blockade of Ca2+ and Na+ entry mitigated contractile arrhythmia in HCM-CMs[19].
Dilated cardiomyopathy: Dilated cardiomyopathy (DCM) represents the final common morphological and functional consequence of various pathological conditions in which a combination of myocyte injury and necrosis associated with tissue fibrosis results in impaired heart mechanical function. Nevertheless, primary familial forms of DCM represent a genetic condition in which the pathological involvement is predominantly limited to the myocardium. The main hallmark of primary DCM is the presence of a left or biventricular dilatation with severely impaired systolic function in the absence of abnormal loading conditions or ischemic heart disease sufficient to cause global systolic impairment[21]. Point mutations in 31 autosomal and 2 X-linked genes have been implicated in causing familial DCM (FDC) but account for only 30% to 35% of genetic causes[22]. One of the more common genes identified in FDC is LMNA, which codes for lamin A/C proteins, intermediate filament proteins of the nuclear lamina[23].
Several animal models of LMNA mutations have been generated to provide initial insights into the pathophysiology of lamin A/C-related DCM[24]. Nevertheless the mechanism that links LMNA mutations with DCM remains uncertain.
Two different LMNA mutations have been modeled using hiPSC-CMs: an autosomal dominant non-sense mutation (R225X) in exon 4 of the lamin A/C and a GCCA insertion at base 50 in LMNA that creates a frameshift and premature stop codon, hence causing lamin A/C haploinsufficiency. LMNAR225X/WT and LMNAFrameshift/WT hiPSC-CMs showed normal phenotypes and basal electrophysiological properties as control hiPSC-CMs. However, when these CMs were subjected to electrical stimulation as in the cardiac environment, they exhibited typical nuclear abnormalities together with increased apoptosis[25]. These two properties were replicated in vitro by shRNA knockdown of LMNA in control hiPSC-CMs, that was ineffective when the MEK1/ERK1/2 pathway was blocked pharmacologically[25], thus identifying this pathway as a potential therapeutic target in LMNA-related DCM.
The same group also modeled a pathogenic phenotype of DCM due to a novel A285V Desmin (DES) mutation identified by whole exome sequencing (WES) using the hiPSC-CM system[26]. Characterization of hiPSC-CMs carrying A285V-DES mutation revealed a poor co-localization of DES with several cytoskeletal proteins, including cardiac troponin-T, α-actinin and F-actin, and diffuse isolated aggregations of DES-positive protein[26]. The electrophysiological analysis revealed that A285V-DES CMs exhibit significant functional abnormalities compared with the control-CMs as demonstrated by the diminished maximum rate of calcium ion re-uptake, slower spontaneous beating rate and failure to respond to the inotropic stress induced by isoproterenol[26]. When control-iPSC-CMs were transduced with a lentivirus carrying the A285V-DES mutation, the resulting CMs simulated the phenotypes of DES-DCM-CMs, thus confirming the idea that abnormal DES-positive protein aggregates due to DES mutation can cause structural and functional abnormalities in cardiomyocytes. The relevance of this study is the demonstration that patient-specific hiPSC-CMs can be used to provide confirmation of a suspected genetic basis for DCM identified by WES.
Lastly, cardiomyocytes derived from patients in a DCM family carrying a point mutation (R173W) in the gene encoding sarcomeric protein cardiac troponin T were analyzed[27]. Compared to healthy individuals in the same family cohort, CMs from DCM patients exhibited altered regulation of Ca2+, decreased contractility, and abnormal distribution of sarcomeric β-actinin. When stimulated with a β-adrenergic agonist, CMs showed characteristics of cellular stress such as reduced beating rates, compromised contraction, and abnormal sarcomeric β-actinin distribution. Treatment with β-adrenergic blockers or overexpression of sarcoplasmic reticulum Ca2+ adenosine triphosphatase rescued the pathological phenotype[27].
Barth syndrome: Barth syndrome (BTHS) is a rare, metabolic, and neuromuscular genetic disorder that occurs exclusively in males. Clinical features include variable combinations of pathologies including DCM, HCM, endocardial fibroelastosis, left ventricular non-compaction, ventricular arrhythmia, sudden cardiac death[28]. The gene responsible for the disorder, tafazzin (TAZ), has recently identified: it is located on the long arm of chromosome X at Xq28, and encodes an acyltransferase that catalyzes the acylation of cardiolipin, the major phospholipid of the mitochondrial inner membrane[28,29]. So far, more than 120 mutations have been described, but no correlation with specific phenotype has been observed[28].
hiPSCs from two unrelated individuals with BTHS were generated, BTH-H and BTH-C, carrying TAZ frameshift (c.517delG) and missense (c.328T > C) mutations, respectively. CMs were generated by treating a cell monolayer with inducers and sorted for the surface marker vascular cell adhesion molecule-1 to obtain preparations highly enriched in CMs. Purified CMs were analyzed morphologically to evaluate sarcomere organization, and to assess mitochondrial functionality. CMs were then seeded onto thin elastomers with patterned lines of fibronectin, obtaining self-organized anisotropic myocardial tissues to study contractility properties[30]. TAZ deficiency in BTHS-CMs impairs mitochondrial functionality as well as sarcomere assembly, generate contractile stress, and markedly increase reactive oxygen species (ROS) production. When linoleic acid (LA), an essential unsaturated fatty acid precursor of mature cardiolipin, was added to CM culture, the metabolic phenotype was corrected, the sarcomere organization and contractile defects were mitigated, and ROS production was strongly reduced[30].
Using the hiPSC-derived model the authors showed that suppression of ROS, not only by LA but also by mitoTEMPO, normalized the metabolic, sarcomerogenesis and contractile phenotypes of BTHS- CMs, thus setting the basis for an effective pharmacological therapy of BTHS patients[30].
Long-QT syndromes: Long-QT syndromes (LQTS) are a group of heritable, usually autosomal dominant disorders with a estimated prevalence of 1:2500, characterized by an abnormally delayed or prolonged ventricular repolarization phase (prolongation of the QT interval on an electrocardiogram) and a propensity toward polymorphic ventricular tachycardia (often termed Torsades de pointes, TdP), syncope and sudden cardiac death in young patients[31]. Clinically LQTS present a broad range of phenotypes even among family members with identical mutations, possibly as a result of genetic modifiers[32]. To date, LQTS have been associated with over 500 different mutations in at least 13 genes encoding cardiac ion channel proteins, but the most prevalent forms are LQT1 and LQT2 caused by potassium channel mutations with a percentage of genotyped cases of > 50% and 30%-40%, respectively, and LQT3 caused by a sodium channel mutation that accounts for 10%-15%[31].
LQT1 patients carry mutations in the KCNQ1 gene (also known as KVLQT1 or Kv7.1), which encodes the pore-forming α-subunits of the channels generating IKs, an adrenergic-sensitive, slow outward potassium current, while LQT2 implicates hERG protein (encoded by KCNH2 gene), which constitutes pore-forming α subunit of the rapidly-activating delayed rectifier potassium current (IKr)[33]. LQT3 is instead associated with gain-of-function mutations of the SCN5A gene, which encodes the α-subunit of the Na+ ion channel NaV1.5[34], while loss-of-function mutations in the same gene are associated with several other genetically heterogeneous disorders including Brugada syndrome, cardiac conduction disease, sick sinus syndrome sudden infant death syndrome and others[35].
LQT1 was the first cardiac disease modeled using hiPSC and since then several hiPSC-CMs from patients carrying mutations in LQTS-associated channels have been considered[36-42]. Indeed, patient-specific hiPSC-CMs represent a platform to investigate the functionality of ion channel mutations expressed in their complex genetic backgrounds and may provide unique insight into therapeutic approaches for disease management[43].
LQT1: A family affected by LQT1 was screened and an autosomal dominant missense mutation R190Q in the KCNQ1 gene was identified[36]. hiPSC from two family members and two healthy controls were generated by retroviral vectors encoding the human transcription factors OCT3/4, SOX2, KLF4, and c-MYC (hOSKM). Using a 3D differentiation protocol, these cells were then differentiated into CMs. Spontaneously beating cells dissociated from LQT1 and control explants responded to pacing and generated three distinct types of APs, designated as “ventricular (V)”, “atrial (A)”, and “nodal (N)” on the basis of their similarity to the APs of human fetal heart CMs[36]. Several disease-specific abnormalities were observed in LQT1-CMs: the duration and the rate adaptation of the AP, a 70%-80% reduction in IKs, as well as vulnerability to catecholaminergic stress[36]. R190Q– KCNQ1 is a dominant mutation and indeed in hiPSC-CMs the mutated protein was absent on cell surface, but still retained in the endoplasmic reticulum. Similar results were obtained by expressing the wild type and the mutated protein in cardiomyoblast H9c2 cell line[36]. Furthermore, electrophysiological studies confirmed the protective effect of β-blockade in the abnormal response to catecholamine stimulation, thus confirming the efficacy of the therapeutic approach for LQT1 patients[36].
LQT2: The voltage-gated inwardly rectifying potassium channel that comprise KCNH2-encoded protein is composed of homo- or heterotetrameric complexes of pore-forming α subunits, like hERG, that associate with modulating β subunits. hERG consists of six transmembrane α helices, a pore helix, and N- and C-termini cytoplasmically located. The channel mediates the rapidly activating component of the IKr in heart[44]. A large number of natural variants have been described, most of them in association with LQT2 syndrome (see http://www.uniprot.org/uniprot/Q12809).
A panel of control and LQT2-related hPSC were generated and characterized by several laboratories. hiPSC-CMs showing five different hERG mutations in genetically unrelated backgrounds were intensely characterized: R176W[42], A561T[38] and A561[45], A614V[37], and N996I[46]. The aminoacids 176 and 996 reside in the N- and C-terminus cytoplasmic domains respectively, while the position 561 is in a transmembrane region of the protein, and the aminoacid 614 is located in the pore-forming segment.
Heterozygous KCNH2 mutations exert a dominant-negative effect on wild-type (WT) hERG channels associated IKr, by impairing trafficking pathways or altering channel kinetics of the resulting co-assembled hERG heterotetramers[31,47]. Due to this behavior, it is possible to transfer them in a WT environment to verify their biological consequences in an unrelated genetic background.
The reprogramming and the following differentiation process were similar for all the laboratories: most of them used a retroviral transduction system, except for Mehta et al[45] that choose a viral-free episomal non-integrating approach, but the following differentiation involving EB formation and spontaneous differentiation was identical for all of them.
It must be pointed out that LQT2 is a disorder with incomplete penetrance where genetic background variations can confound disease traits. For this reason, the KCNH2 N996I mutation was deeply analyzed in (1) hiPSC-CMs from LQT2 patient; (2) hiPSC-CMs from LQT2 patient, previously corrected to wild-type using an homologous recombination system; and (3) NKX2.5eGFP/w hESC-CMs where the N996I-KCNH2 mutation was introduced using the same approach as before[46].
Several observations were commonly reported to all LQT2-CMs: intracellular patch clamp recording of APD or extracellular measurement of field potential duration (FPD) using multi electrode array (MEA) showed prolonged intervals for both A-like and V-like cells[37,38,42,45,46]. A summary of AP characteristic parameters measured by several authors in iPSC- or ESC-derived CMs is reported in Hoekstra et al[48]. Nevertheless, the AP increase was more restricted when isogenic cell lines were compared[46], thus suggesting that a different genetic background can indeed over-estimate the differences between a control versus diseased CMs.
AP was modulated in LQT2-CMs by several drugs: pinacidil, a KATP-channel opener, significantly shortens APD90, while Na-channel blocker, ranolazine, was ineffective[37]; K channel enhancers, nicorandil and PD118057, caused AP shortening and in some cases could abolish early afterdepolarization (EAD)[38]. In most of the papers some arrhythmogenicity of LQT2-CMs was reported, as evidenced by EAD events during spontaneous recordings[37,42,45] or when challenged with the clinically used stressor, isoprenaline[38].
As expected, the measurement of IKr, determined by adding the specific E4031 inhibitor, showed a reduction in LQT2-derived versus control CMs[37,42,45,46].
Definitively, the genetic correction of the N996I-KCNH2 mutation associated with LQT2 restores IKr density and normalizes APD in patient-specific LQT2-CMs, while the introduction of the same mutation in hESC-CMs reduced IKr and prolonged the AP duration[46]. In addition, the molecular defects of hERG A561V and N996I mutants have been analyzed. In the NKX2.5-eGFP+ hESC-N996I cells, as well as in LQT2-N996I CMs, a trafficking defect was identified. Indeed, the 155-kDa protein band, representing the form transported to the cell membrane through the Golgi, was reduced by two-fold compared to the WT protein, while the 135-kDa band, which corresponds to the protein located in the ER, was unaffected[46].
Similarly, hERG mutation A561V causes a reduced membrane localization of glycosylated/mature protein[45]. Treatment of LQT2-A561V CMs with the calpain and proteasome inhibitor ALLN, not only increased membrane localization of mature hERG but also reduced repolarization, increased IKr and reduced arrhythmogenic events, thus suggesting a new therapeutic approach to treat LQT2 patients[45].
LQT3: The α-subunit of the Na(v)1.5 cardiac sodium channel, encoded by SCN5A gene is composed by intracellular N- and C- terminus, and four homologous domains, attached one another by cytoplasmic linkers, forming a pore that conducts Na+ ions across membrane[49]. In LQT3, the gain-of-function SCN5A-mutations cause an increased persistent Na+ influx during depolarization that results in an enhanced late or persistent sodium current due to defective open-state inactivation of the channel[35]. Four different SCN5A alterations have been modeled using hiPSC cellular system: a de novo heterozygous missense mutation F1473C associated with a polymorphism (K897T) in KCNH2, identified in a newborn patient with extreme prolonged QT interval[50]. The second SCN5A modification was again a de novo heterozygous missense V1763M mutation, found in a Chinese girl[51], and the last two V240M and R535Q alterations were from two LQT3-diagnosed patients[52]. The F1473C mutation occurs in the channel inactivation gate of SCN5A while V240M and V1763M reside in transmembrane segments, and R535Q mutation occurs in the first cytoplasmic linker[53].
In the first paper a lentiviral transduction of OSKM was used to reprogram fibroblasts, and a complex protocol rich in developmental modulators was used to achieve CMs differentiation[50]. Ma et al[51] used instead an mRNA-based non viral non integrating reprogramming approach, followed by the spontaneous EB-based CM differentiation. Finally Fatima et al[52] used the classical retroviral infection, followed by a differentiation protocol guided by END-2 co-culture.
The data of hiPSC-CMs from the newborn and from the Chinese girl were compared with those obtained from hiPSC-CMs of healthy parents and of a healthy sister respectively that, because of the common genetic background with the patients, strengthened the obtained results[50,51].
Terrenoire et al[50] reported a mutation-dependent increase in INaL, a right-shifted steady-state channel availability, and faster recovery from inactivation in all clones from the proband and none of the parents’ clones. By using the hiPSC-CM system, the authors easily showed that the KCNH2 heterozygous polymorphism T897 and K897, derived from homozygous parents, had no impact on electrophysiological pathology of the proband[50]. Furthermore, the system allowed the pharmacological evaluation of the current patient’s therapy, not so effective in controlling episodes of arrhythmia. Indeed, a more effective therapy has been identified as a result of the proposed study[50].
Ma et al[51] observed significantly prolonged APD in patient-derived V-like CMs compared with control cells, while Fatima et al[52], showed a tendency to prolonged APD not statistically significant. Relevant to this point Terrenoire et al[50], comment that the relatively depolarized diastolic membrane potentials at this embryonic developmental stage inactivate sodium channels and consequently minimize contributions of sodium channel activity to AP[53]. Relatively to sodium current Ma et al[51] found that TTX-sensitive INaL was significantly larger in patient-derived hiPSC-CMs compared with control hiPSC-CMs[51], while Fatima et al[52] showed that time-to-peak for sodium current and time to 90% of inactivation of the Nav1.5 were significantly longer in the LQT3-CM[52]. In agreement with previous findings[50], mexiletine reduced the late Na+ current and shortened the APD in patient hiPSC-CMs[51].
LQT8/Timothy syndrome: Timothy syndrome (TS) is a multi-system disorder characterized by cardiac, hand, facial and neurodevelopmental features that include QT prolongation, webbed fingers and toes, flattened nasal bridge, low-set ears, small upper jaw, thin upper lip, and characteristic features of autism or autistic spectrum disorders (see http://www.orpha.net).
TS is caused by mutations in the CACNA1C gene and is inherited as autosomal dominant trait. The gene codifies for CaV1.2 channel, the main L-type channel in the mammalian heart that is essential for generating the cardiac action potential and for excitation contraction coupling[54].
To date, just one paper reported the modeling of this syndrome using hiPSC: 16 iPSC lines from two TS patients, and 10 control lines from two unrelated individuals were generated using the classic retroviruses, and CMs were then obtained spontaneously from EBs[55].
A prolonged AP was observed in TS-V-like CMs, while no differences were observed in A-like and N-like cells[55]. The L-type Ca2+ channel current had significantly reduced voltage-dependent inactivation in TS-CMs compared to control cells[55]. In addition, the TS-CMs exhibited a large number of depolarizing events that failed to trigger a full AP, similar to delayed afterdepolarizations (DADs) that arise after ectopic release of Ca2+ from the sarcoplasmic reticulum and which are associated with cardiac arrhythmias[55]. Moreover, the Ca2+ elevations in spontaneously contracting TS-CMs were more prolonged and more irregular than those of control CMs[55]. Finally, roscovitine rescued the electrophysiological properties of TS- CMs by increasing CaV1.2 voltage-dependent inactivation, reducing the APD, and decreasing the frequency of abnormal depolarizing events[55].
Catecholaminergic polymorphic ventricular tachycardia: Another inherited cardiac disorder that was studied using hiPSC-CMs is catecholaminergic polymorphic ventricular tachycardia (CPVT) that is characterized by emotional and physical stress-induced ventricular tachyarrhythmia, syncope and sudden cardiac death in children and young adults. Two type of CPVT have been described: the autosomal dominant form (CPVT1) linked to mutations in the cardiac ryanodine receptor type 2 gene (RYR2) and a rare autosomal recessive form (CPVT2) caused by mutations in the calsequestrin-2 gene (CASQ2)[56].
RYR2 gene encode for the principal Ca2+-releasing channel expressed in the membrane of the sarcoplasmic reticulum (SR). Studies based on in vitro expression of mutant RYR2 in heterologous cell systems and transgenic mice carrying specific RYR2 mutations suggested that arrhythmias in CPVT1 are due to the diastolic Ca2+ leak from the SR following catecholaminergic stimulation[57]. This leakage may lead to develop DADs through activation of the membrane Na+/Ca2+ exchanger, which can eventually result in triggered activity[58].
The CASQ2 gene encodes for a high-capacity, low affinity Ca2+ binding glycoprotein located inside the SR and involve in excitation-contraction coupling process[59]. The functional alterations in intracellular Ca2+ handling resulting from the mutated CASQ2 gene may cause DADs[60]. In 2011, Fatima and his group described the hiPSC generation from a patient with CPVT1 carrying the mutation F2483I in RYR2 gene[61]. The authors differentiated hiPSC-CMs from fibroblasts of the CPVT patient and after 20-30 d of culture the cells were electrophysiologically analyzed. Frequent DADs and arrhythmias in CPVT-CMs exposed to adrenergic agonists were consistently observed. Furthermore, abnormal sensitivity to phosphorylation and cAMP-mediated regulation, together with the tendency for ICa-triggered Ca2+ release to continue following repolarization have been found[61]. These first results using hiPSCs-based cardiac model validated the earlier hypothesis obtained only with animal models. Different groups have obtained similar results: Kujala has demonstrated that in addition to DADs, CPVT-CMs with P2328S RYR2 mutation displayed also EADs which may be involved in cardiac arrhythmogenesis of their patients[62]. Zhang reported specific analyses on calcium current (ICa) and Na+-Ca2+ exchanger current (INCX): in this case RyR2 F2483I mutant CMs have aberrant unitary Ca2+ signaling, smaller INCX reflecting smaller Ca2+-stores, higher ICa-gated Ca2+-release gains, and sensitized adrenergic regulation, consistent with functionally altered Ca2+-release profile of CPVT syndrome[63]. Jung also reported the hiPSC generation of a 24-year-old woman with a diagnosis of familial CPVT1 with S406L missense mutation in the RYR2 gene[64]. In addition to presenting the electrophysiological properties of CPVT-CMs, the rescue capacity of dantrolene, which is believed to stabilize skeletal and cardiac RYRs by binding to a N-terminal sequence, is analyzed. Treatment with dantrolene restored normal Ca2+ spark properties in CPVT-CMs under basal conditions and corrected S406L-RYR2 hyperactivity induced by adrenergic stimulation, with minimal effects in control cells; moreover, the same drug completely abolished DADs and triggered arrhythmias[64].
Two more studies evaluated the effects of different drugs onto CPVT-CMs: Itzhaki reported the positive effects of flecainide, an antiarrhythmic agent, and thapsigargin, a β-blocker on CMs carrying the RYR2 M4109R mutation[65], while Di Pasquale et al[66] reported the rescue of the arrhythmic phenotype induced by catecholaminergic stress by KN-93, an antiarrhythmic drug that inhibits Ca2+/calmodulin-dependent serine–threonine protein kinase II on CMs carrying the RYR2 E2311D mutation.
An additional study focused on the autosomal recessive form of the disease caused by the missense mutation D307H in CASQ2[67]. In agreement with previous observations, isoproterenol stimulation caused DADs, oscillatory arrhythmic prepotentials and after-contractions, and diastolic intracellular calcium rising in CASQ2-derived CPVT model[67]. Most importantly, electron microscopy showed that CMs derived from CPVT patients present an immature morphology with less-organized myofibrils, enlarged SR cisternae, and reduced number of caveolae[67]. These data confirm previous findings derived from knock-out and knock-in Casq2 mice that showed ultrastructural abnormalities in the SR[68,69].
Arrhythmogenic right ventricular cardiomyopathy: Arrhythmogenic right ventricular cardiomyopathy (ARVC) is a heritable primary cardiac disease characterized by the replacement of CMs with fatty or fibrofatty tissue[70]. ARVD has an autosomal dominant trait with reduced penetrance; approximately 40%-50% of ARVD patients have a mutation identified in one of several genes encoding components of the desmosome, which can help confirm a diagnosis of ARVD[71].
Also ARVC has been modeled using hiPSC by three independent groups starting from skin biopsies of five patients: retroviruses were used to transduce patient’s-derived fibroblasts and CMs were differentiated with the spontaneous 3D differentiation protocol[72-74]. The five patients carried different alterations in Plakophilin-2 gene (PKP2), a structural component of desmosome[75]: a heterozygous L614P mutation[72]; a heterozygous insertion resulting in a frame shift from amino acid 324 to a stop codon in position 335 (A324fs335X) and a heterozygous deletion resulting in replacement of threonine by serine in position 50 and in a frame shift leading to a stop codon in position 110 (p.T50SfsX110)[73]; and, finally, a homozygous mutation that causes cryptic splicing with a 7-nucleotide deletion in exon 12, leading to frame-shift of the carboxy-terminal amino acids, whose results were further confirmed with cells carrying a heterozygous c.2013delC in exon 10 of PKP2[74].
Common observations of ARVC-CMs carrying the heterozygous conditions reported specific down-regulation of PKP2 and its interactor plakoglobin both at mRNA and at protein level[72,73]. Transmission electron microscopy (TEM) analysis displayed larger ARVC-CMs with less organized, thicker and more pleomorphic Z-bands compared with the Z-bands in control cells[72], as well as distorted desmosomes associated with accumulation of lipid droplets, that were strongly enhanced in lipogenic media[72,73]. Relevant to this point, the addition of the GSK-3β inhibitor BIO strongly suppress the effects of the lipogenic stress[73].
The phenotypic study of CMs derived from the patient carrying the homozygous mutation led to similar conclusions: an abnormal translocation of plakoglobin proteins associated with very low β-catenin activity[74]. In addition, the culture of these ARVC-CMs, devoid of a correct PKP2 carboxy terminus, in a lipogenic medium additionally containing PPAR-γ activating drugs, demonstrated exaggerated lipogenesis and pronounced apoptosis, that can be prevented by the addition of PPAR-γ antagonists[74]. Similar results were obtained with CMs derived from the patient carrying a heterozygous c.2013delC mutation[74].
The rescue of the pathogenic phenotypes was achieved by introducing the wild-type PKP2 gene back into mutant hiPSC-CMs, thus suggesting that mutation of PKP2 is sufficient to induce the pathological features observed in ARVC-CMs[74].
Interestingly, the same cells were used by Cerrone et al[76] to verify a PKP2-mediated modulation of INa: indeed, as already shown using other cellular systems, ARVC-CMs showed drastically reduced INa and the deficit was restored by re-introducing the wild type PKP2 gene.
Since the discovery of the cellular reprogramming method followed by in vitro CM differentiation, several findings regarding human cardiac cells, especially those exhibiting a pathological phenotype, have been made possible. We can discuss how close or how far is the relation between hiPSC-CMs and the normal counterpart, but surely the possibility to obtain human CMs in a culture dish is adding a missing item to the scientific community. Every model has its own limitations and this is true also for hiPSC-CMs. Indeed, data from the literature reports the most evident and predictable phenotypes, leaving aside those perhaps more interesting but still unexplainable. However, as shown, many patients’ features are found in these hiPSC-CMs, starting from the prolongation of the APD corresponding to the elongation of the cardiac QT interval, or EAD or DAD events equivalent to arrhythmic episodes, or the presence of cellular lipid droplets primarily found in cardiac biopsies of patients with ARVC[77].
Human CMs have been derived also from cells of patients carrying other genetic disorders involving cardiac pathologies such as Leopard syndrome[78], Pompe disease[79], Duchenne muscular dystrophy[80], Friedreich’s ataxia[81], and Fabry disease[82].
In all of the cases, patients-derived CMs showed the main characteristic traits of the related pathology: hypertrophic cells in Leopard[78]; lower β-glucosidase activity, lower markers of metabolism, and higher glycogen content in Pompe[79]; a dystrophic gene expression profile in Duchenne[80]; impaired mitochondrial homeostasis in Friedreich’s ataxia[81]; and globotriaosylceramide accumulation in Fabry disease[82]. Moreover, patients-derived CMs represent a powerful model to test type and dosage of clinically used drugs, laying the basis for a personalized therapy.
Increasing the number and diversifying the type of modeled patients will surely improve the understanding of the biological mechanism that leads to the considered disease. Nevertheless the big ongoing efforts reside in eliminating any kind of interaction of the reprogramming technology with patient’s DNA and in identifying an easily reproducible protocol that leads to CM differentiation. Then, the non integrative Sendai-based reprogramming approach, followed by a chemical defined differentiation medium will surely solve the issue of reproducible cardiomyocyte differentiation.
P- Reviewer: Burlacu A, Salemi VMC S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18552] [Article Influence: 927.6] [Reference Citation Analysis (1)] |
| 2. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14550] [Article Influence: 808.3] [Reference Citation Analysis (0)] |
| 3. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7589] [Cited by in RCA: 7329] [Article Influence: 385.7] [Reference Citation Analysis (0)] |
| 4. | Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1775] [Cited by in RCA: 1616] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 5. | Wobus AM. Potential of embryonic stem cells. Mol Aspects Med. 2001;22:149-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 96] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 6. | Wobus AM, Rohwedel J, Maltsev V, Hescheler J. Development of cardiomyocytes expressing cardiac-specific genes, action potentials, and ionic channels during embryonic stem cell-derived cardiogenesis. Ann N Y Acad Sci. 1995;752:460-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 40] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 7. | Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 327] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 8. | Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855-860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1016] [Cited by in RCA: 1236] [Article Influence: 103.0] [Reference Citation Analysis (0)] |
| 9. | Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2014;114:511-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 656] [Cited by in RCA: 753] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 10. | Sartiani L, Bettiol E, Stillitano F, Mugelli A, Cerbai E, Jaconi ME. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells. 2007;25:1136-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 275] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 11. | Li S, Chen G, Li RA. Calcium signalling of human pluripotent stem cell-derived cardiomyocytes. J Physiol. 2013;591:5279-5290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 12. | Dolnikov K, Shilkrut M, Zeevi-Levin N, Gerecht-Nir S, Amit M, Danon A, Itskovitz-Eldor J, Binah O. Functional properties of human embryonic stem cell-derived cardiomyocytes: intracellular Ca2+ handling and the role of sarcoplasmic reticulum in the contraction. Stem Cells. 2006;24:236-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 144] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 13. | Lieu DK, Liu J, Siu CW, McNerney GP, Tse HF, Abu-Khalil A, Huser T, Li RA. Absence of transverse tubules contributes to non-uniform Ca(2+) wavefronts in mouse and human embryonic stem cell-derived cardiomyocytes. Stem Cells Dev. 2009;18:1493-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 127] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 14. | Ivashchenko CY, Pipes GC, Lozinskaya IM, Lin Z, Xiaoping X, Needle S, Grygielko ET, Hu E, Toomey JR, Lepore JJ. Human-induced pluripotent stem cell-derived cardiomyocytes exhibit temporal changes in phenotype. Am J Physiol Heart Circ Physiol. 2013;305:H913-H922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 15. | Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991-2002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 585] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 16. | Kamakura T, Makiyama T, Sasaki K, Yoshida Y, Wuriyanghai Y, Chen J, Hattori T, Ohno S, Kita T, Horie M. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circ J. 2013;77:1307-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 229] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 17. | Blazeski A, Zhu R, Hunter DW, Weinberg SH, Boheler KR, Zambidis ET, Tung L. Electrophysiological and contractile function of cardiomyocytes derived from human embryonic stem cells. Prog Biophys Mol Biol. 2012;110:178-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 18. | Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet. 2013;381:242-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 778] [Cited by in RCA: 902] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 19. | Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 531] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 20. | Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30-e41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1045] [Cited by in RCA: 996] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 21. | Ku L, Feiger J, Taylor M, Mestroni L. Cardiology patient page. Familial dilated cardiomyopathy. Circulation. 2003;108:e118-e121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Hershberger RE, Siegfried JD. Update 2011: clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2011;57:1641-1649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 257] [Cited by in RCA: 271] [Article Influence: 18.1] [Reference Citation Analysis (1)] |
| 23. | Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Peterson A, Li D, Jakobs P, Litt M, Porter CB. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156:161-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 201] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 24. | Kubben N, Voncken JW, Konings G, van Weeghel M, van den Hoogenhof MM, Gijbels M, van Erk A, Schoonderwoerd K, van den Bosch B, Dahlmans V. Post-natal myogenic and adipogenic developmental: defects and metabolic impairment upon loss of A-type lamins. Nucleus. 2011;2:195-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 25. | Siu CW, Lee YK, Ho JC, Lai WH, Chan YC, Ng KM, Wong LY, Au KW, Lau YM, Zhang J. Modeling of lamin A/C mutation premature cardiac aging using patient‐specific induced pluripotent stem cells. Aging (Albany NY). 2012;4:803-822. [PubMed] |
| 26. | Tse HF, Ho JC, Choi SW, Lee YK, Butler AW, Ng KM, Siu CW, Simpson MA, Lai WH, Chan YC. Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing. Hum Mol Genet. 2013;22:1395-1403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 27. | Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 544] [Article Influence: 38.9] [Reference Citation Analysis (1)] |
| 28. | Clarke SL, Bowron A, Gonzalez IL, Groves SJ, Newbury-Ecob R, Clayton N, Martin RP, Tsai-Goodman B, Garratt V, Ashworth M. Barth syndrome. Orphanet J Rare Dis. 2013;8:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 262] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 29. | Bione S, D’Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet. 1996;12:385-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 546] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 30. | Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616-623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 612] [Cited by in RCA: 666] [Article Influence: 55.5] [Reference Citation Analysis (13)] |
| 31. | Crotti L, Celano G, Dagradi F, Schwartz PJ. Congenital long QT syndrome. Orphanet J Rare Dis. 2008;3:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 190] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 32. | Giudicessi JR, Ackerman MJ. Genotype- and phenotype-guided management of congenital long QT syndrome. Curr Probl Cardiol. 2013;38:417-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 33. | Morita H, Wu J, Zipes DP. The QT syndromes: long and short. Lancet. 2008;372:750-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 215] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 34. | Wang DW, Yazawa K, George AL, Bennett PB. Characterization of human cardiac Na+ channel mutations in the congenital long QT syndrome. Proc Natl Acad Sci USA. 1996;93:13200-13205. [PubMed] |
| 35. | Liu M, Yang KC, Dudley SC. Cardiac sodium channel mutations: why so many phenotypes? Nat Rev Cardiol. 2014;11:607-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 36. | Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 937] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 37. | Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 794] [Cited by in RCA: 782] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 38. | Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J. 2011;32:952-962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 287] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 39. | Yazawa M, Dolmetsch RE. Modeling Timothy syndrome with iPS cells. J Cardiovasc Transl Res. 2013;6:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 40. | Davis RP, Casini S, van den Berg CW, Hoekstra M, Remme CA, Dambrot C, Salvatori D, Oostwaard DW, Wilde AA, Bezzina CR. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125:3079-3091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 194] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 41. | Egashira T, Yuasa S, Suzuki T, Aizawa Y, Yamakawa H, Matsuhashi T, Ohno Y, Tohyama S, Okata S, Seki T. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc Res. 2012;95:419-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 134] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 42. | Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkelä E, Hyttinen J, Kontula K, Swan H, Conklin BR. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5:220-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 218] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 43. | Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol. 2012;13:713-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 334] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 44. | Snyders DJ. Structure and function of cardiac potassium channels. Cardiovasc Res. 1999;42:377-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 123] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 45. | Mehta A, Sequiera GL, Ramachandra CJ, Sudibyo Y, Chung Y, Sheng J, Wong KY, Tan TH, Wong P, Liew R. Re-trafficking of hERG reverses long QT syndrome 2 phenotype in human iPS-derived cardiomyocytes. Cardiovasc Res. 2014;102:497-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 46. | Bellin M, Casini S, Davis RP, D’Aniello C, Haas J, Ward-van Oostwaard D, Tertoolen LG, Jung CB, Elliott DA, Welling A. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013;32:3161-3175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 47. | Zhou Z, Gong Q, Epstein ML, January CT. HERG channel dysfunction in human long QT syndrome. Intracellular transport and functional defects. J Biol Chem. 1998;273:21061-21066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 291] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 48. | Hoekstra M, Mummery CL, Wilde AA, Bezzina CR, Verkerk AO. Induced pluripotent stem cell derived cardiomyocytes as models for cardiac arrhythmias. Front Physiol. 2012;3:346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 154] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 49. | Gellens ME, George AL, Chen LQ, Chahine M, Horn R, Barchi RL, Kallen RG. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc Natl Acad Sci USA. 1992;89:554-558. [PubMed] |
| 50. | Terrenoire C, Wang K, Tung KW, Chung WK, Pass RH, Lu JT, Jean JC, Omari A, Sampson KJ, Kotton DN. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J Gen Physiol. 2013;141:61-72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 152] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 51. | Ma D, Wei H, Zhao Y, Lu J, Li G, Sahib NB, Tan TH, Wong KY, Shim W, Wong P. Modeling type 3 long QT syndrome with cardiomyocytes derived from patient-specific induced pluripotent stem cells. Int J Cardiol. 2013;168:5277-5286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 52. | Fatima A, Kaifeng S, Dittmann S, Xu G, Gupta MK, Linke M, Zechner U, Nguemo F, Milting H, Farr M. The disease-specific phenotype in cardiomyocytes derived from induced pluripotent stem cells of two long QT syndrome type 3 patients. PLoS One. 2013;8:e83005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 53. | Ruan Y, Liu N, Priori SG. Sodium channel mutations and arrhythmias. Nat Rev Cardiol. 2009;6:337-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 212] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 54. | Shaw RM, Colecraft HM. L-type calcium channel targeting and local signalling in cardiac myocytes. Cardiovasc Res. 2013;98:177-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 102] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 55. | Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230-234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 567] [Cited by in RCA: 504] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 56. | Priori SG, Chen SR. Inherited dysfunction of sarcoplasmic reticulum Ca2+ handling and arrhythmogenesis. Circ Res. 2011;108:871-883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 376] [Cited by in RCA: 349] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 57. | Chelu MG, Wehrens XH. Sarcoplasmic reticulum calcium leak and cardiac arrhythmias. Biochem Soc Trans. 2007;35:952-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 58. | Cerrone M, Colombi B, Santoro M, di Barletta MR, Scelsi M, Villani L, Napolitano C, Priori SG. Bidirectional ventricular tachycardia and fibrillation elicited in a knock-in mouse model carrier of a mutation in the cardiac ryanodine receptor. Circ Res. 2005;96:e77-e82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 210] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 59. | Beard NA, Laver DR, Dulhunty AF. Calsequestrin and the calcium release channel of skeletal and cardiac muscle. Prog Biophys Mol Biol. 2004;85:33-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 211] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 60. | Iyer V, Armoundas AA. Unraveling the mechanisms of catecholaminergic polymorphic ventricular tachycardia. Conf Proc IEEE Eng Med Biol Soc. 2006;Suppl:6761-6764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 61. | Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnáiz-Cot JJ, Rosa AO, Nguemo F, Matzkies M, Dittmann S. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell Physiol Biochem. 2011;28:579-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 150] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 62. | Kujala K, Paavola J, Lahti A, Larsson K, Pekkanen-Mattila M, Viitasalo M, Lahtinen AM, Toivonen L, Kontula K, Swan H. Cell model of catecholaminergic polymorphic ventricular tachycardia reveals early and delayed afterdepolarizations. PLoS One. 2012;7:e44660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 63. | Zhang XH, Haviland S, Wei H, Sarić T, Fatima A, Hescheler J, Cleemann L, Morad M. Ca2+ signaling in human induced pluripotent stem cell-derived cardiomyocytes (iPS-CM) from normal and catecholaminergic polymorphic ventricular tachycardia (CPVT)-afflicted subjects. Cell Calcium. 2013;54:57-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 64. | Jung CB, Moretti A, Mederos y Schnitzler M, Iop L, Storch U, Bellin M, Dorn T, Ruppenthal S, Pfeiffer S, Goedel A. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med. 2012;4:180-191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 254] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 65. | Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, Miller L, Belhassen B, Nof E, Glikson M. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol. 2012;60:990-1000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 66. | Di Pasquale E, Lodola F, Miragoli M, Denegri M, Avelino-Cruz JE, Buonocore M, Nakahama H, Portararo P, Bloise R, Napolitano C. CaMKII inhibition rectifies arrhythmic phenotype in a patient-specific model of catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis. 2013;4:e843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 67. | Novak A, Barad L, Zeevi-Levin N, Shick R, Shtrichman R, Lorber A, Itskovitz-Eldor J, Binah O. Cardiomyocytes generated from CPVTD307H patients are arrhythmogenic in response to β-adrenergic stimulation. J Cell Mol Med. 2012;16:468-482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 134] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 68. | Rizzi N, Liu N, Napolitano C, Nori A, Turcato F, Colombi B, Bicciato S, Arcelli D, Spedito A, Scelsi M. Unexpected structural and functional consequences of the R33Q homozygous mutation in cardiac calsequestrin: a complex arrhythmogenic cascade in a knock in mouse model. Circ Res. 2008;103:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 114] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 69. | Knollmann BC, Chopra N, Hlaing T, Akin B, Yang T, Ettensohn K, Knollmann BE, Horton KD, Weissman NJ, Holinstat I. Casq2 deletion causes sarcoplasmic reticulum volume increase, premature Ca2+ release, and catecholaminergic polymorphic ventricular tachycardia. J Clin Invest. 2006;116:2510-2520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 262] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 70. | Basso C, Bauce B, Corrado D, Thiene G. Pathophysiology of arrhythmogenic cardiomyopathy. Nat Rev Cardiol. 2012;9:223-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 187] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 71. | Sen-Chowdhry S, Syrris P, McKenna WJ. Role of genetic analysis in the management of patients with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J Am Coll Cardiol. 2007;50:1813-1821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 167] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 72. | Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X, Oh Y, Tan SH, Ng ML, Shim W. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. 2013;34:1122-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 73. | Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M, Gepstein L. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ Cardiovasc Genet. 2013;6:557-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 74. | Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 388] [Cited by in RCA: 423] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 75. | Garrod D, Chidgey M. Desmosome structure, composition and function. Biochim Biophys Acta. 2008;1778:572-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 441] [Cited by in RCA: 410] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 76. | Cerrone M, Lin X, Zhang M, Agullo-Pascual E, Pfenniger A, Chkourko Gusky H, Novelli V, Kim C, Tirasawadichai T, Judge DP. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 2014;129:1092-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 266] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 77. | Fujita S, Terasaki F, Otsuka K, Katashima T, Kanzaki Y, Kawamura K, Tanaka T, Kitaura Y. Markedly increased intracellular lipid droplets and disruption of intercellular junctions in biopsied myocardium from a patient with arrhythmogenic right ventricular cardiomyopathy. Heart Vessels. 2008;23:440-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 78. | Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808-812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 581] [Cited by in RCA: 517] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 79. | Huang HP, Chen PH, Hwu WL, Chuang CY, Chien YH, Stone L, Chien CL, Li LT, Chiang SC, Chen HF. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum Mol Genet. 2011;20:4851-4864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 80. | Dick E, Kalra S, Anderson D, George V, Ritson M, Laval S, Barresi R, Aartsma-Rus A, Lochmuller H, Denning C. Exon skipping and gene transfer restore dystrophin expression in hiPSC-cardiomyocytes harbouring DMD mutations. Stem Cells Dev. 2013;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 81. | Hick A, Wattenhofer-Donzé M, Chintawar S, Tropel P, Simard JP, Vaucamps N, Gall D, Lambot L, André C, Reutenauer L. Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich’s ataxia. Dis Model Mech. 2013;6:608-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 82. | Itier JM, Ret G, Viale S, Sweet L, Bangari D, Caron A, Le-Gall F, Bénichou B, Leonard J, Deleuze JF. Effective clearance of GL-3 in a human iPSC-derived cardiomyocyte model of Fabry disease. J Inherit Metab Dis. 2014;37:1013-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 83. | Passier R, Mummery C. Cardiomyocyte differentiation from embryonic and adult stem cells. Curr Opin Biotechnol. 2005;16:498-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 84. | Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, Peters A, Mahairaki V, Koliatsos VE, Tung L, Zambidis ET. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 312] [Cited by in RCA: 321] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 85. | Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 868] [Cited by in RCA: 886] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 86. | Wei H, Tan G, Manasi S, Kong G, Yong P, Koh C, Ooi TH, Lim SY, Wong P, Gan SU. One-step derivation of cardiomyocytes and mesenchymal stem cells from human pluripotent stem cells. Stem Cell Res. 2012;9:87-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 78] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 87. | Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 2012;111:1125-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 353] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 88. | Cao N, Liang H, Huang J, Wang J, Chen Y, Chen Z, Yang HT. Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell Res. 2013;23:1119-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 89. | Haraguchi Y, Matsuura K, Shimizu T, Yamato M, Okano T. Simple suspension culture system of human iPS cells maintaining their pluripotency for cardiac cell sheet engineering. J Tissue Eng Regen Med. 2013;Jun 3; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 90. | Lim SY, Sivakumaran P, Crombie DE, Dusting GJ, Pébay A, Dilley RJ. Trichostatin A enhances differentiation of human induced pluripotent stem cells to cardiogenic cells for cardiac tissue engineering. Stem Cells Transl Med. 2013;2:715-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 91. | Pesl M, Acimovic I, Pribyl J, Hezova R, Vilotic A, Fauconnier J, Vrbsky J, Kruzliak P, Skladal P, Kara T. Forced aggregation and defined factors allow highly uniform-sized embryoid bodies and functional cardiomyocytes from human embryonic and induced pluripotent stem cells. Heart Vessels. 2014;29:834-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 92. | Cho SW, Park JS, Heo HJ, Park SW, Song S, Kim I, Han YM, Yamashita JK, Youm JB, Han J. Dual modulation of the mitochondrial permeability transition pore and redox signaling synergistically promotes cardiomyocyte differentiation from pluripotent stem cells. J Am Heart Assoc. 2014;3:e000693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 93. | Karakikes I, Senyei GD, Hansen J, Kong CW, Azeloglu EU, Stillitano F, Lieu DK, Wang J, Ren L, Hulot JS. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl Med. 2014;3:18-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (11)] |