©Author(s) (or their employer(s)) 2026.
World J Stem Cells. Feb 26, 2026; 18(2): 112940
Published online Feb 26, 2026. doi: 10.4252/wjsc.v18.i2.112940
Published online Feb 26, 2026. doi: 10.4252/wjsc.v18.i2.112940
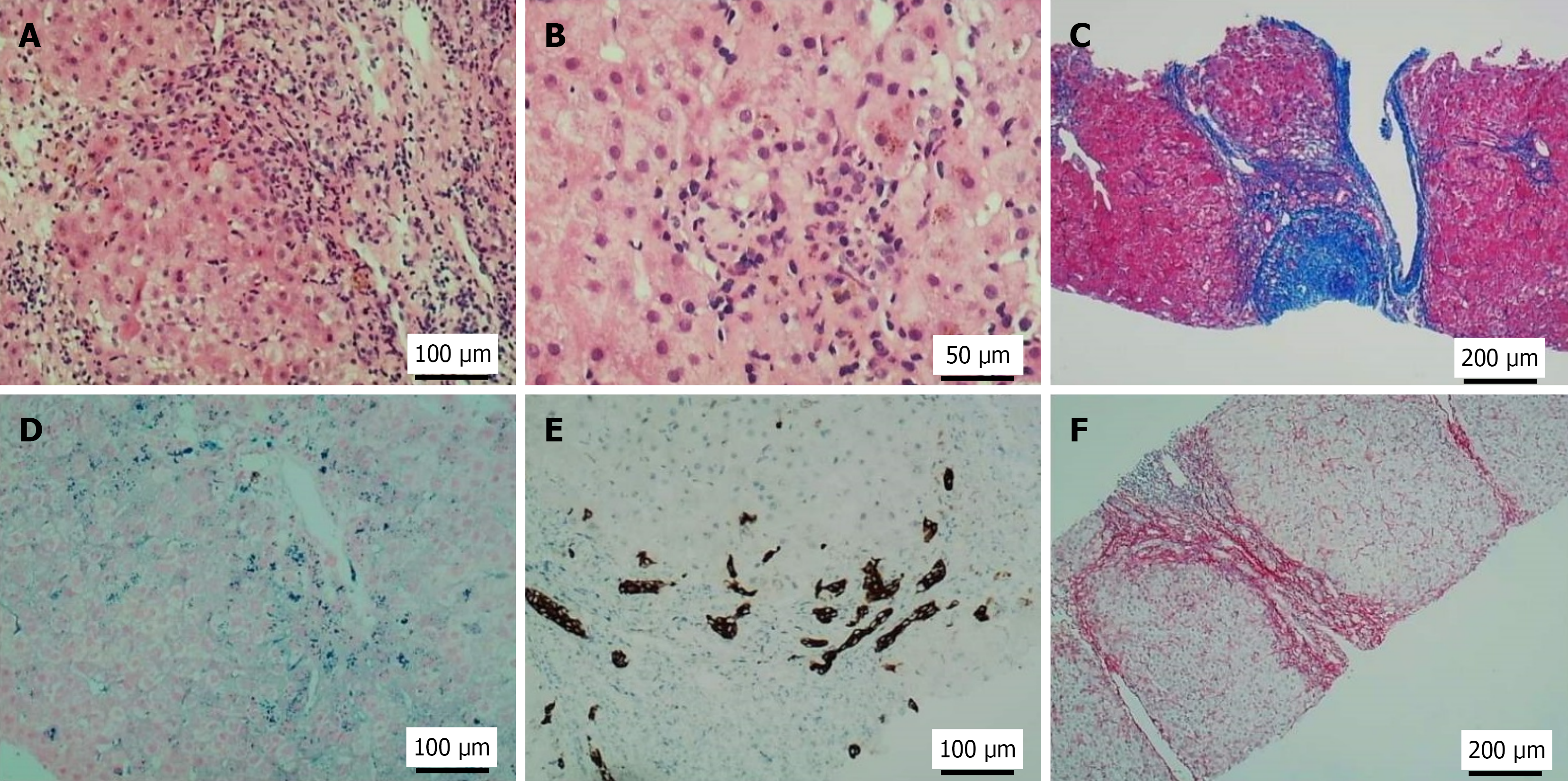
Figure 1 Pathological findings of liver biopsy in the patient.
A: Moderate interface inflammation with infiltration of lymphocytes and neutrophils; B: Hepatocytes containing yellow-brown granules and focal confluent necrosis; C: Masson trichrome stain: Portal-based fibrous expansion and septum (blue collagen); D: Prussian blue stain: Iron granules (blue-black deposits); E: Cytokeratin 7 immunohistochemistry: Biliary differentiation; F: Sirius red stain: Pericellular/periportal fibrosis (red/orange birefringence).
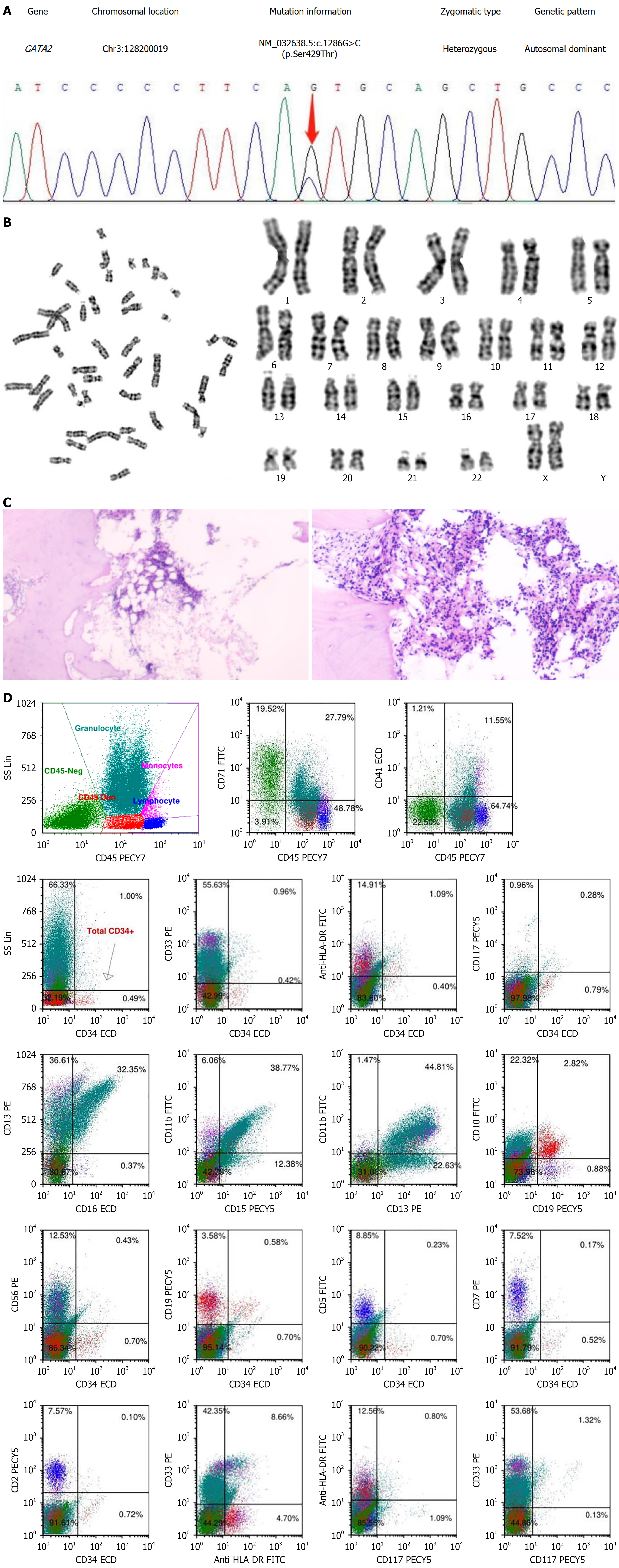
Figure 2 Identification of GATA2 mutation and diagnosis of aplastic anemia in the patient.
A: Sanger sequencing of the gene; B: Karyotype analysis; C: Pathological findings of bone marrow biopsy. The proliferation of bone marrow nucleated cells was low; the erythroid lineage was mainly composed of intermediate and late-stage cells; megakaryocytes were rare; lymphocytes were scattered in small numbers; CD34 small vessels (+); few CD61 megakaryocytes (+); D: Flow cytometric analysis. The proportion of CD34+ cells in nuclear cells was about 0.49%, with no obvious abnormality in the immunophenotype. The relative proportion of granulocytes was normal, and the immunophenotypes CD13, CD16, CD15, and CD11b were disordered. Approximately 2.60% of CD19+CD10+ immature B lymphocytes, consistent with normal B progenitor cell proliferation.
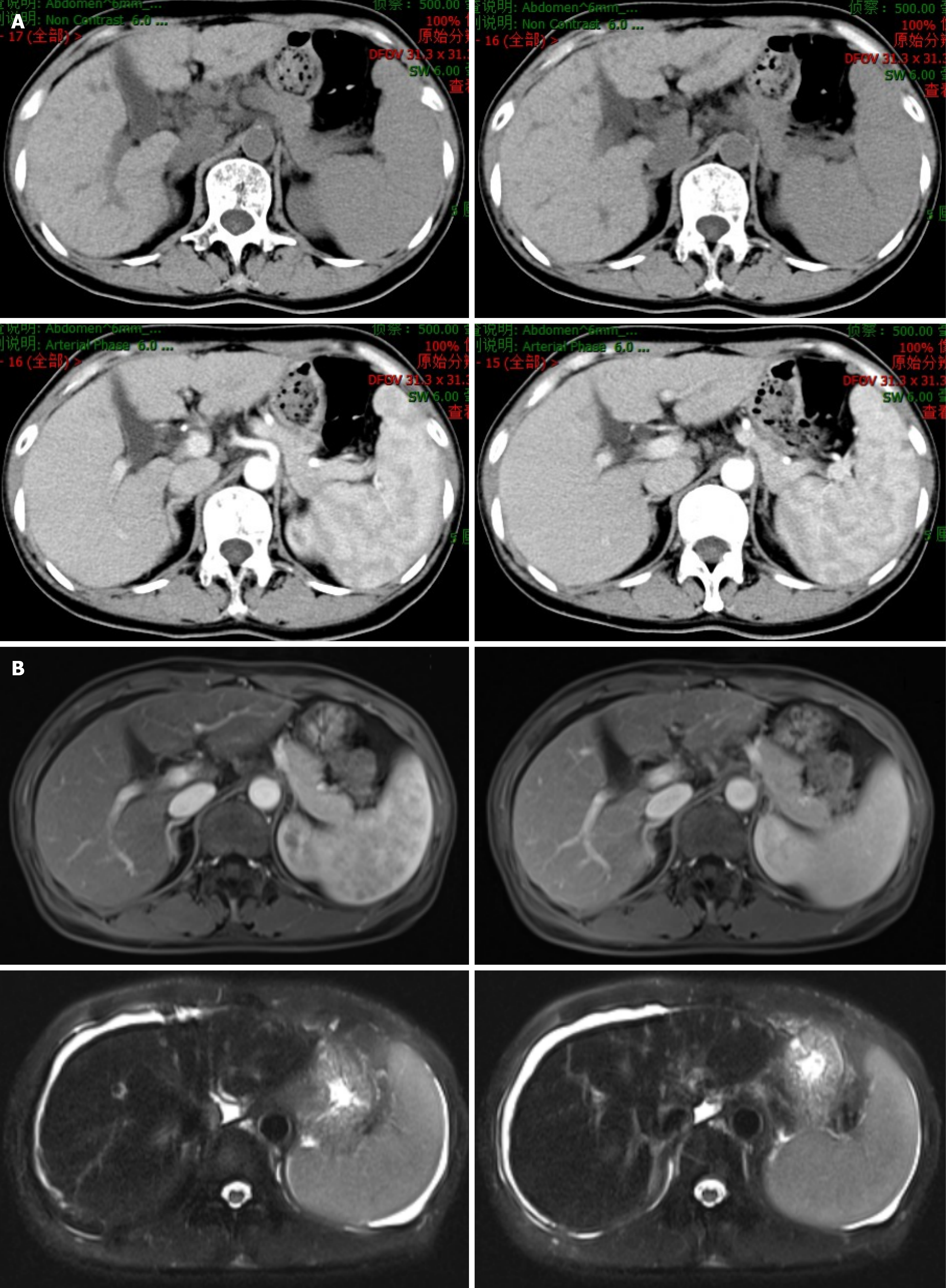
Figure 3 Abdominal magnetic resonance imaging and computed tomography findings of cirrhosis during follow-up.
A: Upper abdominal computed tomography (plain and enhanced) scans revealed irregular margins with slightly widened fissures, and strip low-density shadows around the portal vein in the liver; B: Upper abdominal magnetic resonance imaging revealed heterogeneous signal intensity with an irregular contour, slightly widened fissures, heterogeneous enhancement, and a long T2 signal around the portal vein in the liver. The spleen was enlarged. The upper abdominal magnetic resonance imaging findings indicated cirrhosis with splenomegaly and ascites. No abnormal enhancement was observed on the enhanced scan. The spleen was enlarged.
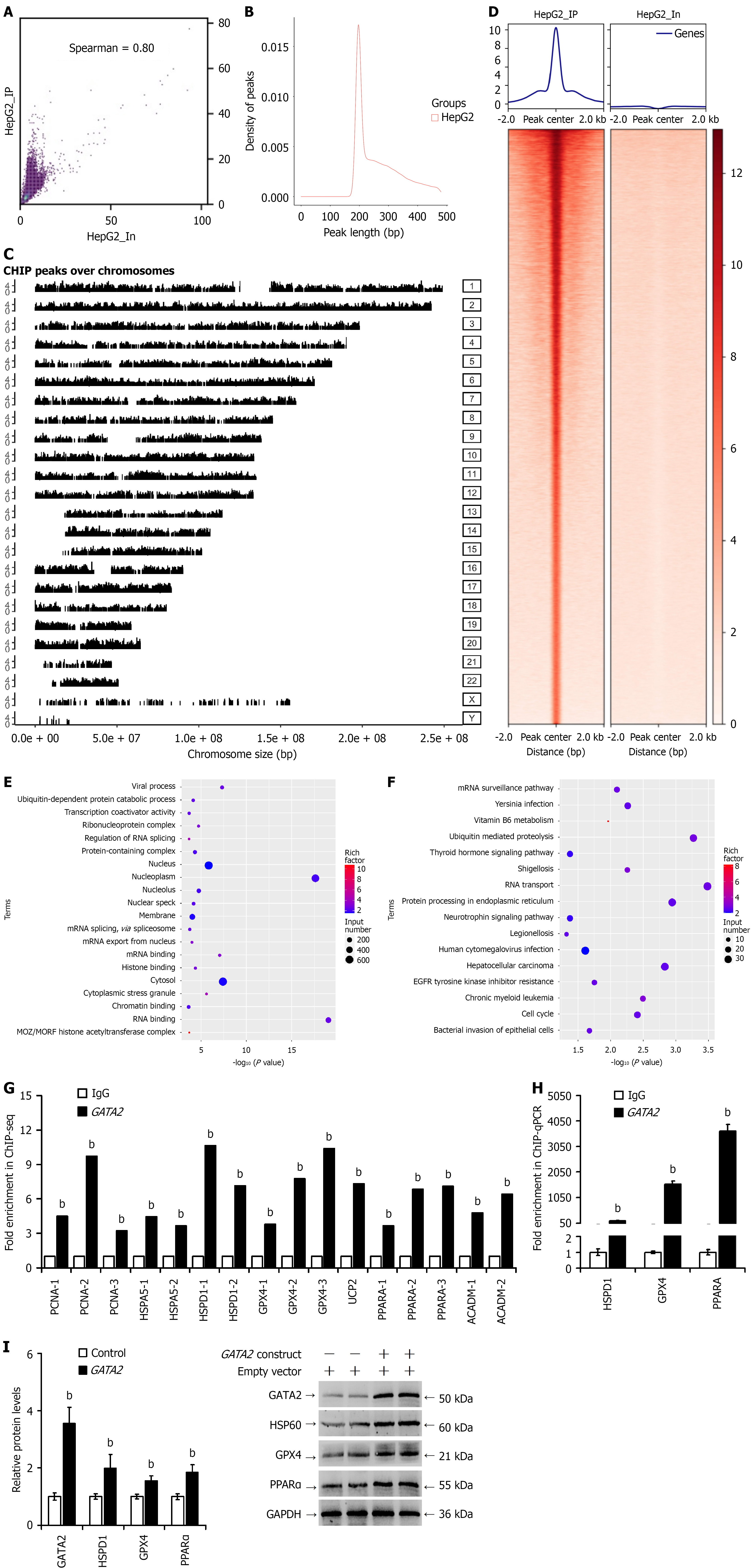
Figure 4 Identification of GATA2 binding sites in gene promoters through chromatin immunoprecipitation sequencing and prediction of its regulatory functions.
A: Scatter plot showing the correlation between biological replicates; B: Histogram depicting the distribution of peak lengths; C: Distribution of peaks across chromosomes (X-axis: Chromosome length; left Y-axis: Peak intensity; right Y-axis: Chromosome numbers); D: Read distribution surrounding transcription start sites (TSS) of peak-associated genes (top: Line chart of average sequencing depth around TSS; bottom: Heatmap of sequencing depth around TSS for peak-associated genes, ranked by average depth); E: Gene Ontology functional enrichment analysis of peak-associated genes; F: Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of peak-associated genes; G: Fold enrichment of selected genes associated with partial regeneration, molecular chaperones, antioxidant response, and fatty acid β-oxidation as determined by chromatin immunoprecipitation sequencing; H: Validation of GATA2 binding to the promoters of heat shock protein family D member 1, glutathione peroxidase 4, and peroxisome proliferator-activated receptor alpha by chromatin immunoprecipitation real-time quantitative polymerase chain reaction. Data are presented as fold enrichment relative to immunoglobulin G; I: Western blot analysis of GATA2, heat shock protein 60, glutathione peroxidase 4, and peroxisome proliferator-activated receptor alpha protein expression following transfection with the Gata2 construct. bP < 0.01 vs immunoglobulin G or control group. ChIP-seq: Chromatin immunoprecipitation sequencing; IgG: Immunoglobulin G; PCNA: Proliferating cell nuclear antigen; HSPD1: Heat shock protein family D member 1; GPX4: Glutathione peroxidase 4; UCP2: Uncoupling protein 2; PPARA: Peroxisome proliferator-activated receptor alpha.
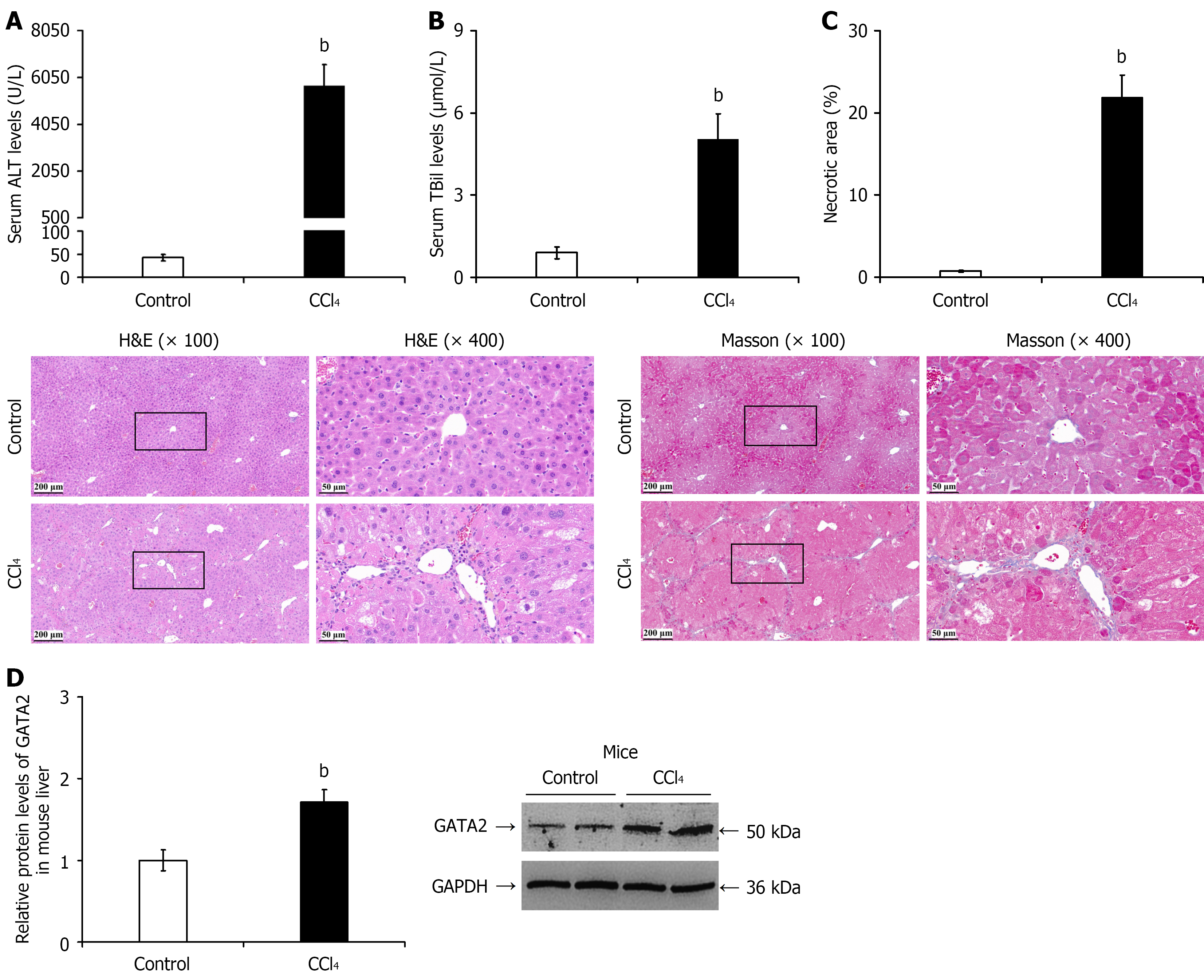
Figure 5 Liver injury assessment and hepatic GATA2 protein expression in chronic liver injury mice.
A: Serum alanine aminotransferase levels; B: Serum total bilirubin levels; C: Representative hematoxylin and eosin and Masson’s trichrome staining images and quantitative analysis of histopathological necrotic areas; D: Western blotting and quantitative analysis of GATA2 protein expression. bP < 0.01 vs control group. ALT: Alanine aminotransferase; TBil: Total bilirubin; H&E: Hematoxylin and eosin.
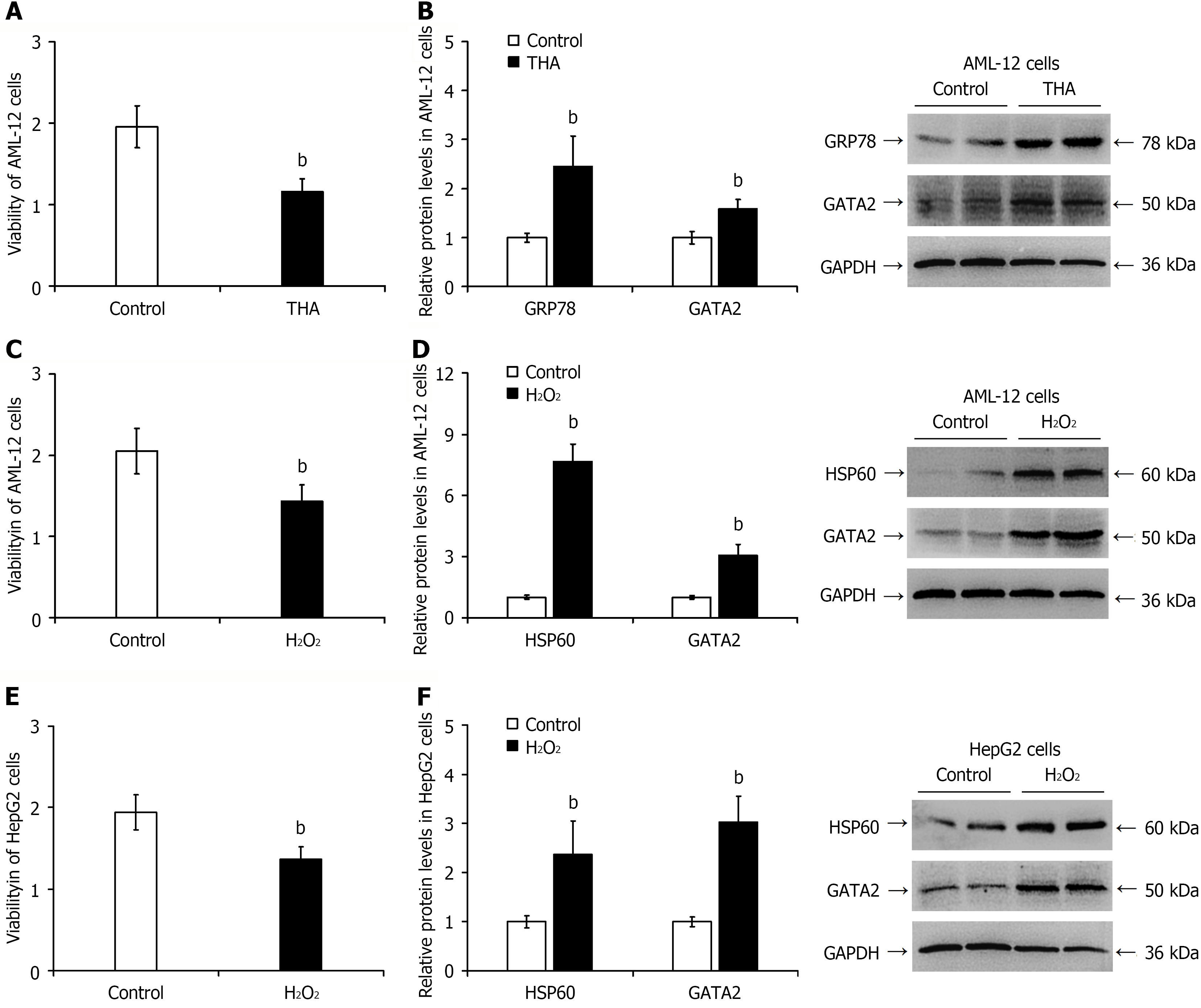
Figure 6 Elevated GATA2 protein expression in hepatocytes under endoplasmic reticulum stress and oxidative stress.
A: Cell viability assessed by Cell Counting Kit-8 assay in thapsigargin-treated AML-12 cells; B: Semi-quantitative analysis of thapsigargin-induced GATA2 protein expression by western blotting (representative blots shown); C and E: Cell viability measured by Cell Counting Kit-8 assay in AML-12 (C) and HepG2 cells (E) treated with H2O2; D and F: Semi-quantitative analysis of H2O2-induced GATA2 protein expression by western blotting in AML-12 (D) and HepG2 cells (F) (representative blots shown). bP < 0.01 vs control group. HSP60: Heat shock protein 60.
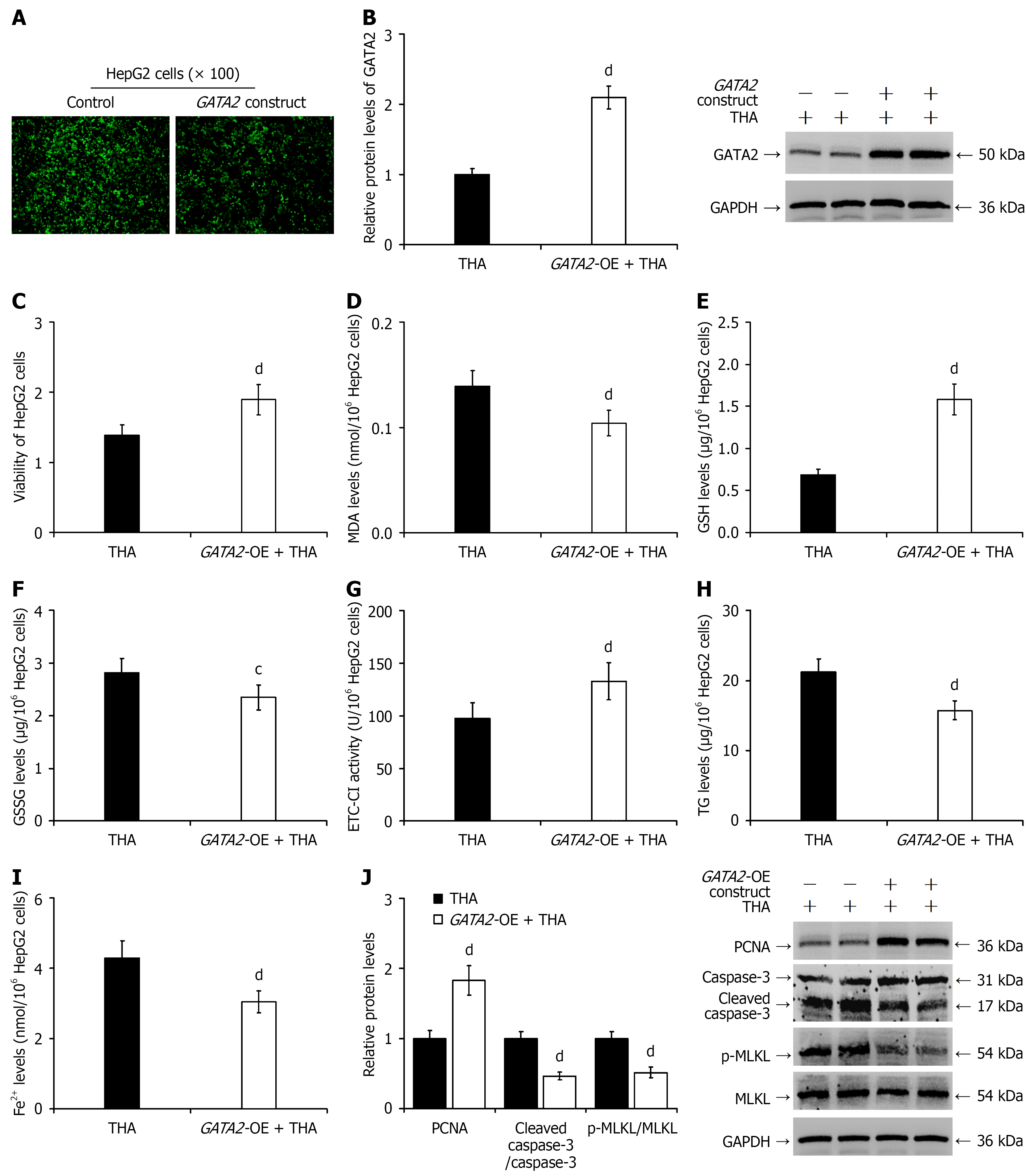
Figure 7 GATA2 overexpression protected HepG2 cells from endoplasmic reticulum stress-induced injury.
A: Fluorescence microscopy images confirming transfection efficiency; B: Western blot analysis of GATA2 protein expression, with representative blots displayed alongside quantitative results; C: Assessment of cell viability using the Cell Counting Kit-8 assay; D: Intracellular malondialdehyde content; E: Glutathione levels; F: Oxidized glutathione levels; G: Electron transfer chain complex I activity; H: Triglyceride levels; I: Ferrous ion levels; J: Western blot analysis of proliferating cell nuclear antigen, cleaved caspase-3, and phosphorylated mixed lineage kinase domain-like protein expression (representative blots shown) with quantification. cP < 0.05 vs the thapsigargin group; dP < 0.01 vs the thapsigargin group. THA: Thapsigargin; MDA: Malondialdehyde; GSH: Glutathione; GSSG: Oxidized glutathione; ETC-CI: Electron transfer chain complex I; TG: Triglyceride; p-MLKL: Phosphorylated mixed lineage kinase domain-like protein; MLKL: Mixed lineage kinase domain-like protein.
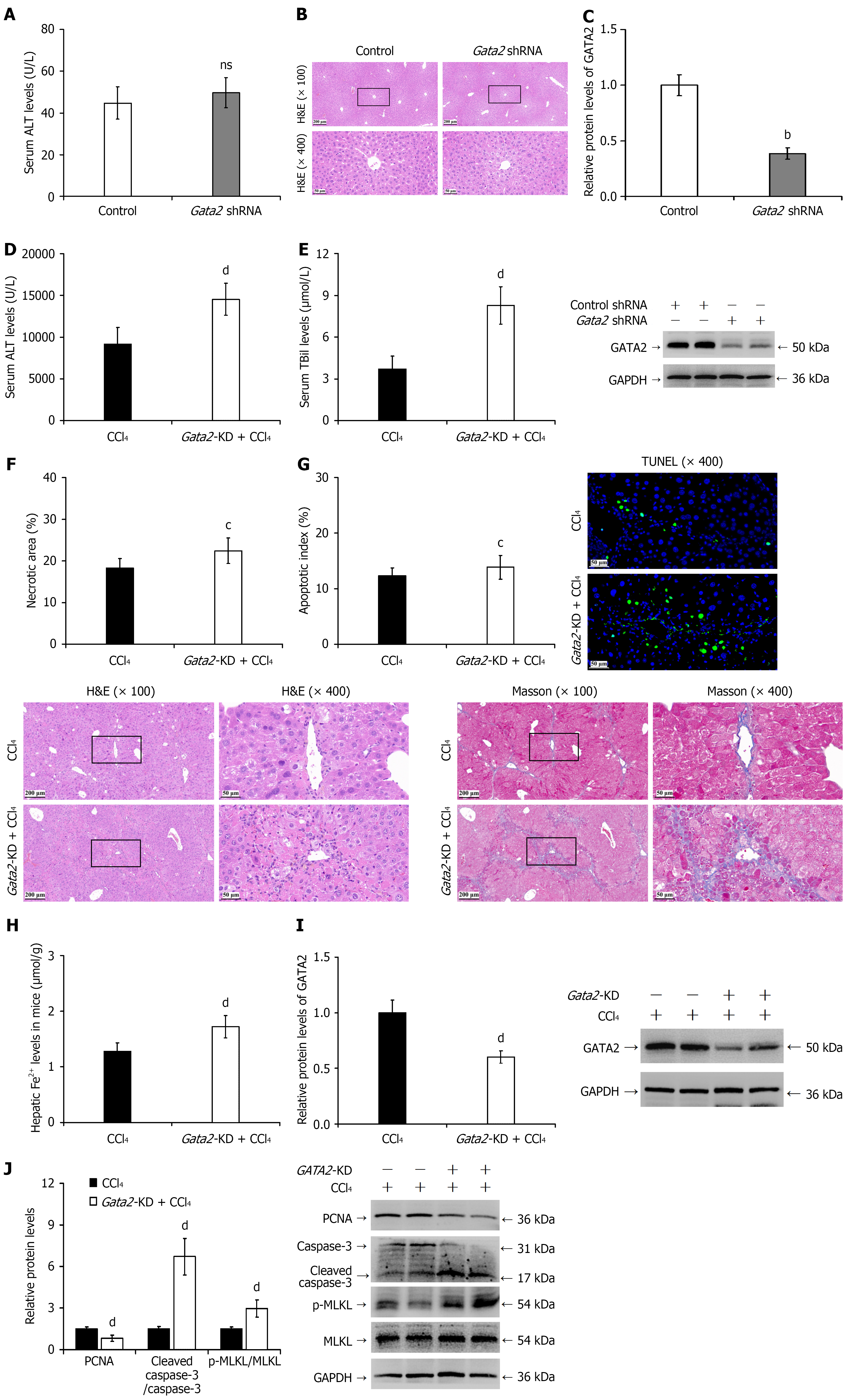
Figure 8 Hepatocyte-specific Gata2 knockdown exacerbates hepatocyte death-regeneration imbalance in chronic liver injury mice.
A-C: Gata2 shRNA-transfected healthy mice: Serum alanine aminotransferase levels (A); hepatic histopathology by hematoxylin and eosin staining (B); western blot analysis of GATA2 protein expression (representative blots shown) with quantification (C); D-I: Gata2 knockdown intervention in chronic liver injury mice: Effect of Gata2 knockdown on serum alanine aminotransferase levels in chronic liver injury mice (D); serum total bilirubin levels (E); quantitative analysis of necrotic areas with representative hematoxylin and eosin and Masson’s trichrome staining images (F); fluorescent Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling staining analysis of hepatocyte apoptosis rate (representative images shown) (G); ferrous ion content (H); western blot analysis of GATA2 protein expression (blots displayed) with quantification (I); J: Western blot analysis of proliferating cell nuclear antigen, cleaved caspase-3, and phosphorylated mixed lineage kinase domain-like protein expression (representative blots shown) with quantification. ns: P > 0.05, bP < 0.01 vs control group; cP < 0.05 vs chronic liver injury group (CCl4), dP < 0.01 vs chronic liver injury group (CCl4). ALT: Alanine aminotransferase; H&E: Hematoxylin and eosin; TBil: Total bilirubin; PCNA: Proliferating cell nuclear antigen; p-MLKL: Phosphorylated mixed lineage kinase domain-like protein; MLKL: Mixed lineage kinase domain-like protein.
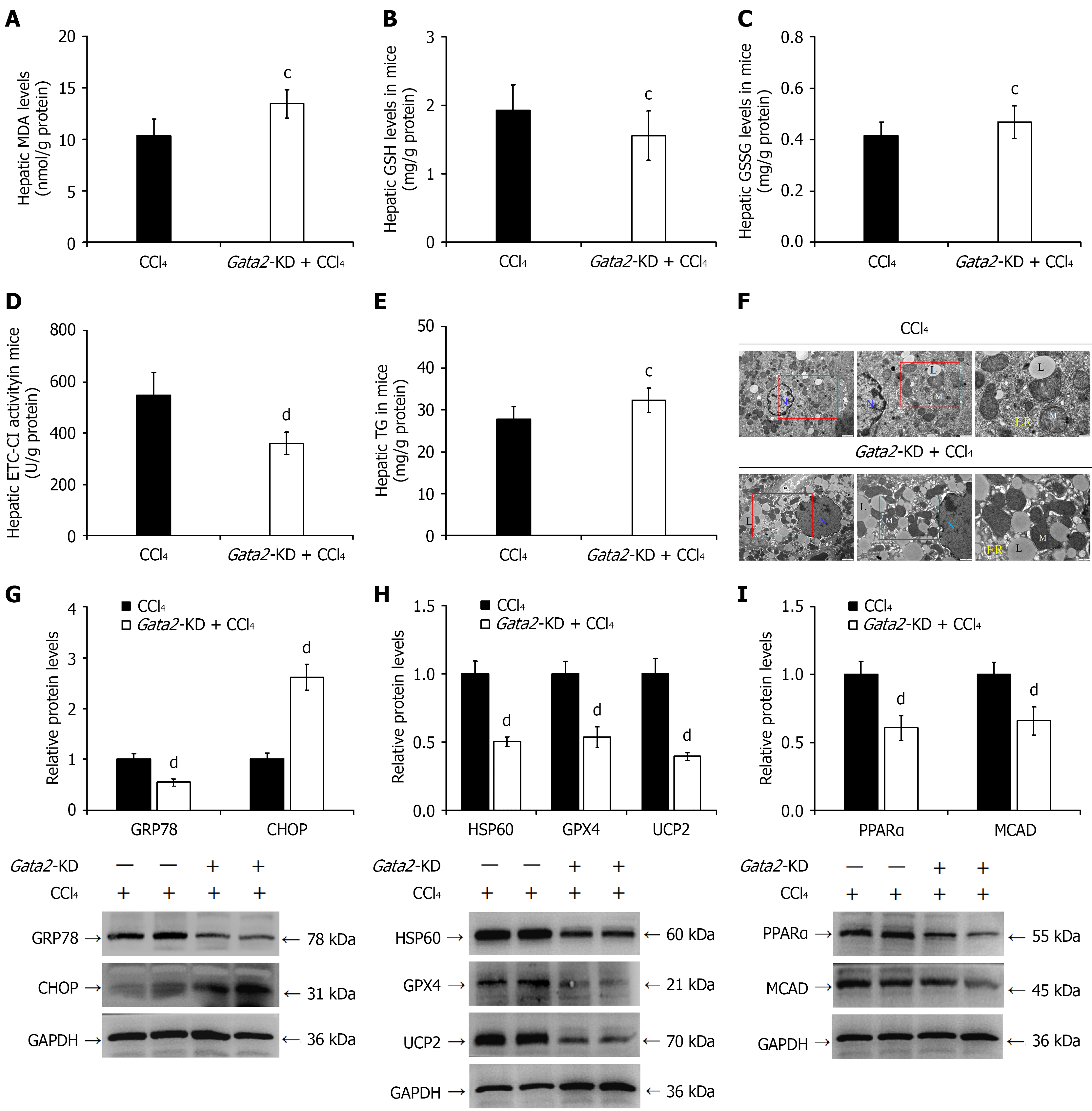
Figure 9 Hepatocyte-specific Gata2 knockdown impairs hepatic stress adaptation and exacerbates triglyceride metabolic dysregulation in chronic liver injury mice.
A: Hepatic malondialdehyde content; B: Glutathione levels; C: Oxidized glutathione levels; D: Electron transfer chain complex I activity; E: Triglyceride levels; F: Representative transmission electron microscope images showing ultrastructural morphology: Nucleus (N), mitochondria (M) with cristae disruption, dilated endoplasmic reticulum (ER), and lipid droplets (L); G: Analysis of endoplasmic reticulum stress-related protein expression; H: Western blot analysis of oxidative stress-related protein expression levels; I: Western blot analysis of triglyceride metabolism-related protein expression levels. cP < 0.05, dP < 0.01 vs chronic liver injury group (CCl4). MDA: Malondialdehyde; GSH: Glutathione; GSSG: Oxidized glutathione; ETC-CI: Electron transfer chain complex I; TG: Triglyceride; p-MLKL: Phosphorylated mixed lineage kinase domain-like protein; MLKL: Mixed lineage kinase domain-like protein; HSP60: Heat shock protein 60; GRP78: Glucose-regulated protein 78; GPX4: Glutathione peroxidase 4; UCP2: Uncoupling protein 2; PPARα: Peroxisome proliferator-activated receptor alpha; MCAD: Medium chain acyl-CoA dehydrogenase.
- Citation: Chen YF, Li SQ, Zhang J, Ma WT, Zhou Y, Rao JX, Yi Y, Cheng QJ, Zhong WW, Chen H, Chen YH, Luo YW, He YH. GATA2 deficiency exacerbates chronic liver injury via disrupting hepatocyte death-regeneration balance: Clinical, histopathological, and molecular evidence. World J Stem Cells 2026; 18(2): 112940
- URL: https://www.wjgnet.com/1948-0210/full/v18/i2/112940.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v18.i2.112940