Published online Jan 26, 2026. doi: 10.4252/wjsc.v18.i1.111348
Revised: August 7, 2025
Accepted: November 18, 2025
Published online: January 26, 2026
Processing time: 202 Days and 18.6 Hours
Breast cancer is one of the most prevalent malignancies affecting women world
To comprehensively characterize the molecular features of BCSCs through multi-omics approaches, construct a prognostic prediction model based on stem cell-related genes, reveal cell-cell communication networks within the tumor microenvironment, and provide theoretical foundation for personalized treatment stra
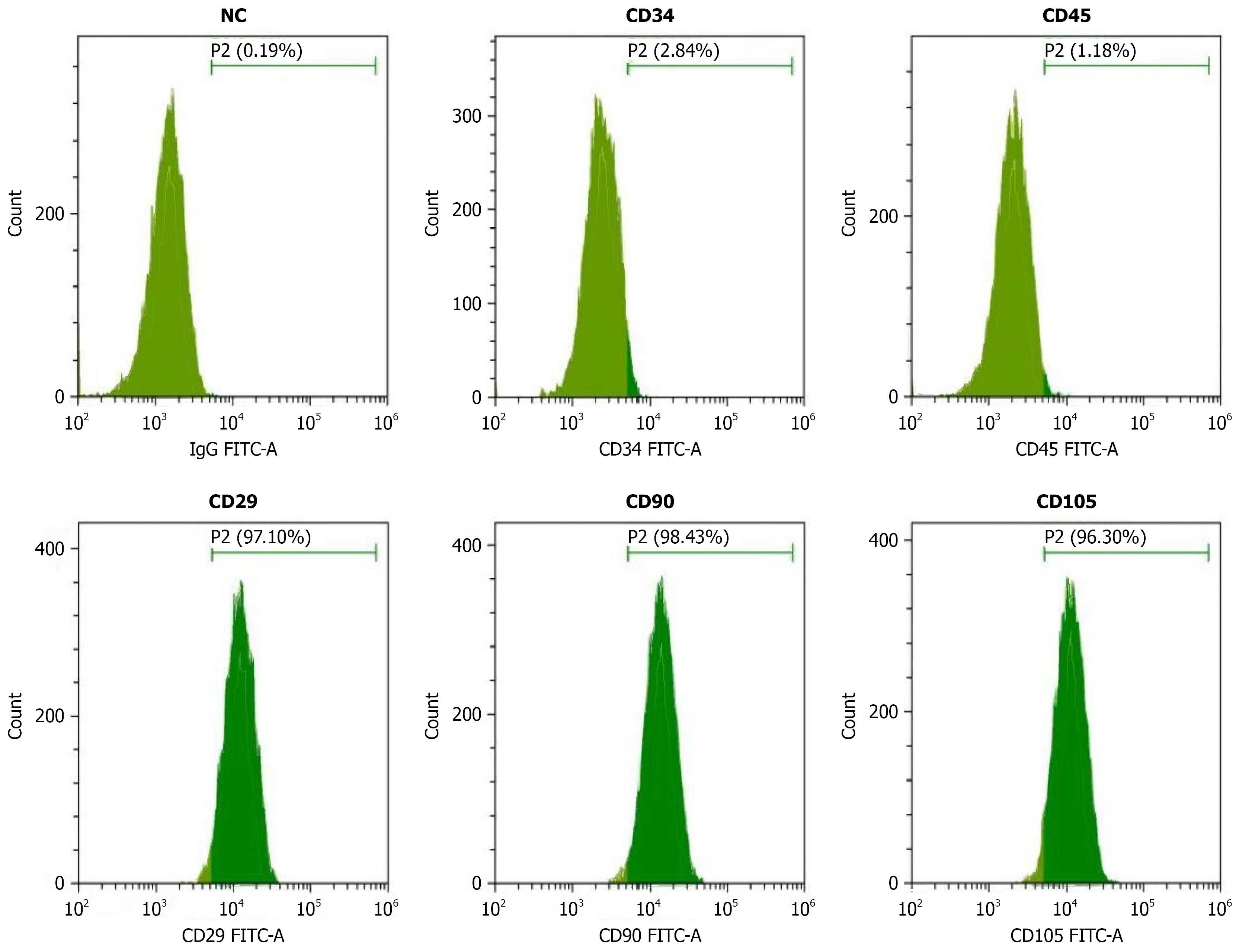
Flow cytometry was employed to detect the expression of BCSC surface markers (CD34, CD45, CD29, CD90, CD105). Transcriptomic analysis was performed to identify differentially expressed genes. Least absolute shrinkage and selection operator regression analysis was utilized to screen key prognostic genes and construct a risk scoring model. Single-cell RNA sequencing and spatial transcriptomics were applied to analyze tumor heterogeneity and spatial gene expression patterns. Cell-cell communication network analysis was conducted to reveal interactions between stem cells and the microenvironment.
Flow cytometric analysis revealed the highest expression of CD105 (96.30%), followed by CD90 (68.43%) and CD34 (62.64%), while CD29 showed lower expression (7.16%) and CD45 exhibited the lowest expression (1.19%). Transcriptomic analysis identified 3837 significantly differentially expressed genes (1478 upregulated and 2359 downregulated). Least absolute shrinkage and selection operator regression analysis selected 10 key prognostic genes, and the constructed risk scoring model effectively distinguished between high-risk and low-risk patient groups (P < 0.001). Single-cell analysis revealed tumor cellular heterogeneity, and spatial transcriptomics demon
This study comprehensively characterized the molecular features of BCSCs through multi-omics approaches, identified reliable surface markers and key regulatory genes, and constructed a prognostic prediction model with clinical application value.
Core Tip: This study integrates flow cytometry, transcriptomics, least absolute shrinkage and selection operator modeling, single-cell and spatial transcriptomics to comprehensively characterize breast cancer stem cells. CD105 was identified as a key surface marker, and a prognostic gene signature including MED18 was developed. MED18 showed strong correlations with tumor, immune, and stromal cells, suggesting a central role in breast cancer stem cell regulation and microenvironment interaction. These findings offer novel insights into tumor heterogeneity and provide potential biomarkers and therapeutic targets for precision treatment in breast cancer.
- Citation: Guo DY, Liu ZY, Yi QC. Breast cancer stem cell activity driven by ME18D gene expression in the tumor microenvironment. World J Stem Cells 2026; 18(1): 111348
- URL: https://www.wjgnet.com/1948-0210/full/v18/i1/111348.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v18.i1.111348
Breast cancer remains one of the most prevalent malignancies affecting women worldwide, with approximately 2.3 million new cases diagnosed annually and representing the leading cause of cancer-related mortality among females globally. Despite significant advances in early detection, surgical techniques, chemotherapy, radiotherapy, and targeted therapies, breast cancer continues to pose substantial clinical challenges, particularly in terms of treatment resistance, disease recurrence, and metastatic progression[1-3]. The heterogeneous nature of breast cancer, encompassing multiple molecular subtypes with distinct biological behaviors and therapeutic responses, underscores the critical need for personalized treatment approaches and improved prognostic stratification methods.
The cancer stem cell (CSC) hypothesis has emerged as a revolutionary paradigm in cancer biology, proposing that a small subpopulation of tumor cells possesses stem cell-like properties including self-renewal capacity, multipotent differentiation potential, and enhanced tumorigenic activity[4-6]. These CSCs have the ability to give rise to a tumor when only a small number of them are transplanted, have unlimited proliferative potential, and are capable of generating the diverse set of cell types that are present in the heterogeneous masses of the tumors. In breast cancer, there is increasing evidence showing that the breast CSCs (BCSCs) have critical contributions to tumorigenesis, tumor evolution, metastasis, resis
BCSCs show unique molecular profiles and surface marker expression compared with the bulk of tumor cells. Among the most studied surface markers for BCSC enrichment, we can mention CD34, a glycoprotein linked to hematopoietic stem cells and endothelial precursor cells; CD29 (integrin β1) involved in cell-matrix adhesion and signaling pathways essential for stem cell survival; CD90 (Thy-1) a glycosylphosphatidylinositol-anchored glycoprotein that mediates cell-cell and cell-matrix interactions and CD105 (endoglin), a transmembrane glycoprotein that plays important roles in angiogenesis and cellular proliferation. The distributions of these markers, such as the high expression of CD90 (97%) and CD105 (60%), supply critical information on the phenotypic profile of BCSCs and their clinical relevance[10,11]. BCSC properties are maintained through a variety of complex molecular networks, including transcription factors, epigenetic regulators, signaling pathways, and the tumor microenvironment. Critical transcription factors, such as NANOG, OCT4, and SOX2 network to establish core regulons that control stemness conservation and self-renewal potential[12,13]. Moreover, Wnt/β-catenin, Notch, Hedgehog and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) signaling pathways are involved in stem cell fate determination and stem cell function and properties[14,15].
Using such multi-level integrative strategy, we aim to identify new biomarkers, understand relevant regulatory mechanisms and build clinically useful prognostic devices that can inform personalized therapeutic decisions and impact patient outcomes. Ultimately, with this research, we hope to translate advances in basic CSC biology into clinical applications, producing a platform for designing next generation and individually targeted therapeutics for breast cancer patients. By profiling breast cancer tumourigenicity-related properties and their molecular networks at the single-cell level, this study can aid in promoting precision oncology and patient care in the personalized medicine era.
Breast cancer cell lines were obtained from the American Type Culture Collection and maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were cultured in a humidified incubator at 37 °C with 5% CO2 atmosphere. Cell passages were performed when cultures reached 80%-90% confluence using 0.25% trypsin-EDTA solution. All experiments were conducted using cells within passages 3-10 to maintain cellular characteristics and experimental consistency.
BCSCs were isolated using established protocols based on sphere formation assays and surface marker expression. Cells were seeded at low density (1000 cells/mL) in serum-free mammosphere medium containing Dulbecco’s Modified Eagle’s Medium/F12, B27 supplement, 20 ng/mL epidermal growth factor, 10 ng/mL basic fibroblast growth factor, and 4 μg/mL insulin. Primary mammospheres were collected after 7-10 days of culture, dissociated into single cells using enzymatic digestion, and replated for secondary sphere formation to enhance stem cell purity. The sphere-forming efficiency was calculated as the percentage of spheres formed relative to the initial number of cells seeded.
Flow cytometry was performed to identify and quantify BCSCs based on surface marker expression. Cells were harvested using trypsin-free cell dissociation solution to preserve surface antigens, washed twice with phosphate-buffered saline, and resuspended at a concentration of 1 × 106 cells/mL. Primary antibodies against stem cell markers including CD34-FITC, CD45-FITC, CD29-FITC, CD90-FITC, and CD105-FITC were added at manufacturer-recommended concentrations and incubated for 30 minutes at 4 °C in the dark[16,17]. Unstained cells served as negative controls (NCs) to determine background fluorescence levels. After antibody incubation, cells were washed three times with phosphate-buffered saline containing 2% fetal bovine serum and analyzed using a flow cytometer equipped with appropriate laser and detector configurations. Data acquisition was performed on at least 10000 events per sample, and analysis was conducted using specialized flow cytometry software. Gates were set based on forward scatter and side scatter parameters to exclude debris and dead cells. The percentage of positive cells for each marker was determined by comparing fluorescence intensity with NC samples.
Protein-protein interaction networks were constructed using differentially expressed genes identified from transcriptomic analysis. Known and predicted protein interactions were retrieved from established databases including STRING, BioGRID, and IntAct. Network visualization and analysis were performed using specialized bioinformatics software to identify key regulatory nodes and functional modules within the network. Hub genes were defined as those with the highest connectivity scores and centrality measures within the interaction network.
Gene Ontology (GO) enrichment analysis was performed to identify biological processes, molecular functions, and cellular components significantly associated with differentially expressed genes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted to map genes onto known signaling pathways and metabolic networks. Enrichment significance was determined using hypergeometric tests with Benjamini-Hochberg correction for multiple testing[18-20]. Pathways with adjusted P values < 0.05 were considered significantly enriched. Functional an
Least absolute shrinkage and selection operator (LASSO) regression analysis was employed to identify key prognostic genes from the pool of differentially expressed genes. The analysis was performed using survival data and gene expression profiles to select genes with the strongest association with patient outcomes. Cross-validation procedures were implemented to determine optimal regularization parameters and prevent overfitting[21-23]. The LASSO algorithm iteratively adjusted regression coefficients, shrinking less important variables toward zero while retaining the most predictive features. Ten-fold cross-validation was performed to identify the optimal lambda (λ) value that minimized prediction error. Genes with non-zero coefficients in the final model were selected as key prognostic markers and used to construct a risk scoring system. A prognostic risk score was calculated for each sample using the formula: Risk score = Σ(coefficient × gene expression level) for all selected genes in the LASSO model. Patients were stratified into high-risk and low-risk groups based on the median risk score as the cutoff value. The prognostic performance of the risk score was evaluated using time-dependent receiver operating characteristic curves and Kaplan-Meier survival analysis.
Single-cell RNA sequencing was performed on breast cancer tissue samples to characterize cellular heterogeneity and identify stem cell populations at the single-cell level. Cells were dissociated from fresh tissue samples using enzymatic digestion protocols optimized to maintain cell viability and RNA integrity[24-26]. Single-cell libraries were prepared using droplet-based or plate-based methods and sequenced to sufficient depth for reliable gene expression quantification. Breast cancer spatial transcriptomic datasets generated using the 10 × Genomics Visium platform were selected based on the following criteria: (1) Tissue sections with > 3000 detected spots; (2) Average of > 4000 genes detected per spot; (3) Tissue coverage > 75%; and (4) Available clinical annotations. Three representative breast cancer tissue samples meeting these criteria were selected for comprehensive analysis. The selected datasets utilized the Visium spatial gene expression platform with 55 μm diameter capture spots and 100 μm center-to-center spacing, providing spatial resolution of approximately 1-10 cells per spot. This resolution enabled detection of tissue-level spatial patterns while maintaining single-cell sensitivity for gene expression profiling.
Cell-cell communication networks were inferred from single-cell RNA sequencing data using computational algorithms that predict ligand-receptor interactions between different cell types. Database resources containing known ligand-receptor pairs were utilized to identify potential communication pathways. The strength and significance of predicted interactions were assessed using statistical methods that account for gene expression levels and cell type abundance. Network visualization tools were employed to display complex communication patterns between different cell po
Statistical analysis were performed using appropriate software packages including R, Python, and specialized bioinformatics tools. Continuous variables were compared using Student’s t-tests or Mann-Whitney U tests depending on data distribution. Categorical variables were analyzed using χ2 tests or Fisher’s exact tests. Survival analysis was conducted using Cox proportional hazards models and log-rank tests. Data visualization included principal component analysis plots, heatmaps, volcano plots, network diagrams, and survival curves. All statistical tests were two-sided, and P values < 0.05 were considered statistically significant. Multiple testing corrections were applied where appropriate using false discovery rate control methods. All experiments were performed in biological triplicates with technical replicates to ensure reproducibility and statistical power.
Flow cytometry was performed to detect and identify key stem cell surface markers in breast cancer cells. The NC showed only 0.19% positivity, confirming the specificity and reliability of the detection system. Among the stem cell markers analyzed, CD34 demonstrated 62.64% positive expression, indicating a substantial proportion of tumor cells with stem cell-like characteristics, while CD45 showed low positivity (1.19%), confirming minimal immune cell contamination. Further stem cell marker analysis revealed CD29 positivity at 7.16%, CD90 at 68.43%, and CD105 at 96.30%. The high expression rates of CD90 and CD105 suggest these markers may serve as important indicators for identifying BCSCs. Particularly, the extremely high CD105 positivity (96.30%) indicates its universal expression in BCSCs and potential key role in maintaining stem cell properties and promoting tumor angiogenesis. These flow cytometric results provide important experimental evidence for BCSC phenotypic characteristics and establish a foundation for subsequent functional studies and targeted therapeutic strategies (Figure 1).
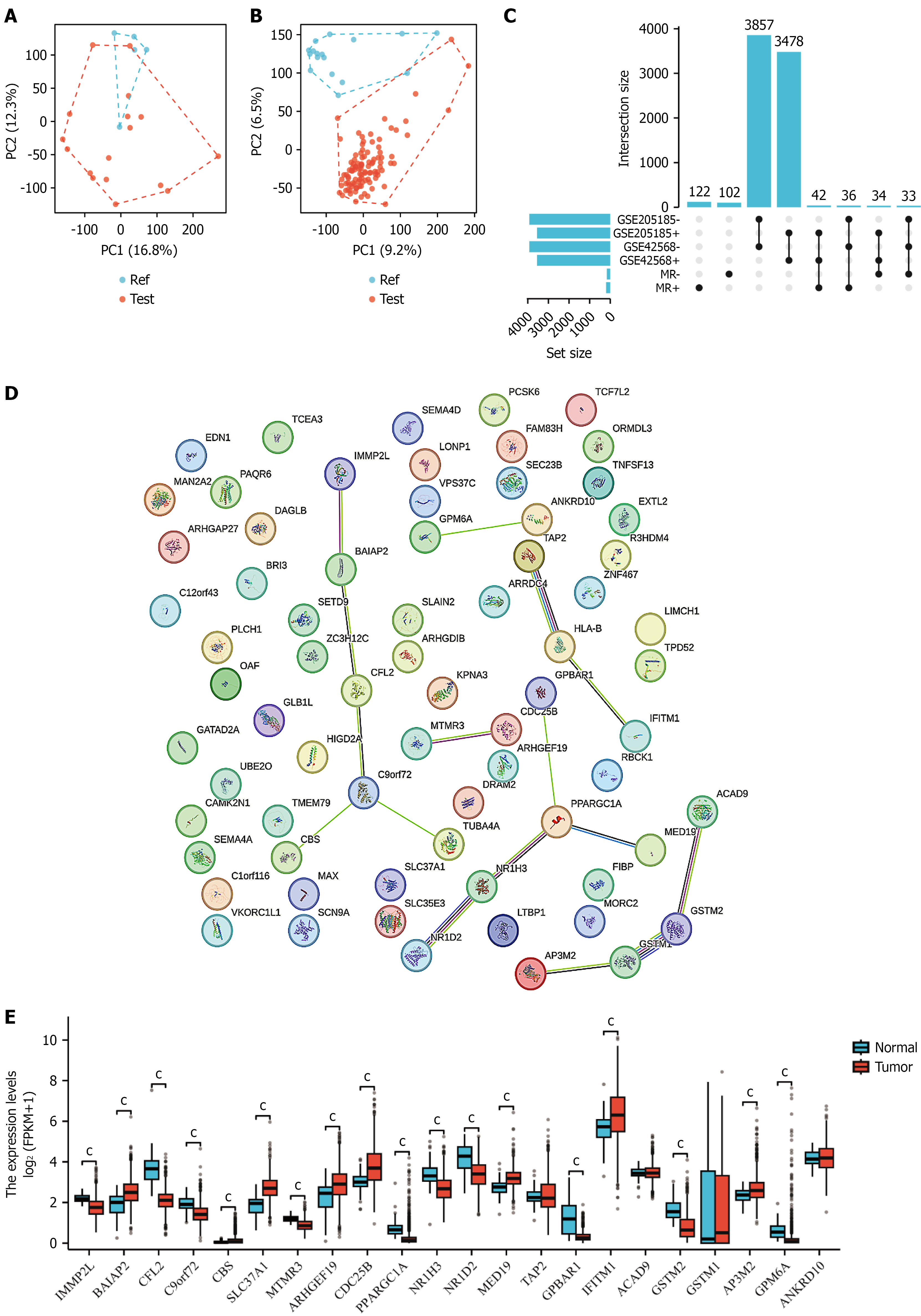
Principal component analysis revealed distinct separation between BCSC samples and control samples at the transcriptomic level. In the PC1-PC2 dimensional space (Figure 2A and B), stem cell samples (blue dots) and control samples (red dots) formed two independent clusters, with PC1 and PC2 explaining 16.8% and 9.2% of variance respectively, confirming that BCSCs possess unique molecular characteristics. Differential expression analysis identified 3837 significantly altered genes, including 1478 upregulated and 2359 downregulated genes (Figure 2C). These genes are involved in stem cell biological functions, cell cycle regulation, and signal transduction pathways. The expression heatmap demonstrated clear pattern differences between the two groups, supporting the existence of specific transcriptional networks in BCSCs. Protein-protein interaction network analysis (Figure 2D) revealed complex functional associations among differentially expressed genes, highlighting key regulatory nodes potentially crucial for maintaining stem cell characteristics. Comparative analysis of gene expression in normal vs tumor tissues (Figure 2E) showed abnormal expression patterns of stem cell-related genes in breast cancer, providing molecular insights into stem cell regulatory network disruption during tumorigenesis.
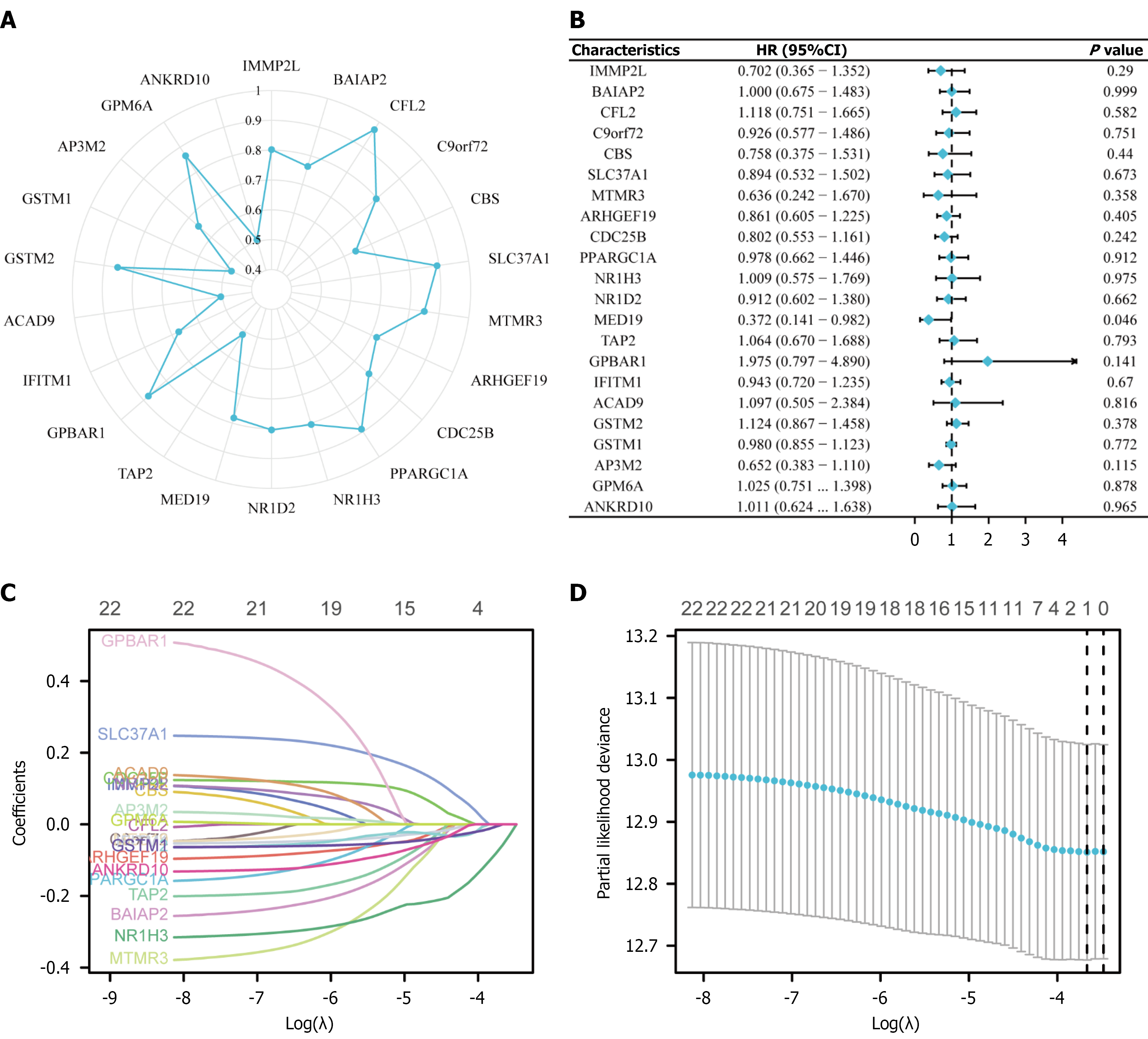
To identify key genes associated with BCSC characteristics, LASSO regression analysis was performed. Protein-protein interaction network analysis (Figure 3A) revealed core regulatory genes including ANKHD1, DMAP1, BAIAP2, GSTM1, PPARGC1A, and others that play crucial roles in stem cell function regulation. Forest plot analysis (Figure 3B) demonstrated the prognostic value of these genes, with most functioning as protective factors (hazard ratio < 1), including BAIAP2, TXNIP, and PPARGC1A, while genes like GPSM1 and ANKHD1 acted as risk factors. LASSO coefficient trajectory analysis (Figure 3C) showed how gene coefficients changed with increasing penalty parameter λ, with most coefficients approaching zero during regularization. Ten-fold cross-validation (Figure 3D) identified the optimal λ value at logλ ≈ -5.5, retaining 10 key genes for the final prognostic model. This LASSO-derived gene signature provides both biological relevance to stem cell characteristics and strong prognostic prediction capability for breast cancer patients.
The LASSO-based risk score model was successfully validated across independent datasets. Risk score distribution analysis (Figure 4A) showed significant differences between high-risk and low-risk groups (P < 0.001), confirming effective patient stratification. Clinical correlation analysis (Figure 4B) revealed associations with tumor staging, molecular subtypes, and metastatic status, demonstrating clinical applicability. Gene expression validation (Figure 4C and D) confirmed significant differential expression of core model genes between risk groups (P < 0.001), supporting their reliability as prognostic markers. Notably, stem cell-associated genes showed abnormal expression in high-risk patients, suggesting their role in regulating stem cell activity and patient outcomes. Single-cell RNA sequencing analysis (Figure 4E-G) revealed tumor heterogeneity through t-SNE visualization, identifying distinct cell subpopulations with unique transcriptional profiles. Cell type annotation showed some subpopulations exhibited typical stem cell characteristics with high expression of pluripotency and self-renewal genes, providing single-cell evidence for the biological foundation of the risk score model.
To understand the biological functions of key genes in the BCSC risk score model, systematic pathway enrichment and functional network analyses were performed. GO enrichment analysis (Figure 5A) revealed that differentially expressed genes were primarily enriched in cell cycle regulation, DNA repair, protein folding, and apoptosis control, which are closely related to stem cell self-renewal and fate determination. Protein-protein interaction network analysis (Figure 5B) demonstrated complex functional associations among key genes, revealing important regulatory hubs including transcription factors, signaling proteins, and metabolic enzymes. KEGG pathway analysis (Figure 5C) identified cancer-related signaling pathways such as cell cycle, p53, PI3K/Akt, and mitogen activated protein-kinase pathways. Pathway network heatmaps (Figure 5D and E) displayed enrichment levels with red indicating upregulated gene pathways and green indicating downregulated gene pathways. Comprehensive functional enrichment results (Figure 5F) integrated GO and KEGG findings, highlighting significant enrichment in cell cycle checkpoints, DNA damage response, and stem cell pluripotency maintenance. These findings validate the biological rationale of the risk score model and provide systematic insights into BCSC molecular regulation mechanisms.
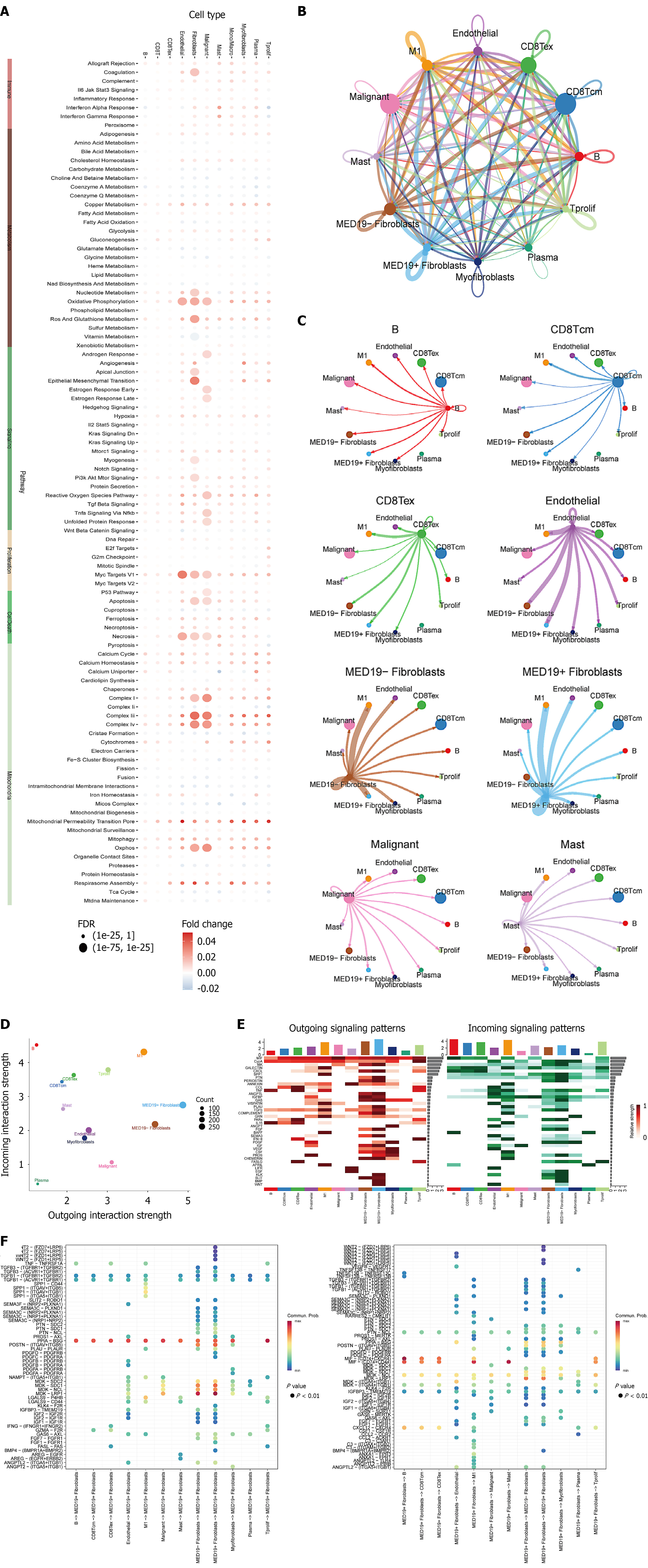
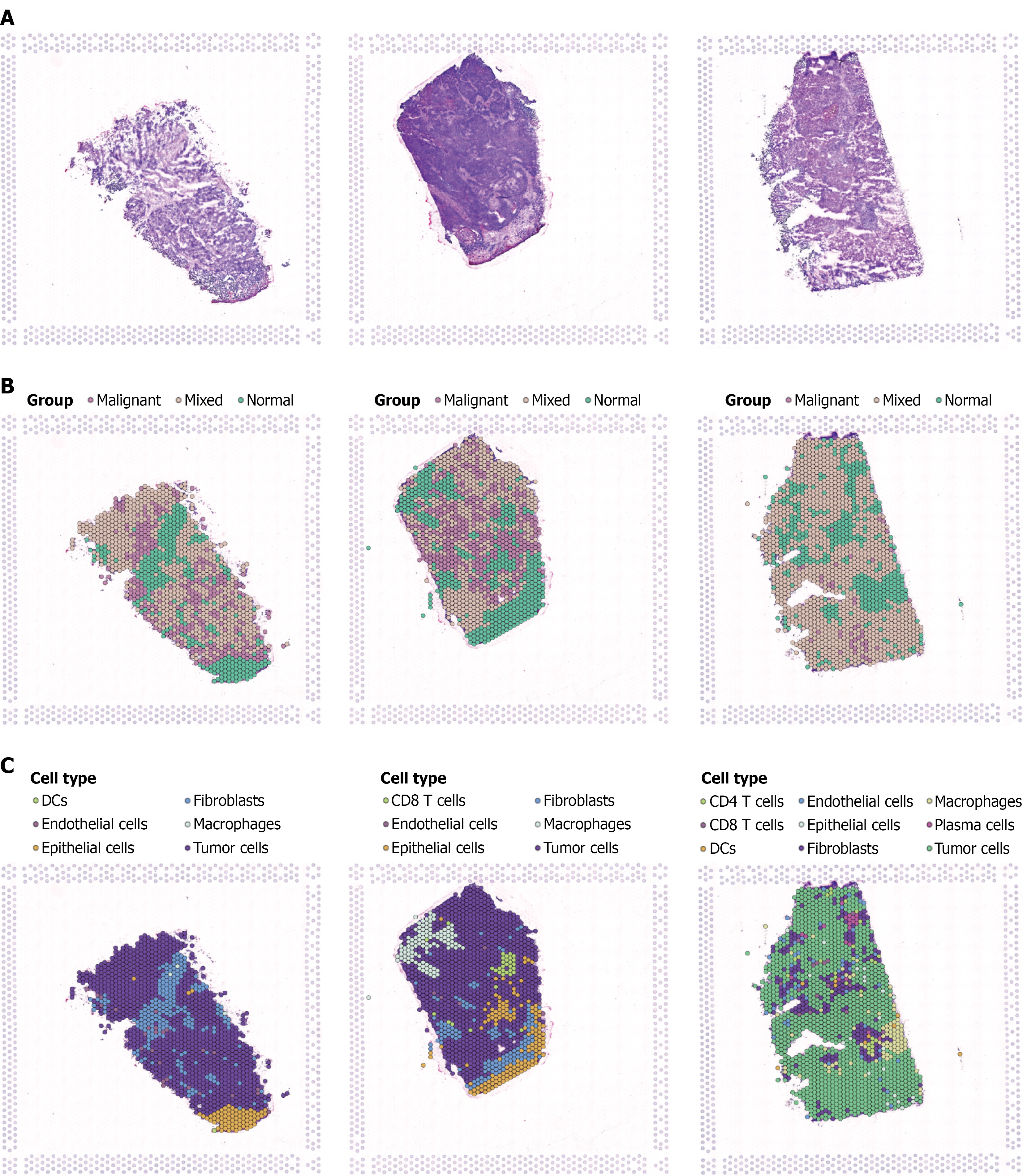
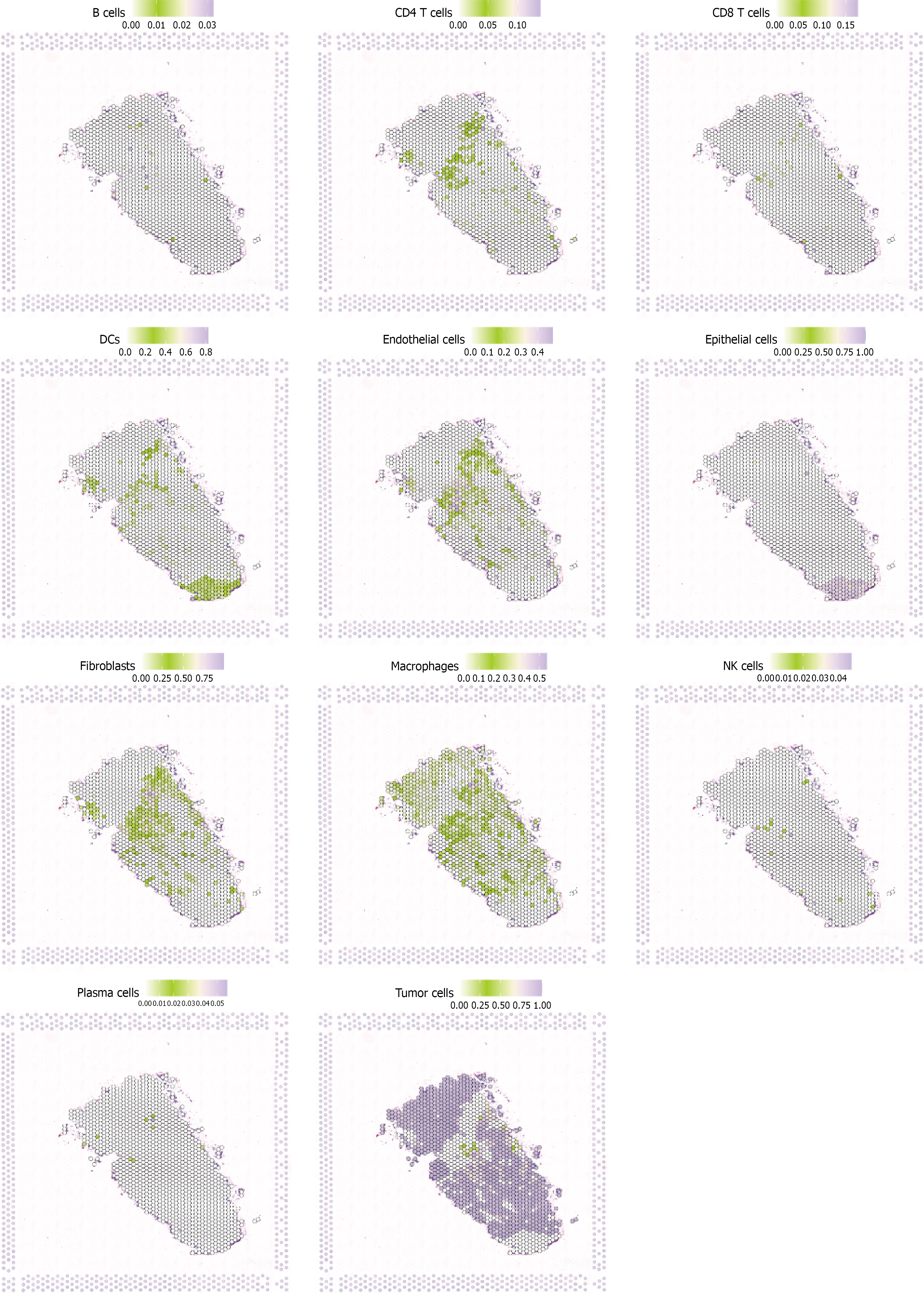
Single-cell spatial transcriptomic analysis was performed on three breast cancer tissue samples to examine cellular composition and spatial distribution. Tissue morphological analysis (Figure 6A) showed typical tumor architecture with cancer cell aggregation areas and stromal tissue distribution, providing histological foundation for cellular analysis. Risk score spatial distribution (Figure 6B) revealed spatial heterogeneity within tumor tissues, with high-risk regions (red/orange) and low-risk regions (green/blue) showing distinct clustering patterns. This heterogeneity indicates the presence of cell subpopulations with different prognostic risks that are spatially clustered, reflecting local tumor microenvironment influences on stem cell properties. Cell type annotation (Figure 6C) identified major cellular components including cancer cells, immune cells, fibroblasts, and endothelial cells. Cancer cells clustered in parenchymal regions while immune and stromal cells distributed in margins and interstitial areas. The spatial proximity of different cell types suggests potential cell-cell interactions that may influence stem cell behavior and tumor progression.
To understand the spatial distribution characteristics of key genes selected by LASSO regression, spatial transcriptomic analysis was performed on 11 core genes in breast cancer tissues. Results revealed highly heterogeneous spatial expression patterns of these stem cell-related genes within tumor tissues. Genes such as BCL6, COL7A2, and COL15A1 showed relatively localized expression in specific tumor regions, suggesting their role in maintaining local stem cell properties. In contrast, genes like macrophages and MXRA8 exhibited higher expression levels at tissue margins and stromal areas, correlating with immune infiltration and matrix remodeling processes in the tumor microenvironment. Spatial expression analysis revealed that fibronectin, macrophage, and MXRA8 genes formed distinct expression gradients from tumor center to periphery, potentially reflecting spatial differences in oxygen, nutrients, and growth factors that influence stem cell biological activity. Tumor cells-related genes showed expression patterns highly consistent with tumor cell density, validating their association with stem cell property maintenance. These spatial expression findings provide important spatial biological evidence for understanding BCSC function within the complex three-dimensional tumor environment (Figure 7).
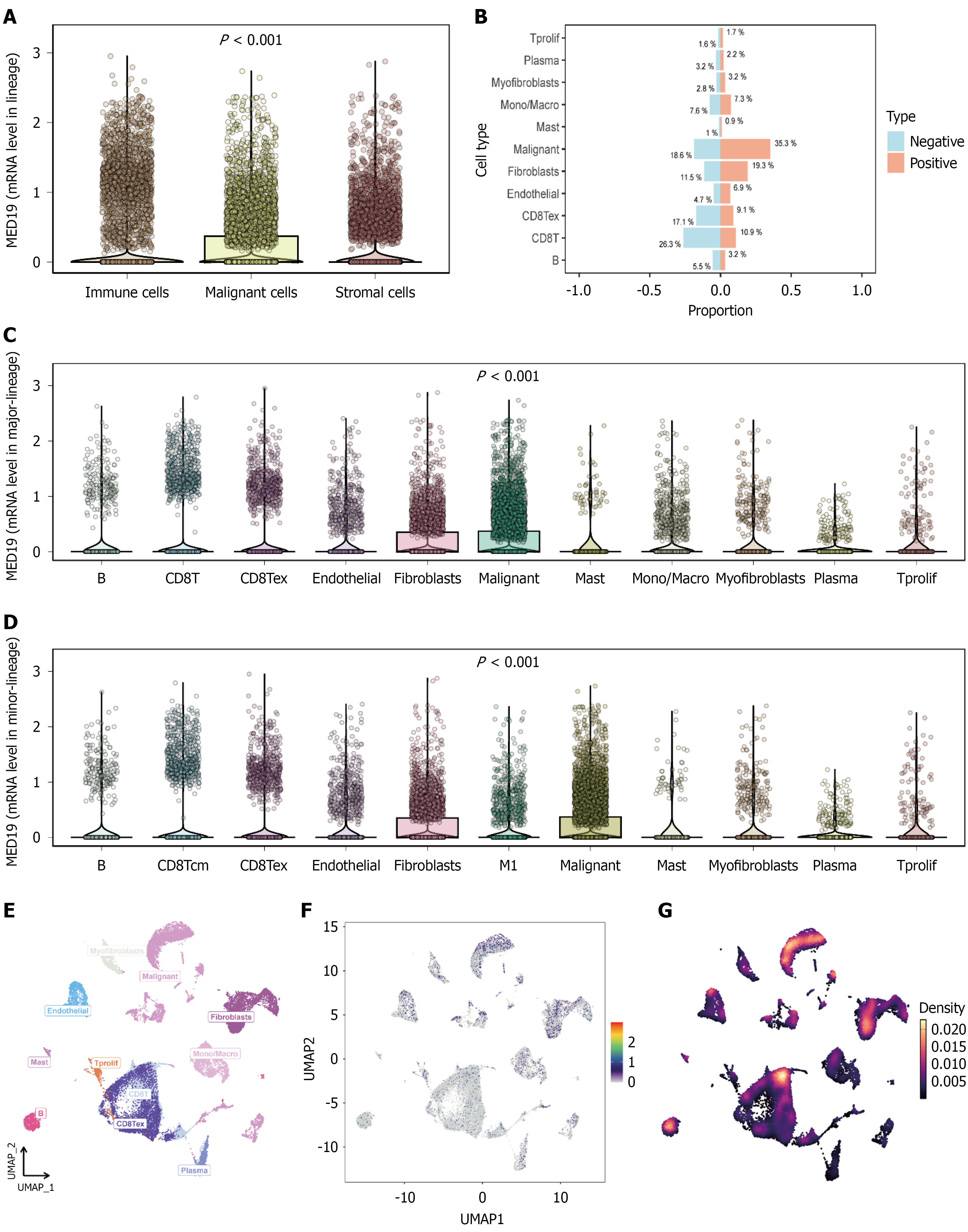
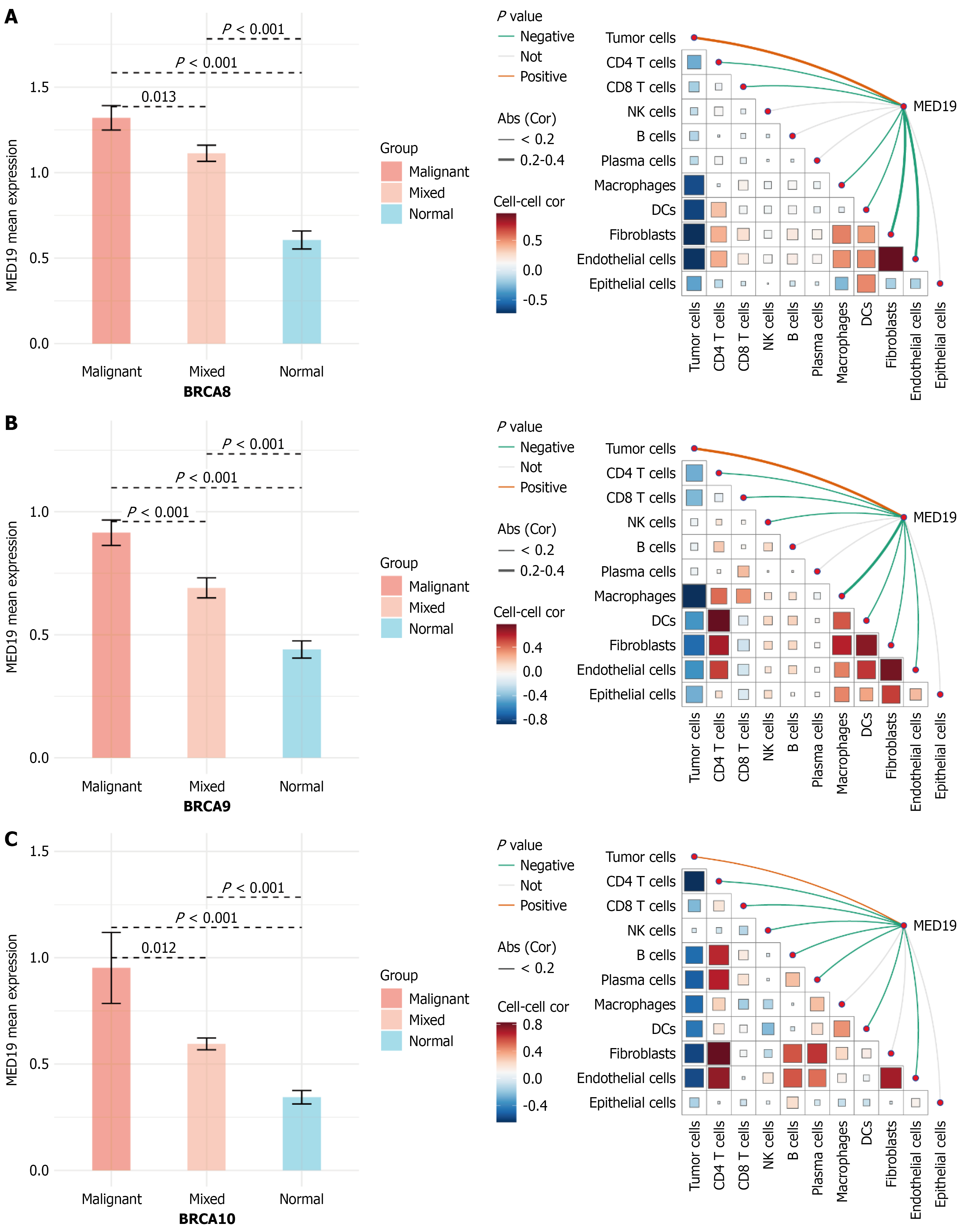
To validate the clinical significance of key genes from the LASSO regression model, MED18 gene expression and its relationship with cell-cell communication were analyzed in breast cancer tissues. Consistent patterns were observed across three independent breast cancer samples (Figure 8). MED18 expression was significantly higher in malignant tissues compared to normal controls (P < 0.001), and also showed significant upregulation in breast cancer subtypes including BRCA8/BRCA9/BRCA10 (P < 0.001), validating its importance as a BCSC-related gene. Cell-cell communication network analysis revealed the relationship between MED18 expression and interactions among different cell types in the tumor microenvironment. Correlation heatmaps showed significant associations between MED18 and multiple cell types, particularly strong positive correlations with tumor cells, macrophages, fibroblasts, and endothelial cells, suggesting MED18 may maintain stem cell properties through regulating intercellular signaling networks. Network analysis positioned MED18 at the core of complex cell-cell communication networks with close functional connections to multiple cell types. The strong correlations with immune cells and stromal cells indicate MED18’s role in tumor immune microenvironment regulation and matrix remodeling processes, confirming its reliability as a BCSC marker gene and revealing its key regulatory function in constructing complex tumor microenvironment networks.
BCSCs play pivotal roles in tumor initiation, progression, metastasis, therapeutic resistance, and disease recurrence[9,27]. Understanding the molecular characteristics and regulatory mechanisms of BCSCs is crucial for developing more effective therapeutic strategies and improving patient outcomes[28,29]. CD90 (Thy-1) demonstrated substantial expression at 68.43% positivity, while CD34 showed 62.64% positive expression, both indicating significant proportions of tumor cells with stem cell-like characteristics. In contrast, CD29 (integrin β1) showed relatively lower expression at 7.16%, and CD45 exhibited minimal positivity at 1.19%, confirming the absence of immune cell contamination in the stem cell population. The differential expression patterns of these markers provide valuable insights into the heterogeneous nature of BCSCs and establish a foundation for developing targeted therapeutic strategies.
The exceptionally high expression of CD105 is particularly noteworthy, as it suggests this marker may serve as the most reliable identifier for BCSCs. Its role in angiogenesis aligns with the known ability of CSCs to promote tumor vascularization and metastatic spread. This finding has important implications for both diagnostic applications and therapeutic targeting, as CD105 could serve as both a biomarker for stem cell identification and a potential therapeutic target for anti-angiogenic treatments. While our flow cytometric identification of surface markers provides compelling evidence for BCSC populations, future functional validation studies are essential to definitively establish the stemness hierarchy. Comparative analyses of sphere-forming capacity, self-renewal potential, and in vivo tumorigenicity between CD105+ and CD105- populations would strengthen our phenotypic characterization. Such functional assays would validate whether high CD105 expression correlates with enhanced tumorigenic capacity and confirm its utility as a definitive BCSC marker.
The application of LASSO regression analysis has enabled the identification of core regulatory genes with prognostic significance. The analysis revealed key genes including ANKHD1, DMAP1, BAIAP2, GSTM1, and PPARGC1A that play crucial roles in stem cell function regulation. Forest plot analysis demonstrated that most of these genes function as protective factors (hazard ratio < 1), including BAIAP2, TXNIP, and PPARGC1A, while genes such as GPSM1 and ANKHD1 acted as risk factors. The LASSO coefficient trajectory analysis showed how gene coefficients changed with increasing penalty parameter λ, with most coefficients approaching zero during regularization. Ten-fold cross-validation identified the optimal λ value at logλ ≈ -5.5, ultimately retaining 10 key genes for the final prognostic model. This refined gene signature provides both biological relevance to stem cell characteristics and robust prognostic prediction capability for breast cancer patients.
The validation of MED18 as a key stem cell-related gene has provided important insights into the regulatory mechanisms governing BCSCs. MED18 expression was significantly higher in malignant tissues compared to normal controls (P < 0.001) and showed consistent upregulation across breast cancer subtypes, validating its importance as a BCSC-related gene.
As a core component of the mediator complex, MED18 likely orchestrates BCSC properties through integration with canonical stemness pathways. The mediator complex serves as a crucial bridge between transcription factors and RNA polymerase II, suggesting that MED18 may facilitate the transcriptional programs downstream of Wnt/β-catenin, Notch, and Hedgehog signaling. In the context of Wnt signaling, MED18 could enhance β-catenin/T-cell factor-mediated transcription by stabilizing enhancer-promoter loops at target genes such as c-MYC and CCND1. Similarly, for Notch signaling, MED18 may facilitate RBPJ-mediated transcription of stemness genes like HES1 and HEY1. The PI3K/Akt pathway, known to regulate BCSC self-renewal, might also require MED18-mediated chromatin remodeling to activate downstream effectors like SOX2 and NANOG. This positions MED18 not merely as a stem cell marker, but as a central chromatin-level coordinator that integrates multiple stemness-maintaining pathways. The strong correlations with immune cells and stromal cells indicate MED18’s involvement in tumor immune microenvironment regulation and matrix remodeling processes. This multifaceted role positions MED18 as a key orchestrator of the complex tumor microenvironment networks that support stem cell maintenance and tumor progression. These findings confirm MED18’s reliability as a BCSC marker and reveal its potential as a therapeutic target for disrupting tumor microenvironment interactions. The comprehensive characterization of BCSCs has significant implications for clinical practice and therapeutic development[30,31]. The identification of reliable surface markers, particularly CD105 and CD90, provides tools for stem cell detection and monitoring in clinical samples. The high expression of CD105 (96.30%) makes it an attractive target for both diagnostic applications and therapeutic interventions. The LASSO-derived prognostic model offers a practical tool for patient risk stratification that could guide treatment decisions. The model’s ability to distinguish between high-risk and low-risk patients based on stem cell-related gene expression patterns provides a foundation for personalized therapeutic approaches. This could enable clinicians to identify patients who may benefit from more aggressive treatment regimens or novel targeted therapies. The spatial transcriptomic findings reveal important insights into tumor heterogeneity that could inform surgical planning and targeted therapy delivery. Understanding the spatial distribution of stem cell populations within tumors could guide precision medicine approaches, including targeted drug delivery systems that specifically target stem cell-enriched regions.
Despite significant progress in BCSC research, several important challenges remain. The heterogeneous nature of CSCs, both within individual tumors and across different patients, complicates the development of universal therapeutic targets and biomarkers[32]. The dynamic plasticity of CSCs, whereby non-stem cancer cells can acquire stem-like properties under certain conditions, adds another layer of complexity that must be addressed in therapeutic development. The lack of standardized methods for CSC identification and isolation continues to hinder progress in the field. While surface marker-based approaches provide valuable insights, they may not capture the full spectrum of CSC diversity and functional states. Future research should focus on developing more comprehensive and standardized approaches for stem cell characterization. The translation of laboratory findings to clinical applications remains a critical challenge requiring robust validation studies and the development of clinically applicable assays and therapeutic protocols. Large-scale clinical trials will be necessary to validate the prognostic models and therapeutic targets identified through preclinical research.
While we identified MED18 as a central regulator in BCSC networks, our study lacks detailed mechanistic investigation of how MED18 integrates with established stemness pathways (Wnt/β-catenin, Notch, Hedgehog) or its specific role in transcriptional regulation of stem cell programs. Chromatin immunoprecipitation sequencing and co-immunoprecipitation experiments would be needed to map MED18’s direct targets and protein interactions.
The comprehensive characterization of BCSCs through multi-omics approaches has provided unprecedented insights into their molecular signatures, spatial organization, and functional properties.
| 1. | Ferrigno Guajardo A, Salazar-Alejo M, Mesa-Chavez F, Gutierrez-Ornelas J, Platas A, Verduzco-Aguirre H, Villarreal-Garza C. Effectiveness of an online mindfulness-based stress-reduction intervention to reduce anxiety in breast cancer survivors: a randomized-controlled trial. Support Care Cancer. 2025;33:623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Guo R, Wang X, Yang Y, Zou J, Li M, Li Z, Yan Y, Lan N, Nie J, Tang Y, Zhang G. Prognostic value of a lncRNA signature in early-stage invasive breast cancer patients. Cancer Cell Int. 2025;25:233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 3. | Huang M, Zhang J, Yan C, Ling R, Wang T. KLF13 promotes breast cancer progression through the HTRA1 and the Hedgehog signaling pathway. Breast Cancer. 2025;32:1088-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 4. | Margarit DH, Reale MV, Paccosi G, Romanelli LM. Decoding cancer dynamics: Hypergraph analysis of stem cell marker interactions through O-information and community detection. Phys Rev E. 2025;111:054410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 5. | Nakano K, Oki E, Yamazaki M, Suzuki M, Kawai S, Fujita T, Kato A, Zaitsu Y, Jogo T, Kato C, Watanabe T, Hashimoto E, Nishime C, Fujii E, Ando K, Nagae G, Harimoto N, Ota M, Saeki H, Aburatani H, Maehara Y, Yamazaki T. Colorectal cancer cell line-derived organoid model with stem cell properties captures the regrowing state of residual cancer cells after neoadjuvant chemotherapy. Cell Death Discov. 2025;11:282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 6. | Shibasaki M, Ikeda H, Mimura A, Hashimoto J, Suo T, Kuranaga T, Shin-Ya K, Imoto M, Kakeya H. Isolation and structure determination of streptospherins B-F, novel cancer stem cell inhibitors, produced by Streptomyces sp. KUSC-240. J Antibiot (Tokyo). 2025;78:535-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Castagnoli L, Franceschini A, Cancila V, Dugo M, Bigliardi M, Chiodoni C, Toneguzzo P, Regondi V, Corsetto PA, Pietrantonio F, Mazzucchelli S, Corsi F, Belfiore A, Vingiani A, Pruneri G, Ligorio F, Colombo MP, Tagliabue E, Tripodo C, Vernieri C, Triulzi T, Pupa SM. Correction: CD36 enrichment in HER2-positive mesenchymal stem cells drives therapy refractoriness in breast cancer. J Exp Clin Cancer Res. 2025;44:165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 8. | Cecil DL, Herendeen D, Slota M, O'Meara MM, Dang Y, Corulli L, Disis ML. Identification and Validation of Th1-Selective Epitopes Derived from Proteins Overexpressed in Breast Cancer Stem Cells. Vaccines (Basel). 2025;13:525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 9. | Gairola K, Singh G, Bahuguna A, Pujari R, Kesharwani RK, Dubey SK. Computational simulation guided prediction of the inhibitory effect of curcumin, diallyl sulfide and its conjugates on ALDH1A1 to target breast cancer stem cells (BCSCs). J Biomol Struct Dyn. 2025;1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 10. | Liu G, Zhu G, Wu X, Tang Z, Shao W, Wang M, Xia H, Sun Q, Yan M. Thy-1 knockdown promotes the osteogenic differentiation of GMSCs via the Wnt/β-catenin pathway. J Cell Mol Med. 2023;27:3805-3815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 11. | Valdivia A, Avalos AM, Leyton L. Thy-1 (CD90)-regulated cell adhesion and migration of mesenchymal cells: insights into adhesomes, mechanical forces, and signaling pathways. Front Cell Dev Biol. 2023;11:1221306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 12. | Çordan İ, Günler T. CD105 (Endoglin) Expression as a Prognostic Marker in Aggressive Papillary Thyroid Carcinoma. Clin Endocrinol (Oxf). 2025;103:596-604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 13. | Liu J, He C, Zhao J. Analysis of Serum Soluble Endoglin and s-CD105 Levels in Relation to Postoperative Recurrence and Metastasis in Breast Cancer Patients Treated with Modified Radical Mastectomy. Int J Womens Health. 2025;17:1597-1604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 14. | Guo C, Li S, Liu J, Ma Y, Liang A, Lou Y, Liu H, Wang H. FBF1 maintains stem cell-like properties in breast cancer via PI3K/AKT/SOX2 axis. Stem Cell Res Ther. 2025;16:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 15. | Ismail DF, El-Keey MM, Elgendy SM, Hessien M. Impregnation of mesenchymal stem cell conditioned media with wortmannin enhanced its antiproliferative effect in breast cancer cells via PI3K/Akt/mTOR pathway. BMC Res Notes. 2025;18:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 16. | Hejji MAA, Al-Sukruwah MA, Hussen J. Flow cytometric analysis of lymphocyte subsets in the peripheral blood of two local goat breeds in Saudi Arabia. Open Vet J. 2025;15:1765-1770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 17. | Karkala F, de Bosscher I, Windster JD, Stroebel S, van Zanten L, Alves MM, Sacchetti A. Flow Cytometric Analysis and Sorting of Murine Enteric Nervous System Cells: An Optimized Protocol. Int J Mol Sci. 2025;26:4824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 18. | Abulfaraj AA, Shami AY, Alotaibi NM, Alomran MM, Aloufi AS, Al-Andal A, AlHamdan NR, Alshehrei FM, Sefrji FO, Alsaadi KH, Abuauf HW, Alshareef SA, Jalal RS. Exploration of genes encoding KEGG pathway enzymes in rhizospheric microbiome of the wild plant Abutilon fruticosum. AMB Express. 2024;14:27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 19. | Huang F, Fu M, Li J, Chen L, Feng K, Huang T, Cai YD. Analysis and prediction of protein stability based on interaction network, gene ontology, and KEGG pathway enrichment scores. Biochim Biophys Acta Proteins Proteom. 2023;1871:140889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 48] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 20. | Robben M, Nasr MS, Das A, Veerla JP, Huber M, Jaworski J, Weidanz J, Luber J. Comparison of the Strengths and Weaknesses of Machine Learning Algorithms and Feature Selection on KEGG Database Microbial Gene Pathway Annotation and Its Effects on Reconstructed Network Topology. J Comput Biol. 2023;30:766-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 21. | Ghanem M, Ghaith AK, Tsai SHL, Yeh YC, Akinduro OO, Michaelides L, El-Hajj VG, Saad H, Tfaily A, Nieves AB, Quiñones-Hinojosa A, Abode-Iyamah K, Bydon M. Machine learning-driven national analysis for predicting adverse outcomes in intramedullary spinal cord tumor surgery. Eur Spine J. 2025;34:3863-3875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 22. | Li K, Tang H, Wang Y, Wang X. Identification of hub genes involved in the pathogenesis of diabetic nephropathy: A multi-omics study integrating machine learning, mendelian randomization and mediation analysis. Diabetes Obes Metab. 2025;27:4927-4941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 23. | Yang Y, Zhang G. Comprehensive analysis and experimental validation of BST1 as a novel diagnostic biomarker for pediatric sepsis using multiple machine learning algorithms. Eur J Pediatr. 2025;184:441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 24. | Lv W, Ma Y, Li D, Xiong K, Cui J, Pan S, Zhang N, Li Y, Chen Y, Guo D. Single-Cell RNA Sequencing and Spatial Transcriptomics Reveal a Novel Mechanism of Oligodendrocyte-Neuron Interaction in Cognitive Decline After High-Altitude Cerebral Edema. CNS Neurosci Ther. 2025;31:e70485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 25. | Xu W, Liang T, Fang H, Fu L, Deng D, Tan X, Liu L, Tang D, Zheng H, Ding Q, Hou X, Feng D, Tao T, Wu S. Single-Cell RNA Sequencing Identifies MMP11(+) Cancer-Associated Fibroblasts as Drivers of Angiogenesis and Bladder Cancer Progression. Adv Sci (Weinh). 2025;12:e02774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Yao J, Li S, Ban W, Zeng L, Cui H, Chen K, Xu H. Single-Cell RNA Sequencing Delineates the Atlas and Cell Interactions of the Testicular Cells in Chinese Soft-Shelled Turtle (Pelodiscus sinensis). Mar Biotechnol (NY). 2025;27:101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 27. | Ali K, Nabeel M, Mohsin F, Iqtedar M, Islam M, Rasool MF, Hashmi FK, Hussain SA, Saeed H. Recent developments in targeting breast cancer stem cells (BCSCs): a descriptive review of therapeutic strategies and emerging therapies. Med Oncol. 2024;41:112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 28. | Zekri AN, Bahnassy A, Mourad M, Malash I, Ahmed O, Abdellateif MS. Genetic profiling of different phenotypic subsets of breast cancer stem cells (BCSCs) in breast cancer patients. Cancer Cell Int. 2022;22:423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 29. | Zhang C, Xu S, Yin C, Hu S, Liu P. The role of the mTOR pathway in breast cancer stem cells (BCSCs): mechanisms and therapeutic potentials. Stem Cell Res Ther. 2025;16:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 30. | Xuan Y, Wang Y. Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2019;10:694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 31. | Xuan Y, Wang Y. Correction to: Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2022;13:226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 32. | Damaševičius R. Explainable Artificial Intelligence Methods for Breast Cancer Recognition. Innov Discov. 2024;1:25. [DOI] [Full Text] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/