Published online Oct 26, 2020. doi: 10.4252/wjsc.v12.i10.1171
Peer-review started: June 11, 2020
First decision: June 20, 2020
Revised: July 3, 2020
Accepted: August 16, 2020
Article in press: August 16, 2020
Published online: October 26, 2020
Processing time: 137 Days and 2.3 Hours
Retinal organoids serve as excellent human-specific disease models for conditions affecting otherwise inaccessible retinal tissue from patients. They permit the isolation of key cell types affected in various eye diseases including retinal ganglion cells (RGCs) and Müller glia.
To refine human-induced pluripotent stem cells (hiPSCs) differentiated into three-dimensional (3D) retinal organoids to generate sufficient numbers of RGCs and Müller glia progenitors for downstream analyses.
In this study we described, evaluated, and refined methods with which to generate Müller glia and RGC progenitors, isolated them via magnetic-activated cell sorting, and assessed their lineage stability after prolonged 2D culture. Putative progenitor populations were characterized via quantitative PCR and immunocytochemistry, and the ultrastructural composition of retinal organoid cells was investigated.
Our study confirms the feasibility of generating marker-characterized Müller glia and RGC progenitors within retinal organoids. Such retinal organoids can be dissociated and the Müller glia and RGC progenitor-like cells isolated via magnetic-activated cell sorting and propagated as monolayers.
Enrichment of Müller glia and RGC progenitors from retinal organoids is a feasible method with which to study cell type-specific disease phenotypes and to potentially generate specific retinal populations for cell replacement therapies.
Core Tip: Retinal organoids derived from human-induced pluripotent stem cells are excellent tools for enriching specific subpopulations for subsequent studies of cell type-specific disease phenotypes affecting the eye. Here, we describe the generation of retinal organoids and the harvest as well as 2D maintenance of retinal ganglion cells and Müller glia progenitors.
-
Citation: Freude KK, Saruhanian S, McCauley A, Paterson C, Odette M, Oostenink A, Hyttel P, Gillies M, Haukedal H, Kolko M. Enrichment of retinal ganglion and Müller glia progenitors from retinal organoids derived from human induced pluripotent stem
cells - possibilities and current limitations. World J Stem Cells 2020; 12(10): 1171-1183 - URL: https://www.wjgnet.com/1948-0210/full/v12/i10/1171.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v12.i10.1171
Neurodegenerative eye conditions are poorly understood and treatment options are often insufficient to prevent blindness. One of the most common neurodegenerative eye diseases is glaucoma, characterized by asymptomatic loss of peripheral vision. This spectrum of eye diseases are all defined by a progressive loss of the inner-most retinal neurons, the retinal ganglion cells (RGCs), and their axons[1]. Glaucoma can be subdivided into primary and secondary forms. The most common subtype of primary glaucoma is open-angle glaucoma (POAG) which can, in turn, be further subdivided into high-tension glaucoma and normal tension glaucoma.
Irrespective of the type, all patients with diagnosed glaucoma are treated with strategies that lower intraocular pressure (IOP). Despite IOP-lowering treatments, 15% of glaucoma patients become blind, while as many as 42% have been shown to lose sight in at least one eye[2]. Thus, research to more comprehensively understand the pathophysiology of glaucoma is warranted to develop novel neuroprotective treatments.
The RGCs lost in glaucoma are, similar to other neurons, post-mitotic, thereby lacking or at least having limited regenerative capacity to compensate for cell death caused by various pathological processes. The most recognized risk factors for glaucoma are IOP, age, and genetics[3]. It is predicted that the number of glaucoma patients will increase to over 110 million by 2040 due to the aging of our societies[4]. In addition to glaucoma, a similar type of damage to RGCs has been observed in the two most common irreversible inherited optic neuropathies: Dominant optic atrophy (DOA)[5] and Leber's hereditary optic neuropathy (LHON)[6]. The loss of RGCs in these disorders occurs as a result of mitochondrial dysfunction and is unrelated to IOP. The most frequent mutations in DOA affect OPA1 Mitochondrial Dynamin Like GTPase (OPA1). OPA1 localizes to the inner mitochondrial membrane and regulates mitochondrial stability and energy output. The damage and eventual loss of RGCs in DOA are caused by the progressive impairment of mitochondrial oxidation and the lack of energy production in the form of ATP through the mitochondrial respiratory chain. Causative mutations for LHON affect members of the gene family encoding the mitochondrially encoded NADH: Ubiquinone oxidoreductase core subunits (MT-ND1, MT-ND4, MT-ND5 and MT-ND6)[7]. These mutations affect complex 1 of the electron transport chain of the mitochondria and manifest in failure of oxidative pho-sphorylation accompanied by impaired or absent ATP production.
Consequently, mitochondrial dysfunction in both diseases leads to apoptosis of the RGCs which in turn leads to loss of vision[8]. It has been reported that Müller glia - the glial cells of the retina - provide trophic and energetic support to adjacent RGCs[9,10]. In addition to this function, Müller glia span the width of the retina and thus play an important role in maintaining its structural integrity. They also function in retinal homeostasis. One of the most important roles of the Müller glia is believed to be the uptake of the neurotransmitter glutamate, thereby preventing RGC excitotoxicity[11]. This protective role of Müller glia on RGCs is further highlighted by cell culture studies showing increased survival of RGCs when co-cultured with Müller cells[12]. It has therefore been proposed that neurodegenerative diseases of the eye characterized by RGC loss are driven by failure of Müller glia to sufficiently support them. Should this hypothesis be supported, potential intervention strategies might be aimed at targeting Müller glia, thereby slowing or even preventing RGC loss by restoring their trophic support.
However, to test whether RGC atrophy results from loss of trophic support from Müller glia in patients, human-specific and cell type (RGCs and Müller glia)-specific disease paradigms are required.
Human-induced pluripotent stem cells (hiPSCs) hold great potential in modeling disease, both to acquire insights into cell type-specific pathogenesis and etiology and to enable the development and testing of new treatment strategies. In the case of modeling retinal diseases, several protocols are available for differentiating hiPSC into RGCs (reviewed in[13] and[14]). These protocols differ in terms of their efficiency and reproducibility, but they all yield a proportion of mature, functional RGCs. In contrast, only a very limited number of studies have robustly generated Müller glia in vitro from heterogeneous populations of three-dimensional (3D) spontaneous retinal organoids[15,16]. In vitro-grown retinal organoids closely recapitulate embryonic eye development including the emergence of specific cell types, although their maturation is limited. During development, invagination of the optic vesicles forms the optic cups. Within those optic cups the outer wall develops into retinal pigment epithelium (RPE), while the inner wall gives rise to the neurosensory layers called neural retina (NR), containing the retinal visual cells. The NR includes both Müller glia and RGCs. RGCs emerge as early as day 25 during retinal organoid differentiation, whilst evidence of Müller glia is not apparent before day 50[17]. Due to the complexity of the retina and our limited understanding of the pathology of these neurodegenerative diseases, there is a need to implement 3D differentiation protocols to obtain RGCs and Müller glia in vitro. In this study, we showed that: (1) Differentiating hiPSCs into 3D retinal organoid structures yields sufficient numbers of RGCs and Müller glia progenitors; (2) These cells can be isolated via magnetic cell separation and cultured in 2D; and (3) Such cells express characteristic markers of RGCs and Müller glia. We propose that these enriched cultures are ideal for dissecting RGC and Müller glia interactions in a human- and disease-specific context in a clinically inaccessible tissue.
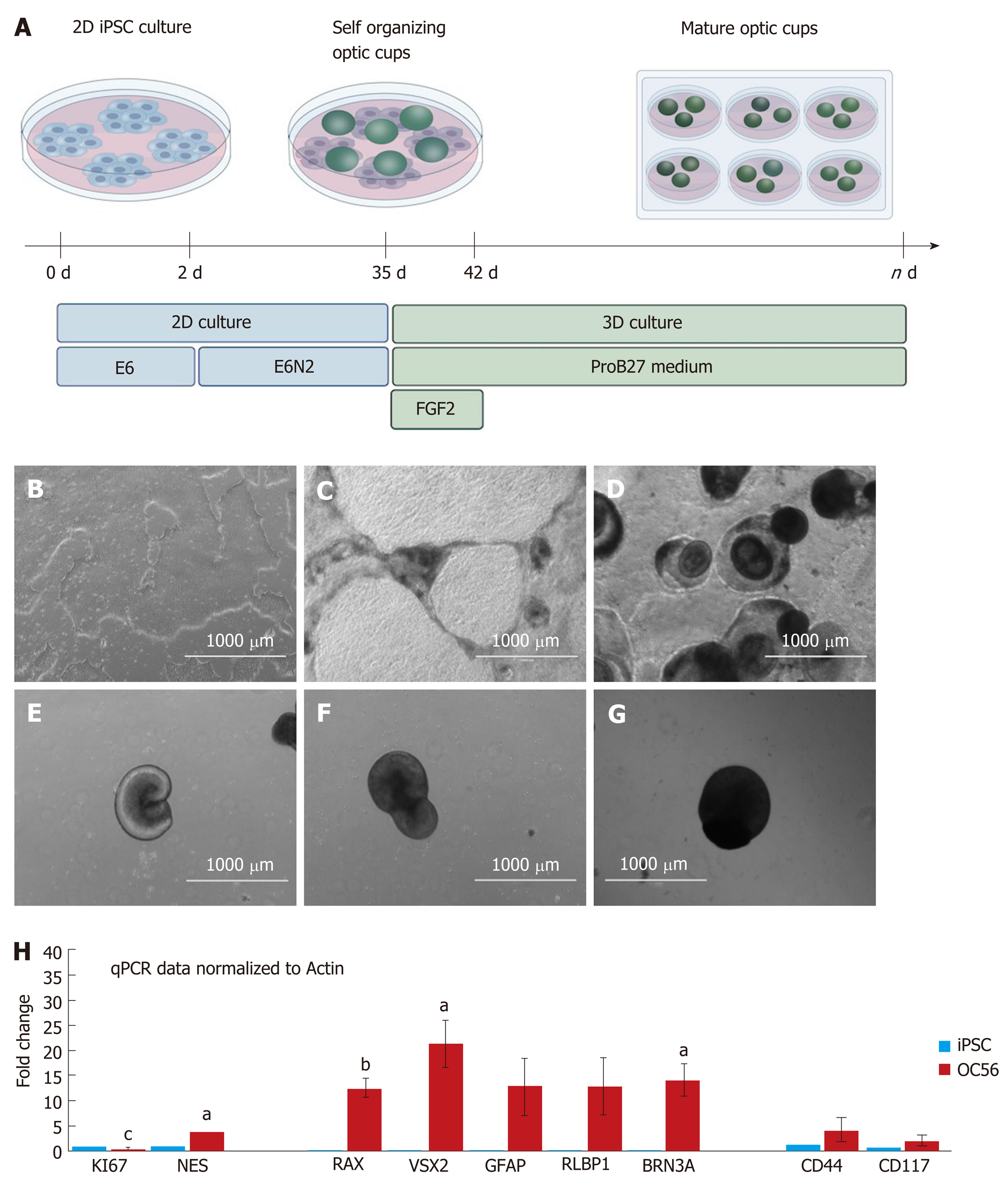
Refinement of iPSC differentiated into 3D retinal organoids to generate sufficient numbers of RGCs and Müller glia progenitors.
The study was approved by the Ethics Committee of the Capital Region of Denmark (Protocol No. H-19038704). The Human iPSC cell line “BIONi010-C-19” at passage 35 was thawed on Matrigel (cat. 7643022, Th. Geyer)-coated cell culture dishes and maintained in Essential 8 (E8) medium (cat. A1517001; Thermo Fisher Scientific, Waltham, MA, United States) containing 0.1% penicillin-streptomycin (pen-strep) (cat. P0781; Sigma-Aldrich). Upon thawing, to increase cell viability, RevitaCell™ (cat. A2644501; Thermo Fisher Scientific) was added and subsequently removed after 24 h with the next media change.
Retinal organoid differentiation was adapted from[18]. Differentiation was initiated once hiPSCs had reached 70%-80% confluency in a 6 cm dish in E8 medium (day 0). On day 0, the medium was changed for Essential 6 (E6) medium (cat. A1516501; Thermo Fisher Scientific) with 0.1% pen-strep. On day 2, 1% Cell Therapy Systems (CTS) N2 supplement (cat. A1370701; Thermo Fisher Scientific) was added to the medium (E6N2). This E6N2 medium was changed every other day for approximately 4 wk. On day 28, the organoids were manually isolated using a needle and scalpel and approximately 10 organoids were placed in each well of a non-adherent 96-well plate in DMEM/F12 1:1, L-glutamine 1% (cat. D8437; Sigma-Aldrich) MEM non-essential amino acids (cat. M7145, Sigma-Aldrich), supplemented with 2% CTS (cat. A1370701; Thermo Fisher Scientific) and B27 (cat. 12587010; Thermo Fisher Scientific), 0.1% pen-strep (cat. P0781; Sigma-Aldrich) and 10 ng/mL FGF2 (cat. Cyt-557; Prospec, Rehovot, Israel). This medium is referred to as ProB27 medium + FGF2. Five days after isolating the organoids, the plate was placed on a shaker within the incubator. At day 35, FGF2 was removed from the medium and the media changes continued every other day until day 87. At this point magnetic-activated cell sorting was performed.
RNA was extracted using the RNeasy® Plus Mini Kit (cat. 74134; Qiagen, Hilden, Germany) according to the manufacturer’s protocol. cDNA was synthesized from 1 μg total RNA in a 20 μL reaction using the iScript™ cDNA synthesis Kit (cat. 1708890; Bio-Rad, Hercules, CA, United States). Following synthesis, the cDNA was diluted four times with double-distilled water and stored at -20°C. Quantitative PCR (qPCR) reactions were performed in triplicate using FastStart LightCycler® 480 SYBR Green I Master (cat. 04707516001; Roche, Basel, Switzerland) in conjunction with the LightCycler® 480 real-time PCR system (Roche). cDNA samples (n = 5) were subjected to PCR amplification with the primers listed in Table 1.
| Primer | Forward sequence 5’-3’ | Reverse sequence 5’-3’ | Accession number |
| GAPDH_1 | AGGGCTGCTTTTAACTCTGGT | CCCCACTTGATTTTGGAGGGA | NM_001289745.2 |
| GAPDH_2 | GAAGGTGAAGGTCGGAGTCAA | GGAACATGTAAACCATGTAGTTGAG | NM_002046 |
| Ki67 | GGGCGAAGTTCACAGTCAAT | CTCCTTCACTGGGGTCTTGA | NM_001145966.1 |
| Nestin | CAGGAGAAACAGGGCCTACA | AAGCTGAGGGAAGTCTTGGA | NM_006617.1 |
| RAX | GAACAGCCCAAGAAAAAGCA | GCTTCATGGAGGACACTTCC | NG_013031.1 |
| VSX2 | AAGGATGGCATCATGGACTC | TCAGTTCCTCCTGGGAGATG | NG_013092.1 |
| GFAP | AAGAGATCCGCACGCAGTAT | AGGTCAAGGACTGCAACTGG | NM_001242376.1 |
| RLBP1 | CAACTGGAGCAGCTCACAAC | TCCTTCTCTTGCACCCTCTC | XM_011521870.2 |
| CD44 | CCCAGATGGAGAAAGCTCTG | CCTGAAGTGCTGCTCCTTTC | NM_001001391 |
| CD117 | GCAAATACACGTGCACCAAC | GCACCCCTTGAGGGAATAAT | NM.001093772 |
| BRN3A | 5'CGGGGAGCCATAATCTGCAA3’ | TTTCGACTCAGTTCGTGCGT3 | NM_006237.4 |
Retinal organoids were cultured in suspension culture until day 49-56 and fixed in 3% glutaraldehyde (cat. 104239; Merck, Kenilworth, NJ, United States) in 0.1 M Na-phosphate buffer (pH 7.2) at 4°C for 1 h. Retinal organoids (n = 15) were embedded in 4% Agarose and cut into 1-2 mm3 blocks under a stereomicroscope, and then washed with 0.1 M Na-phosphate buffer. This was followed by post-fixation in 1% osmium tetroxide (cat. 124505; Merck) in 0.1 mol/L Na-phosphate buffer for 1 h at room temperature. After washing in double-distilled water, retinal organoids were dehydrated to ethanol in a stepwise fashion. Propylene oxide (cat. 807027) was employed as an intermediate allowing for infiltration with Epon. The following day, retinal organoids were embedded in Epon, and incubated at 60°C for 48 h. Semi-thin (2 μm) sections were cut on a microtome with glass knives (Leica, Reichert Ultracut UTC, Wien, Austria), and then stained with 1% Toluidine blue in 1% Borax. Ultra-thin (50-70 nm) sections were cut on the microtome with a diamond knife, collected onto grids, and stained with 2% uranyl acetate (cat. 21447; Polyscience, Niles, IL, United States) and lead citrate[19]. Imaging and analysis were performed using the Philips CM100 transmission electron microscope with a Morada digital camera and iTEM software system.
Magnetic-activated cell sorting (MACS) was performed using equipment and solutions from Miltenyi Biotec (Bergisch Gladbach, Germany). Specifically, dissociation of retinal organoids was conducted using the Multi Tissue Dissociation Kit 2 (cat. 130-110-203; Miltenyi Biotec) and the gentleMACS Octo Dissociator with Heaters (cat. 130-096-427; Miltenyi Biotec) with gentleMACS C Tubes (constituents of the gentleMACS Octo Dissociator Kit). Separation of RGCs and Müller glia was achieved using the OctoMACS Starter Kit (cat. 130-042-108; Miltenyi Biotec), human anti-CD90 MicroBeads (cat. 130-096-253; Miltenyi Biotec), and human anti-CD40-Biotin (cat. 130-094-142; Miltenyi Biotec) in combination with the human anti-Biotin Microbeads UltraPure (cat. 130-090-485; Miltenyi Biotec). Dissociation of cells within the retinal organoids was achieved using the gentleMACS Octo Dissociator using the 37C_ABDK_1 program. Cells were subsequently centrifuged (300 rpm for 10 min) and resuspended in B27 medium (DMEM/F12, 2% B27, 1% Non-essential Amino Acid Solution and 0.1% pen-strep), and then separated via MACS using either human anti-CD 90 microbeads or human anti-CD40-Biotin and human anti-Biotin Microbeads. Conducting the separation procedure with the B27 medium instead of the provided buffers was crucial for cell survival. CD90 was used to enrich cultures for RGCs, while CD40 was used to enrich for Müller glia. Following sorting of 20 retinal organoids for both conditions, positively isolated cells were plated on polyornithine-coated acid-treated coverslips (cat. P4957; Sigma-Aldrich) and laminin-coated acid-treated coverslips (cat. L2020; Sigma-Aldrich) and maintained on them for 14 d. Subsequently, cell identity was assessed by immunocytochemistry (ICC).
Retinal organoids were incubated in MeOH:DMSO:H2O2 (30%) 4:1:1 for 2 h and then washed in 100% MeOH. The tissue was equilibrated in PBT (PBS with 0.1% Tween-20) through a series of descending MeOH concentrations in PBT (75%, 50%, 25%, 0%) for 30 min at each concentration. After equilibration, the tissues were blocked in 0.5% TNB (cat. NEL704A001KT, a proprietary TSA-block supplied by Perkin Elmer, Waltham, MA, United States) with 5% DMSO for at least 2 h. Primary antibodies were anti-NESTIN (cat. MAB5326; Millipore), anti-CD44 (cat. MA4405; Thermo Fisher Scientific), anti-PAX6 (cat. 901301; Biolegend, San Diego, CA, United States) and anti-CHX10 (cat. PA1-12566; Thermo Fisher Scientific) diluted 1:10 in 0.5% TNB with 5% DMSO. Incubation was performed overnight at 4°C with gentle agitation. The following day, tissues were washed extensively in PBT with one change of PBT per hour for 8 h. Secondary antibodies were diluted 1:500 in 0.5% TNB with 5% DMSO and incubation was conducted overnight at 4°C with gentle nutation. Donkey anti-mouse Alexa Fluor 488 (cat. A-21202; Thermo Fisher Scientific) was used to detect NESTIN, donkey anti-rat Alexa Fluor 488 (cat. A-21208; Thermo Fisher Scientific) was used to detect CD44, and donkey anti-sheep Alexa Fluor 488 (cat. A-11015; Thermo Fisher Scientific) was used to detect CHX10. The following day, the tissues were washed for at least 1 h in PBT with a couple of changes of the PBT. Clearing of the retinal organoids was performed using either fructose glycerol or ethyl cinnamate. Briefly, for fructose glycerol clearing, tissues were equilibrated to 100% MeOH through a series of ascending MeOH concentrations in PBT (0%, 25%, 50%, 75%). Prior to microscopy, 80 μL fructose glycerol clearing solution was added to organoids using a 200 μL tip with the end cut off. Ethyl cinnamate clearing was subsequently used since it proved to be superior to fructose glycerol clearing. In this method, PBT was removed and tissues were equilibrated to 100% EtOH through a series of ascending EtOH concentrations (30%, 70%, 100%, 100%) with 2% Tween-20 for approximately 2 h per step. Prior to microscopy, ethyl cinnamate was added to the tissues for 2 h.
For the qPCR experiments, data are presented as the mean ± standard error of the mean. Statistical analysis was conducted using GraphPad Prism 7.03 and determined using the Student’s t test, one-way analysis of variance (ANOVA) with Tukey’s post-test, or two-way ANOVA with Bonferroni post hoc test for differences between each group, as indicated. Statistically significant differences are labeled in the figures as aP < 0.05, bP < 0.01, and cP < 0.001.
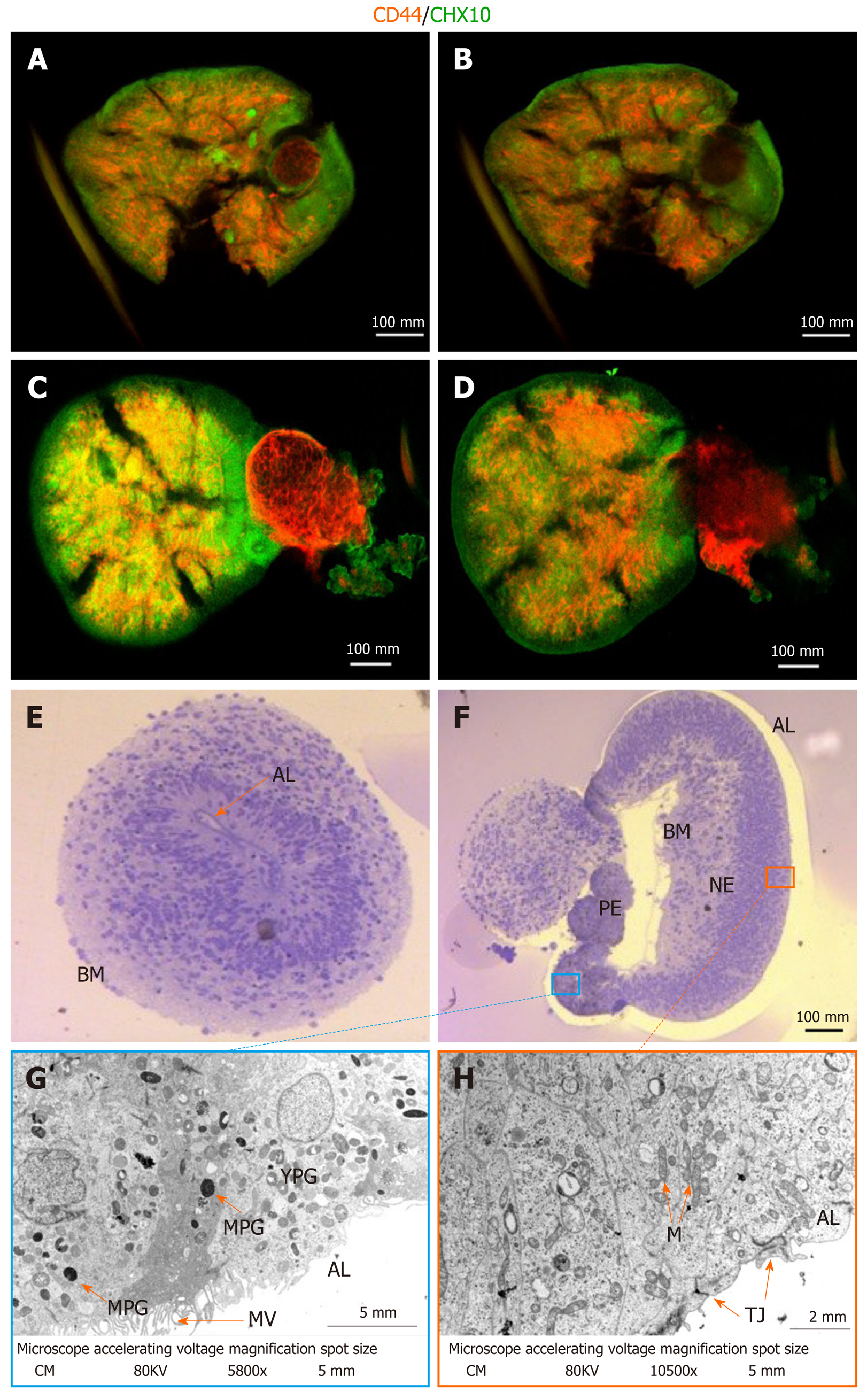
Following a modified protocol to yield retinal epithelial cells[18], we generated retinal organoids and analyzed their potential to generate RGCs and Müller glia progenitors (Figure 1A; refer to the Materials for specific details). Human iPSCs were grown as a monoculture in 2D until a confluency of 70%-80% (Figure 1B). Subsequently, they were transferred to E6 media and supplemented with N2 to promote neuroepithelial differentiation. From day 6, dark pigmented regions became apparent (Figure 1C), and from day 14, the pigmented areas formed 3D structures on top of the cell monolayer (Figure 1D). From day 20 onwards, hollow vesicles with pigmented retinal epithelial-like cells emerged from the 2D layer (Figure 1E), and these were manually isolated on day 28. Further culturing proceeded as 3D optic vesicles (Figure 1F and G) which were necessarily maintained in a shaking incubator: Static culturing resulted in optic vesicle aggregation, followed by necrotic changes within the optic vesicles. From day 28, optic vesicles were propagated in ProB27 and FGF2: FGF2 was removed after day 35, following which the structures were considered to be differentiated into retinal organoids, having transitioned from open optic vesicles to closed optic cup structures (Figure 1A). Retinal organoids did not increase in size further after removal of FGF2 (Figure 1G), but a continuous process of maturation was observed. Maturity of the generated retinal organoids was assessed via gene expression profiling, whole-mount ICC and TEM. All experimental analyses were carried out in triplicate on a total of 45 retinal cups. The optic cup presented in Figure 2A to 2D is a representative specimen of all optic cups investigated in this study. RGC progenitor cells appeared to be evenly distributed throughout the optic cups at day 49, as indicated by staining for CHX10/VSX2[20]. Putative Müller glia progenitors expressing CD44[21] appeared to arise in an aggregate adjacent to the primary optic cup structure (Figure 2C and D). In addition, Müller glia progenitors were detected within the 3D structures albeit in lower numbers compared to RGC progenitors (Figure 2C and D). This result was obtained in conjunction with fructose glycerol clearing following ICC for CHX10/VSX2 (to visualize RGC progenitors) and CD44 (for Müller glia progenitors) and it was evident that only superficial cell layers could be imaged since tissue clearing was incomplete. Nevertheless, assessment via qPCR revealed that optic cups at day 56 in maturation expressed genes consistent with the presence of both RGCs and Müller glia (and 1H). Specifically, we assessed proliferative activity within the optic cups using primers detecting Ki67, which was significantly reduced in expression compared to that in hiPSCs (1.044 ± 0.1091 vs 0.2368 ± 0.048; P < 0.001). The early neural progenitor marker NESTIN, was significantly upregulated in optic cups compared to hiPSCs, indicating the desired differentiation towards the neural lineage to obtain RGCs and Müller glia (0.7906 ± 0.147 vs 3.774 ± 0.8007; P < 0.05). Moreover, the expression of Nestin is indicative of a residual population of neural progenitors, reflecting the early maturation state of the optic cup compared to the adult eye. Confirming the progenitor status of RGCs, we detected significantly elevated expression of Retina and Anterior Neural Fold Homeobox (RAX) (0.00464 ± 0.00102 vs 12.63 ± 1.82; P < 0.01) and Visual system homeobox 2 (VSX2) (0.0192 ± 0.00506 vs 21.34 ± 4.703; P < 0.05) in optic cups compared to hiPSCs. Expression of both genes is associated with differentiation towards mature RGCs[17]. RAX is expressed early in eye primordia and is required for retinal cell fate determination. VSX2, a transcription factor inducing Sonic Hedgehog (SHH) expression, is involved in differentiation and proliferation of RGCs[22]. The higher expression of VSX2 compared to RAX is consistent with optic cups at this stage (day 56) containing RGCs representing a late progenitor status, indicating a later stage of optic cup maturity[23]. This is concordant with both the observed decrease in proliferation and arrested overall growth of the optic cups at this stage. Indicative of the presence of maturing or even mature RGCs, we detected increased expression of brain-specific homeobox/POU domain protein 3A (BRN3A) (0.0195 ± 0.0175 vs 14 ± 3.296, P < 0.05). BRN3A has been recognized as a very robust ex vivo marker for RGCs[24] and is widely employed to determine the abundance and presence of in vitro RGCs including those derived from iPSCs[25]. In vitro differentiation of Müller glia is more challenging than the generation of RGCs. Strikingly, we observed increased expression of retinaldehyde binding protein 1 (RLBP1), a well-recognized marker for Müller glia[26] in our optic cups (0.0382 ± 0.0145 vs 12.92 ± 5.391; P > 0.05). However, further validation is required since RLBP1 is also expressed in RPE. Taken together with the increased expression of Glial Fibrillary Acidic Protein (GFAP) (0.1199 ± 0.1388 vs 12.83 ± 5.649; P > 0.05), CD44 (1.119 ± 0.5062 vs 4.159 ± 2.542; P > 0.05) and CD117 (0.8989 ± 0.1672 vs 1.976 ± 0.7269; P > 0.05), these findings suggest the expression of bona fide Müller glia markers. CD44 and CD117 are both considered to be markers of mature Müller glia[27], which explains their relatively low expression in our still-maturing optic cups. Nevertheless, CD44 is not an exclusive Müller glia marker since it is also expressed by immature retinal pigment epithelial cells[28]. GFAP, conversely, is a classical glial marker often employed to detect astrocytes. In combination with expression of RLBP1, CD44 and CD117, GFAP expression is indicative of the presence of immature Müller glia in the day 56 optic cups.
Given the fact that fructose glycerol clearing following whole-mount ICC was suboptimal for imaging the proximal-most cells of the optic cups, we applied and optimized ethyl cinnamate clearing. This procedure was superior in labelling and detecting the RGCs and Müller glia in the inner structures of the optic cups (Figure 2A-D). It is clear from this optimized procedure that Müller glia are interspersed with RGCs as would be expected in the human retina (Figure 2B and D).
Further analyzes of the maturity and detailed cellular composition of the optic vesicles and cups was performed using light microscopy (LM) of semi-thin sections and transmission electron microscopy (TEM) of ultra-thin sections. At the LM level, optic vesicles with the basal membrane either outside (as expected from in vivo embryonic development) (Figure 2E) or inside (inverted) were detected (Figure 2F). Furthermore, initial invagination required for formation of an optic cup was noted in one of the inverted optic vesicles (Figure 2F). In this forming optic cup, two distinct cellular epithelial compartments could be distinguished: Retinal pigmented epithelium (RPE) and retinal neural epithelium (RNE). The pigmented epithelium was invaginating into the nervous epithelium with the basement membranes of the two epithelia on the inside and the apical surfaces on the external surface (inverted). At the TEM level, the apical layer (AL) of both epithelia presented tight junctions (TJ) and the RPE was, in addition, covered by abundant microvilli (MV) (Figure 2G). Further, the RPE was characterized by the presence of pigmented granules at various stages of maturation, from more electron-lucent immature (YPG) to electron-dense mature granules (MPG) (Figure 2G). Retinal pigmented granules with the highest expression were detected around the pigmented epithelium on the inner region of the invagination (Figure 2G). Such presence of pigmented granules is indicative of retinal epithelial (RPE) maturation marked by melanogenesis. Progressive RPE maturation following melanogenesis is characterized by formation of an apical and basolateral layer with clear polarity between both layers. The appearance of tight junctions and microvilli is a further indicator of proceeding maturation status. In conclusion, the process of invagination as well as the formation of distinct RPE and retinal neural epithelia suggests successful optic cup formation.
These optic cups contained sufficient numbers of RGCs and Müller glia (Figure 2A-D) for extraction and analysis or implementation in further experimental analyses.
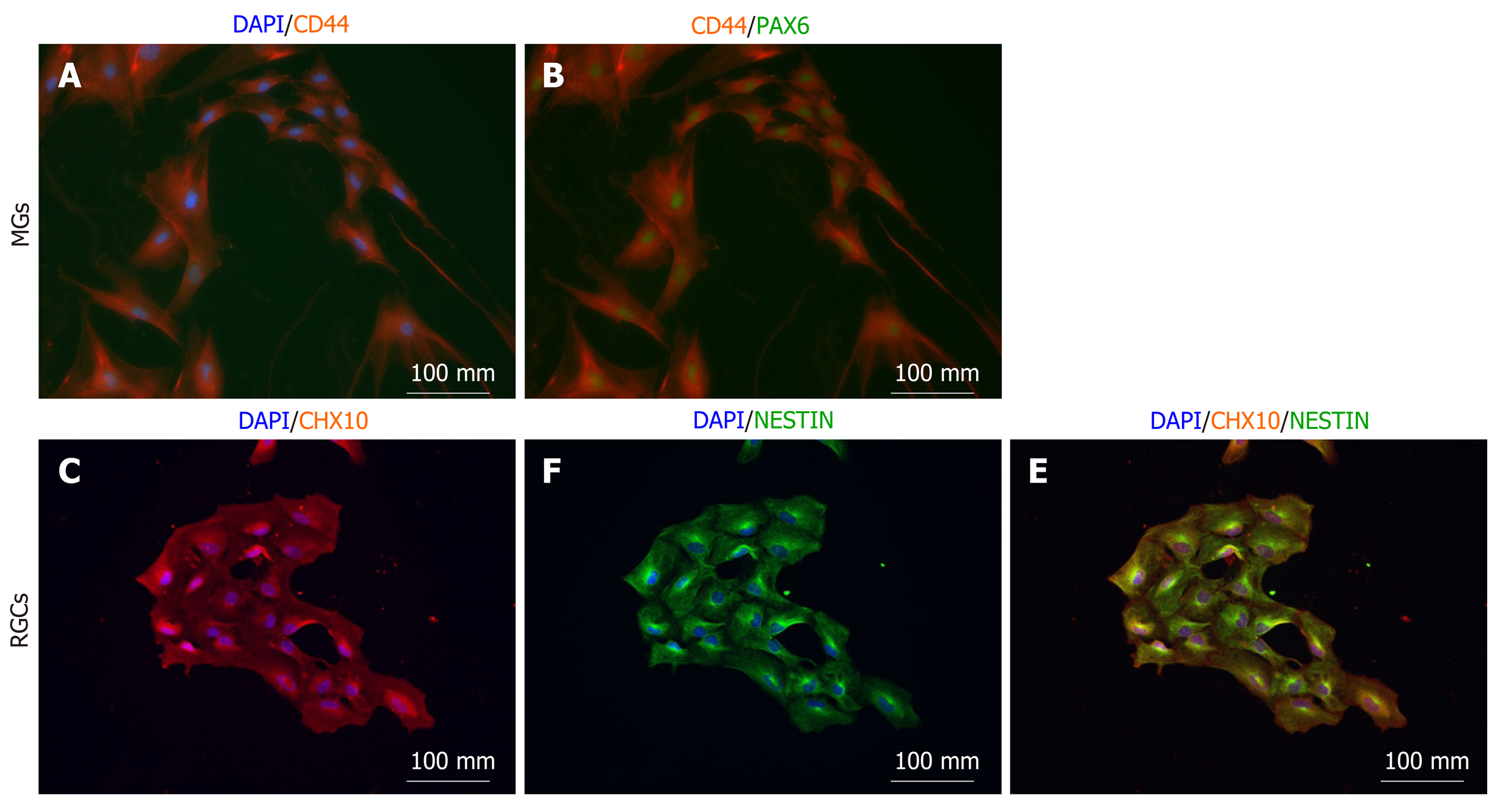
In order to study cell-specific consequences and interactions in eye disorders such as glaucoma it is essential to possess the ability to isolate specific cell types to establish selective co-cultures. Compelling evidence indicates the supportive and detrimental role of Müller glia on RGCs in healthy and diseased eyes, respectively. In order to pave the way for complex analyses of cell-specific interactions, we isolated Müller glia and RGCs from optic cups using the cell integrity-preserving magnetic-associated cell sorting (MACS) approach detailed in the Materials. We isolated Müller glia using CD40 beads and RGCs using CD90 beads. Retinal organoids are on average comprised of 50.000 cells at day 56, but this number can vary based on size, structure and cellular composition (± 10.000). On average we were able to isolate 200 (± 100) cells using the CD90 beads and 50 (± 25) using the CD40 beads per retinal organoids. Due to the low numbers we combined a total of 20 retinal organoids per condition. These cells were cultured as 2D layers for 14 d. At this stage, cells were fixed and their identity assessed. Even though the morphology of the isolated and 2D-cultured cells does not recapitulate their in vivo complexity, characteristic cell type-specific markers were expressed and detectable via ICC.
Isolated putative Müller glia showed consistent co-expression of the Müller glia markers CD44 (Figure 3A) and PAX6 (Figure 3B), suggesting that the isolation of immature Müller glia was successful. CD44 has been identified as a cell surface marker on Müller glia[21]. PAX6, a key transcription factor regulating eye development[29], is expressed by mature Müller glia[30].
Isolated putative RGCs were consistently positive for CHX10 (also known as VSX2) (Figure 3C), even though the cytosolic expression pattern was not expected due to the fact that CHX10 is a nuclear transcription marker. Further marker analyses as elaborated in the discussion is needed to validate RGC lineage identity. CHX10 is expressed in proliferating retinal progenitor cells and becomes progressively downregulated during RGC maturation. All isolated cells were co-positive for Nestin (Figure 3D and 3F), confirming the neuronal identity of the isolated RGC precursors. It remains to be determined whether these cells proceed to become mature RGCs or develop into bipolar cells.
Retinal neurodegeneration is one of the most frequent causes of blindness. The cause of progressive RGC loss, which often characterizes retinal neurodegeneration, is unknown and researchers worldwide are struggling to find new treatment strategies that can prevent RGC loss and thus save vision. As the retina is an inaccessible tissue in patients, research into retinal diseases has been conducted using retinal animal models, which are often physiologically very distinct from humans[31]. Thus, there is an indispensable need for novel methods to improve treatment options for sight-threatening retinal diseases.
Here, we have presented evidence that both RGCs and Müller glia can be generated at sufficient numbers for subsequent analyses using a modified retinal organoid protocol[18]. We confirmed by qPCR and selective ICC that expression of both RGC and Müller glia markers can be detected in the retinal organoids, confirming sufficient generation within these structures (Figure 1H and 2A-D). Moreover, we provided evidence that MACS sorting allows isolation of progenitors with RGCs and Müller glia characteristics and their subsequent 2D expansion without detectable changes in lineage identity (Figure 3). In order to confirm these results future experiments should investigate the long term culturing effect on the isolated RGCs and Müller glia, as well as inclusion of several lineage specific markers. Combinational assessment of BRN3A/B, ISL1, NEFL, NEFM, vGLUT2, EBF3, EYA2, GAP43, SRPX and STM2 expression on mRNA and protein level can be used to confirm bona fide mature RGCs in long-term 2D cultures after MACS[32]. In order to confirm bona fide Müller glia a combinational assessment of Vimentin, Glutamate synthetase, CRALBP, AQP4 and GFAP on mRNA and protein level can be applied[32]. Currently, the numbers of studies investigating human-derived RGCs and Müller glia are limited even though these in vitro systems provide a tremendous tool with which to study the pathophysiological mechanisms underlying blinding retinal conditions. Although it is a breakthrough that one can differentiate retinal cells from hiPSC, the next significant step will be to develop reliable and reproducible methods for achieving this.
When isolating human retinal cells from iPSC cell lines, it is important to consider their lineage stability and maturation level. In the present study, we provide evidence that MACS sorting using cell-type-specific surface antibodies in our hands proved successful in isolating RGC and Müller glia progenitors. Importantly, lineage-specific marker analyzes via ICC performed 14 d after isolation, showed lineage stability of the isolated cells. Although these findings are limited due to the maturation level and numbers of cells isolated, it confirms that the pathway for initial differentiation in retinal organoids and subsequent isolation of RGCs and Müller glia is a feasible protocol.
Another important aspect of our study is our finding that standard fructose glycerol clearing is suboptimal for imaging whole-mount ICC-stained retinal organoids. Instead, we show that clearing with ethyl cinnamate successfully facilitates imaging of the inner-most structures of retinal organoids (Figure 2A-D).
Our current work complements studies implementing the retinal organoid differentiation protocol, followed by Müller glia isolation, 2D culture and maturation in rodent eyes via intravitreal injections[33]. Other interesting approaches include transdifferentiation of human Müller glia into RGCs[34], which might constitute a cell replacement strategy for RGCs. The ability of Müller glia to differentiate into RGCs has been demonstrated in non-mammalian species such as zebrafish, indicating a conserved ability of Müller glia to replace RGCs under specific circumstances such as injury or disease[35]. Even though cell replacement therapies comprise an important downstream application of such protocols, in vitro models of specific cell types are required to understand the etiology of retinal neurodegeneration to allow earlier therapeutic interventions. Such cell type-specific models can consist of homogenous cultures of either RGCs or Müller glia to study the cell type-specific pathologies, or they can be co-cultured to study the intercellular dependencies and importance of secretory factors. Notably, homogenous cultures of one particular cell type are challenging to maintain and often the maturity of these cells can be a limiting factor. In such cases it should be considered whether subsequent transplantation and integration of human iPSC-derived Müller glia would be an advantage. It has been shown previously that RGCs can be generated from hiPSC without the need to implement the retinal organoid differentiation protocol[32]. It remains to be seen whether the 2D differentiation protocol and retinal organoid protocols with MACS isolation of RGCs results in a different outcome in terms of maturity and gene expression profiles.
Several challenges remain to be solved in order to make retinal organoid approaches successful. One of such is the size and cell type variability between the different retinal organoids. Such variabilities can have a great impact on reproducibility and reliability of functional readouts and disease phenotype analyses as seen in the field of organoids[36]. Another challenge is the long term culture without creating a necrotic core in the center of the organoids, which will negatively impact cell survival and gene expression of surrounding cells[37]. If those hurdles can be addressed by appropriate culture conditions organoids are the preferred starting material to extract otherwise difficult to generate progenitor cells from specific lineages, including Müller glia.
In summary, the present study provides important information on the generation and characteristics of human-derived RGCs and Müller glia. Human models to study retinal degenerative conditions are in great demand as an increasing number of patients are losing sight due to an aging population. Retinal tissue is not accessible in patients and animal models often fail to fully recapitulate the human pathological processes. The presented in vitro model system is particularly suitable for studying the genetic implications and significance of specific single nucleotide polymorphisms in glaucoma, implementing CRISPR/Cas9-targeted gene editing and subsequent analyses of changes in biological pathways in a cell type-specific approach. Moreover, cellular interactions and secreted molecules can be studied to identify novel druggable targets to prevent blindness. Differentiated RGCs and Müller glia from human-derived iPSCs will enable the identification of patient-specific differences and potentially pave the way for personalized treatment strategies to slow the rate of progression or to cure blinding retinal neurodegenerative conditions via tailored in vitro drug screens.
Degenerative eye diseases, such as glaucoma, are characterized by a progressive loss of retinal ganglion cells (RGCs), neurons within the eye required for image and non-image transformation. RGCs have been shown to interact with, and be dependent upon, Müller glia. Both cell types are central in the pathology of glaucoma.
The development of in vitro differentiation protocols for generating retinal cells and tissues is crucial to identify disease-relevant cellular changes and to test potential intervention strategies in a human context.
In this study we investigated the potential of retinal organoids to produce RGC and Müller glia progenitors and the efficiency with which these cell types can be isolated and expanded as enriched monolayer cultures.
Retinal organoids were differentiated from human induced pluripotent stem cells (hiPSCs), then magnetic-activated cell sorting applied to isolate RGC and Müller glia progenitors. Gene and protein expression were assessed in both retinal organoids and monolayers of RGC and Müller glia progenitors via qPCR and immunocytochemistry respectively. Furthermore, retinal organoid ultrastructure was examined via transmission electron microscopy.
Retinal organoids were grown successfully for at least 56 d and the presence of RGC and Müller glia progenitors validated via optimized whole mount immuno-cytochemistry. Furthermore, pure populations of RGC and Müller glia progenitors could be isolated using magnetic-activated cell sorting and successfully propagated in enriched monolayers for up to 2 wk.
Retinal organoids generated from hiPSCs recapitulate eye development in terms of their cellular heterogeneity and tissue architecture. Key cell types, such as RGC and Müller glia progenitors, can be isolated from the retinal organoids and expanded for subsequent cell-specific phenotype characterizations in disease-relevant contexts.
In vitro generation of the relevant cell types affected by degenerative eye diseases is important in order to study specific cell type disease vulnerability and selective cellular interactions. The generation of retinal organoids with subsequent isolation of RGC and especially Müller glia progenitors is an important step for in vitro disease modeling.
| 1. | Jonas JB, Aung T, Bourne RR, Bron AM, Ritch R, Panda-Jonas S. Glaucoma. Lancet. 2017;390:2183-2193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 634] [Cited by in RCA: 974] [Article Influence: 108.2] [Reference Citation Analysis (0)] |
| 2. | Heijl A, Buchholz P, Norrgren G, Bengtsson B. Rates of visual field progression in clinical glaucoma care. Acta Ophthalmol. 2013;91:406-412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 155] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 3. | McMonnies CW. Glaucoma history and risk factors. J Optom. 2017;10:71-78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 244] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 4. | Kolko M, Horwitz A, Thygesen J, Jeppesen J, Torp-Pedersen C. The Prevalence and Incidence of Glaucoma in Denmark in a Fifteen Year Period: A Nationwide Study. PLoS One. 2015;10:e0132048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 5. | Yu-Wai-Man P, Bailie M, Atawan A, Chinnery PF, Griffiths PG. Pattern of retinal ganglion cell loss in dominant optic atrophy due to OPA1 mutations. Eye (Lond). 2011;25:596-602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 6. | Majander A, Robson AG, João C, Holder GE, Chinnery PF, Moore AT, Votruba M, Stockman A, Yu-Wai-Man P. The pattern of retinal ganglion cell dysfunction in Leber hereditary optic neuropathy. Mitochondrion. 2017;36:138-149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 7. | Dai Y, Wang C, Nie Z, Han J, Chen T, Zhao X, Ai C, Ji Y, Gao T, Jiang P. Mutation analysis of Leber's hereditary optic neuropathy using a multi-gene panel. Biomed Rep. 2018;8:51-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 8. | Chun BY, Rizzo JF. Dominant Optic Atrophy and Leber's Hereditary Optic Neuropathy: Update on Clinical Features and Current Therapeutic Approaches. Semin Pediatr Neurol. 2017;24:129-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 9. | Bringmann A, Iandiev I, Pannicke T, Wurm A, Hollborn M, Wiedemann P, Osborne NN, Reichenbach A. Cellular signaling and factors involved in Müller cell gliosis: neuroprotective and detrimental effects. Prog Retin Eye Res. 2009;28:423-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 588] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 10. | Vecino E, Rodriguez FD, Ruzafa N, Pereiro X, Sharma SC. Glia-neuron interactions in the mammalian retina. Prog Retin Eye Res. 2016;51:1-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 605] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 11. | Bringmann A, Grosche A, Pannicke T, Reichenbach A. GABA and Glutamate Uptake and Metabolism in Retinal Glial (Müller) Cells. Front Endocrinol (Lausanne). 2013;4:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 133] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 12. | Skytt DM, Toft-Kehler AK, Brændstrup CT, Cejvanovic S, Gurubaran IS, Bergersen LH, Kolko M. Glia-Neuron Interactions in the Retina Can Be Studied in Cocultures of Müller Cells and Retinal Ganglion Cells. Biomed Res Int. 2016;2016:1087647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Ji SL, Tang SB. Differentiation of retinal ganglion cells from induced pluripotent stem cells: a review. Int J Ophthalmol. 2019;12:152-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 14. | Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao LH, Peters A, Park TS, Zambidis ET, Meyer JS, Gamm DM, Yau KW, Canto-Soler MV. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun. 2014;5:4047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 571] [Cited by in RCA: 738] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 15. | Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 955] [Cited by in RCA: 1087] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 16. | Meyer JS, Shearer RL, Capowski EE, Wright LS, Wallace KA, McMillan EL, Zhang SC, Gamm DM. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA. 2009;106:16698-16703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 468] [Cited by in RCA: 481] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 17. | Reichman S, Slembrouck A, Gagliardi G, Chaffiol A, Terray A, Nanteau C, Potey A, Belle M, Rabesandratana O, Duebel J, Orieux G, Nandrot EF, Sahel JA, Goureau O. Generation of Storable Retinal Organoids and Retinal Pigmented Epithelium from Adherent Human iPS Cells in Xeno-Free and Feeder-Free Conditions. Stem Cells. 2017;35:1176-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 172] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 18. | Reichman S, Terray A, Slembrouck A, Nanteau C, Orieux G, Habeler W, Nandrot EF, Sahel JA, Monville C, Goureau O. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc Natl Acad Sci USA. 2014;111:8518-8523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 228] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 19. | REYNOLDS ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208-212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17689] [Cited by in RCA: 17323] [Article Influence: 618.7] [Reference Citation Analysis (0)] |
| 20. | Tanaka T, Yokoi T, Tamalu F, Watanabe S, Nishina S, Azuma N. Generation of retinal ganglion cells with functional axons from human induced pluripotent stem cells. Sci Rep. 2015;5:8344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 21. | Shinoe T, Kuribayashi H, Saya H, Seiki M, Aburatani H, Watanabe S. Identification of CD44 as a cell surface marker for Müller glia precursor cells. J Neurochem. 2010;115:1633-1642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 22. | Sigulinsky CL, Green ES, Clark AM, Levine EM. Vsx2/Chx10 ensures the correct timing and magnitude of Hedgehog signaling in the mouse retina. Dev Biol. 2008;317:560-575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Phillips MJ, Perez ET, Martin JM, Reshel ST, Wallace KA, Capowski EE, Singh R, Wright LS, Clark EM, Barney PM, Stewart R, Dickerson SJ, Miller MJ, Percin EF, Thomson JA, Gamm DM. Modeling human retinal development with patient-specific induced pluripotent stem cells reveals multiple roles for visual system homeobox 2. Stem Cells. 2014;32:1480-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 24. | Nadal-Nicolás FM, Jiménez-López M, Sobrado-Calvo P, Nieto-López L, Cánovas-Martínez I, Salinas-Navarro M, Vidal-Sanz M, Agudo M. Brn3a as a marker of retinal ganglion cells: qualitative and quantitative time course studies in naive and optic nerve-injured retinas. Invest Ophthalmol Vis Sci. 2009;50:3860-3868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 448] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 25. | Gill KP, Hewitt AW, Davidson KC, Pébay A, Wong RC. Methods of Retinal Ganglion Cell Differentiation From Pluripotent Stem Cells. Transl Vis Sci Technol. 2014;3:7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 26. | Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Müller glial cells. J Comp Neurol. 2008;509:225-238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 327] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 27. | Too LK, Gracie G, Hasic E, Iwakura JH, Cherepanoff S. Adult human retinal Müller glia display distinct peripheral and macular expression of CD117 and CD44 stem cell-associated proteins. Acta Histochem. 2017;119:142-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 28. | Liu NP, Roberts WL, Hale LP, Levesque MC, Patel DD, Lu CL, Jaffe GJ. Expression of CD44 and variant isoforms in cultured human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 1997;38:2027-2037. [PubMed] |
| 29. | Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. Pax6: a multi-level regulator of ocular development. Prog Retin Eye Res. 2012;31:351-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 186] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 30. | Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J Neurosci. 2007;27:7028-7040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 522] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 31. | Baden T, Euler T, Berens P. Understanding the retinal basis of vision across species. Nat Rev Neurosci. 2020;21:5-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 202] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 32. | Gill KP, Hung SS, Sharov A, Lo CY, Needham K, Lidgerwood GE, Jackson S, Crombie DE, Nayagam BA, Cook AL, Hewitt AW, Pébay A, Wong RC. Enriched retinal ganglion cells derived from human embryonic stem cells. Sci Rep. 2016;6:30552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 33. | Eastlake K, Wang W, Jayaram H, Murray-Dunning C, Carr AJF, Ramsden CM, Vugler A, Gore K, Clemo N, Stewart M, Coffey P, Khaw PT, Limb GA. Phenotypic and Functional Characterization of Müller Glia Isolated from Induced Pluripotent Stem Cell-Derived Retinal Organoids: Improvement of Retinal Ganglion Cell Function upon Transplantation. Stem Cells Transl Med. 2019;8:775-784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 34. | Singhal S, Bhatia B, Jayaram H, Becker S, Jones MF, Cottrill PB, Khaw PT, Salt TE, Limb GA. Human Müller glia with stem cell characteristics differentiate into retinal ganglion cell (RGC) precursors in vitro and partially restore RGC function in vivo following transplantation. Stem Cells Transl Med. 2012;1:188-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 35. | Lenkowski JR, Raymond PA. Müller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res. 2014;40:94-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 252] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 36. | Huch M, Knoblich JA, Lutolf MP, Martinez-Arias A. The hope and the hype of organoid research. Development. 2017;144:938-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 270] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 37. | Wang H. Modeling Neurological Diseases With Human Brain Organoids. Front Synaptic Neurosci. 2018;10:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 124] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
Manuscript source: Invited manuscript
Specialty type: Cell biology
Country/Territory of origin: Denmark
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
P-Reviewer: Kiselev SL, Mariappan I, Politi L S-Editor: Liu JH L-Editor: Filipodia P-Editor: Zhang YL