©The Author(s) 2026.
World J Stem Cells. Jan 26, 2026; 18(1): 111348
Published online Jan 26, 2026. doi: 10.4252/wjsc.v18.i1.111348
Published online Jan 26, 2026. doi: 10.4252/wjsc.v18.i1.111348
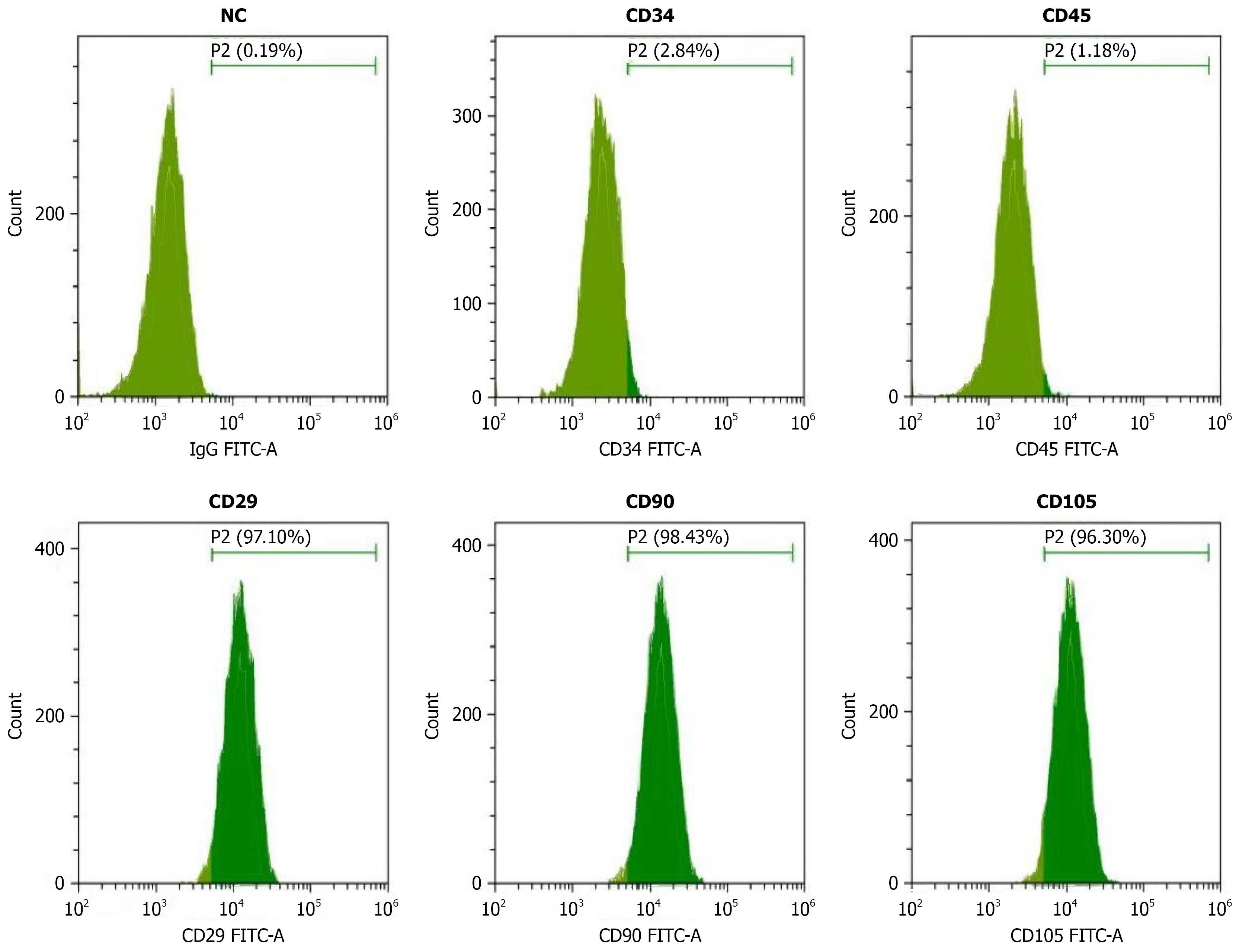
Figure 1 Flow cytometric analysis of breast cancer stem cell surface markers.
Flow cytometry histograms showing the expression of stem cell markers in breast cancer cells. Negative control (0.19% positive), CD34 (62.64% positive), CD45 (1.19% positive), CD29 (7.16% positive), CD90 (68.43% positive), and CD105 (96.30% positive). X-axis represents fluorescence intensity, Y-axis represents cell count. Percentage values indicate the proportion of positive cells for each marker, demonstrating distinct expression patterns of stem cell-associated surface antigens. NC: Negative control.
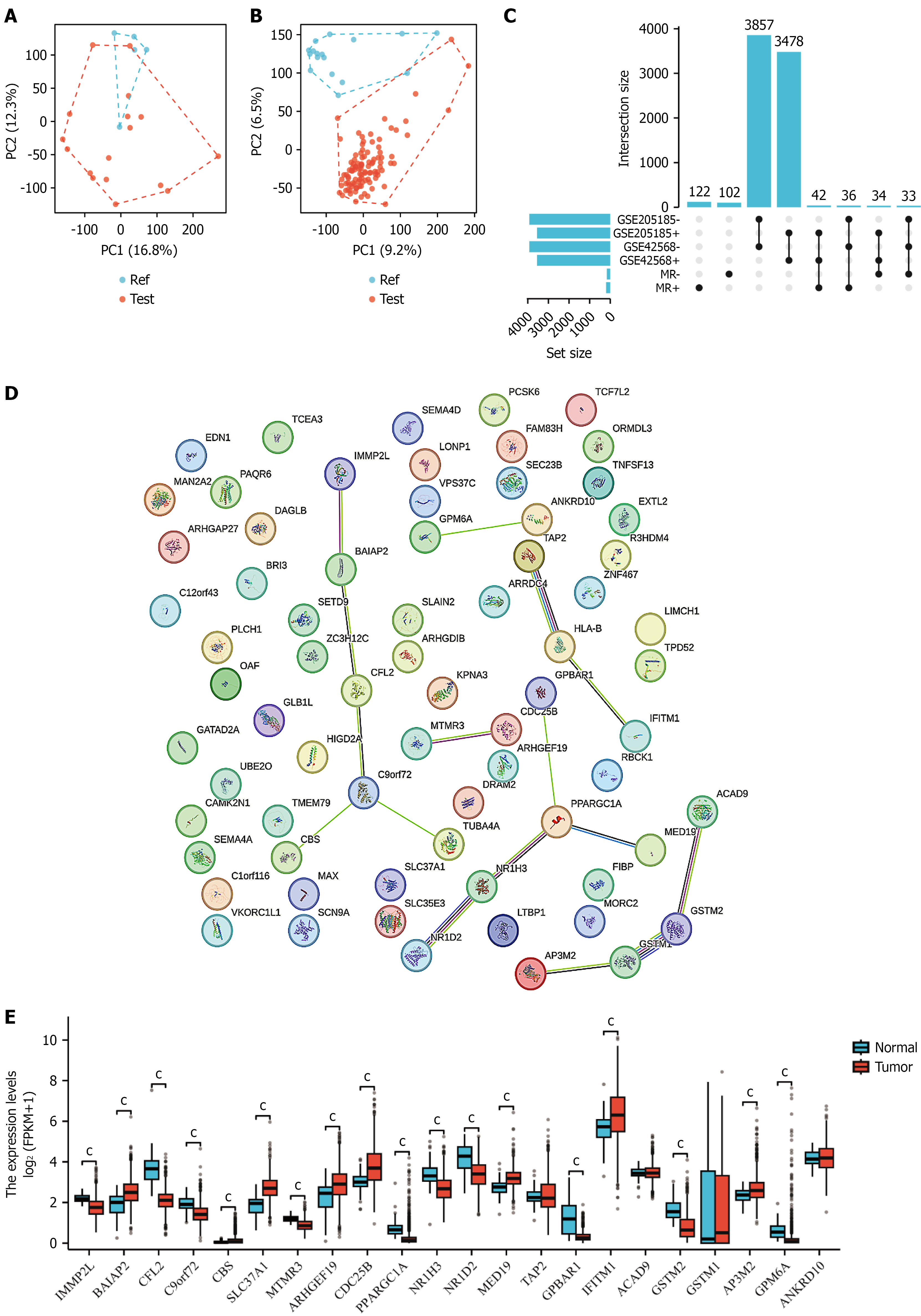
Figure 2 Transcriptomic characterization of breast cancer stem cells.
A and B: Principal component analysis plots showing distinct clustering of breast cancer stem cell samples (blue) and control samples (red) based on gene expression profiles. PC1 and PC2 explain 16.8% and 9.2% of variance, respectively; C: Differential gene expression analysis revealing 3837 significantly altered genes (1478 upregulated, 2359 downregulated) with corresponding heatmap showing expression patterns; D: Protein-protein interaction network of differentially expressed genes displaying functional associations and key regulatory nodes; E: Expression levels of stem cell-related genes in normal tissues vs breast cancer tissues, showing statistical significance (cP < 0.001).
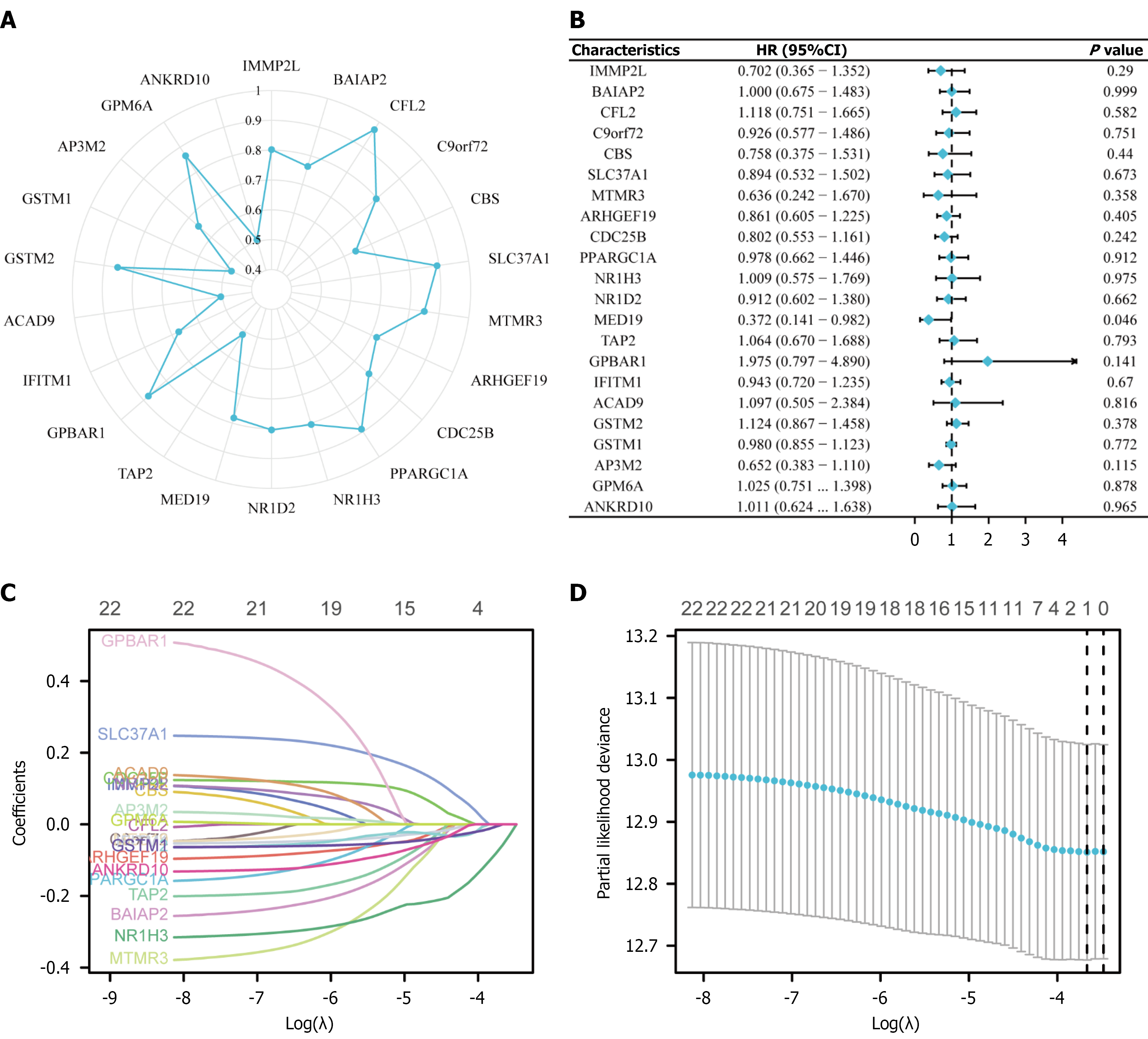
Figure 3 Least absolute shrinkage and selection operator regression analysis for breast cancer stem cell prognostic model construction.
A: Protein-protein interaction network showing key regulatory genes including ANKHD1, DMAP1, BAIAP2, GSTM1, and PPARGC1A; B: Forest plot displaying hazard ratios and 95% confidence intervals for candidate genes, with most genes showing protective effects (hazard ratio < 1); C: Least absolute shrinkage and selection operator coefficient trajectory plot showing gene coefficient changes as penalty parameter λ increases, with gene numbers indicated at the top; D: Ten-fold cross-validation plot identifying optimal λ value (logλ ≈ -5.5) with minimum partial likelihood deviance, retaining 10 genes for the final prognostic model.
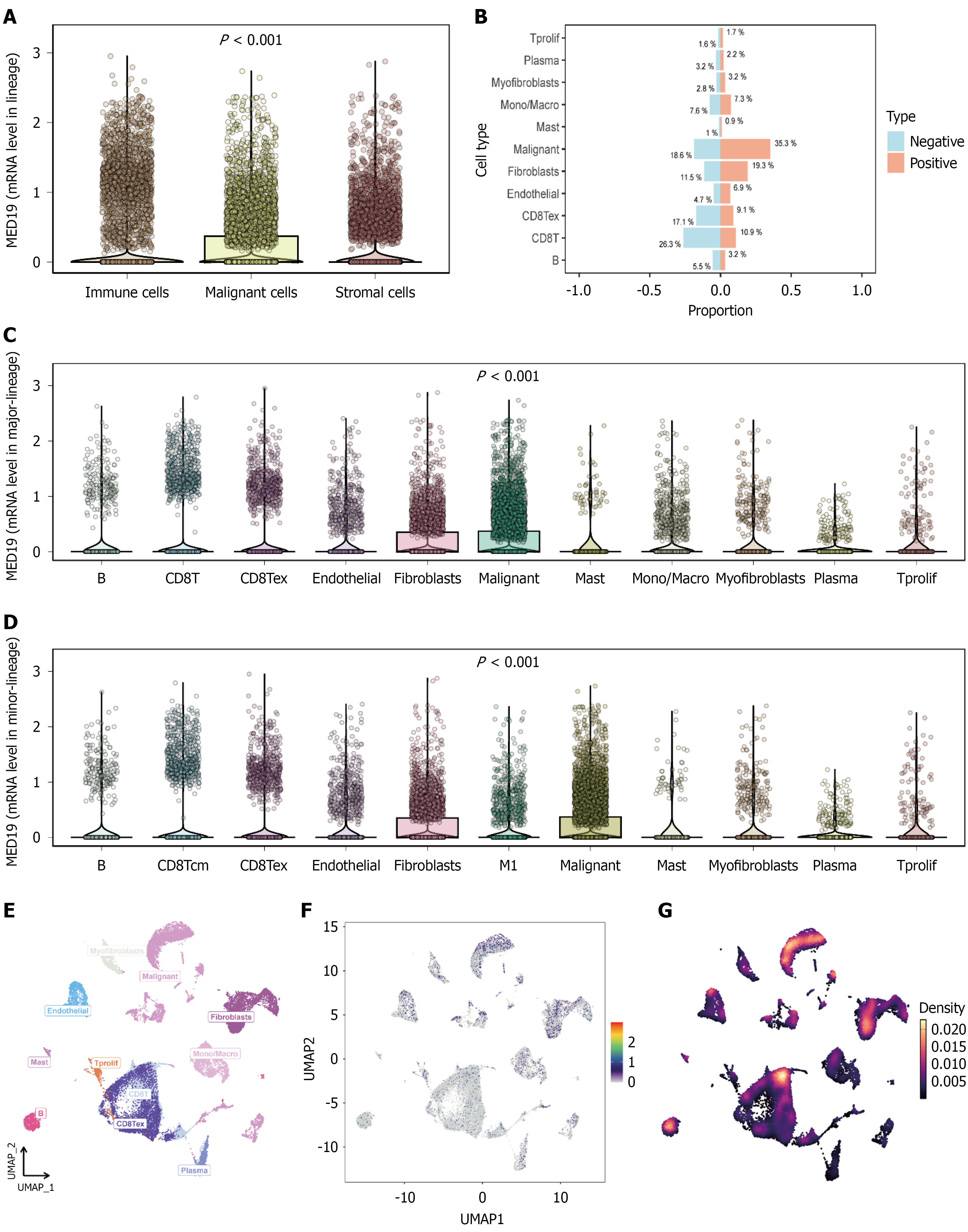
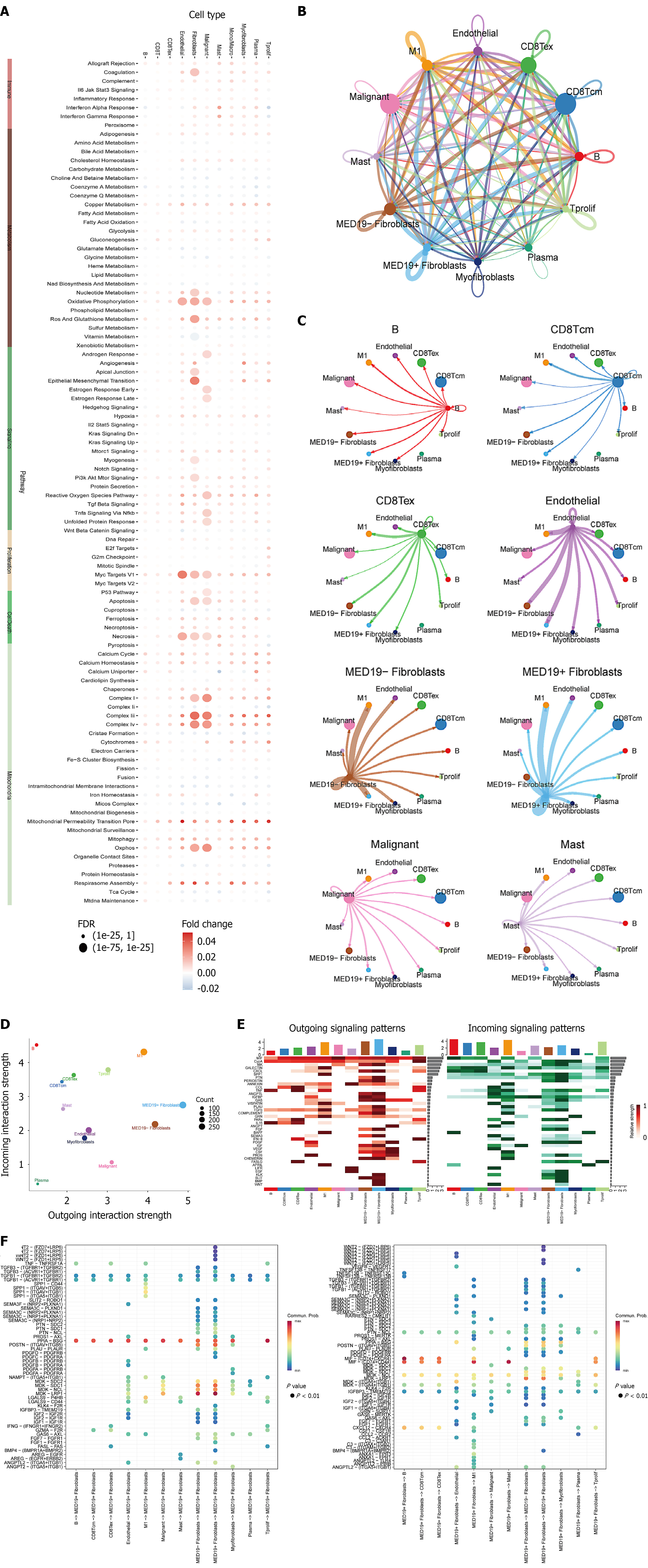
Figure 4 Validation of risk score model and single-cell transcriptomic analysis.
A: Risk score distribution showing significant differences between high-risk and low-risk groups (P < 0.001); B: Clinical feature correlation analysis displaying associations between risk scores and various clinicopathological characteristics; C and D: Expression validation of core model genes demonstrating significant differential patterns between risk groups (P < 0.001); E and F: T-SNE visualization of single-cell RNA sequencing data revealing distinct cell subpopulations with different transcriptional characteristics; G: Cell type annotation showing subpopulations with stem cell-like features characterized by high expression of pluripotency and self-renewal genes.
Figure 5 Pathway enrichment analysis and functional network of breast cancer stem cell-related genes.
A: Gene Ontology enrichment analysis showing key biological processes including cell cycle regulation, DNA repair, and apoptosis control; B: Protein-protein interaction network revealing functional associations among key genes with regulatory hubs; C: Kyoto Encyclopedia of Genes and Genomes pathway enrichment identifying cancer-related signaling pathways (cell cycle, p53, phosphatidylinositol 3-kinase/protein kinase B, mitogen activated protein-kinase); D and E: Pathway network heatmaps displaying enrichment levels with red indicating upregulated pathways and green indicating downregulated pathways; F: Comprehensive functional enrichment results integrating Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses, highlighting significant enrichment in cell cycle checkpoints, DNA damage response, and stem cell pluripotency maintenance.
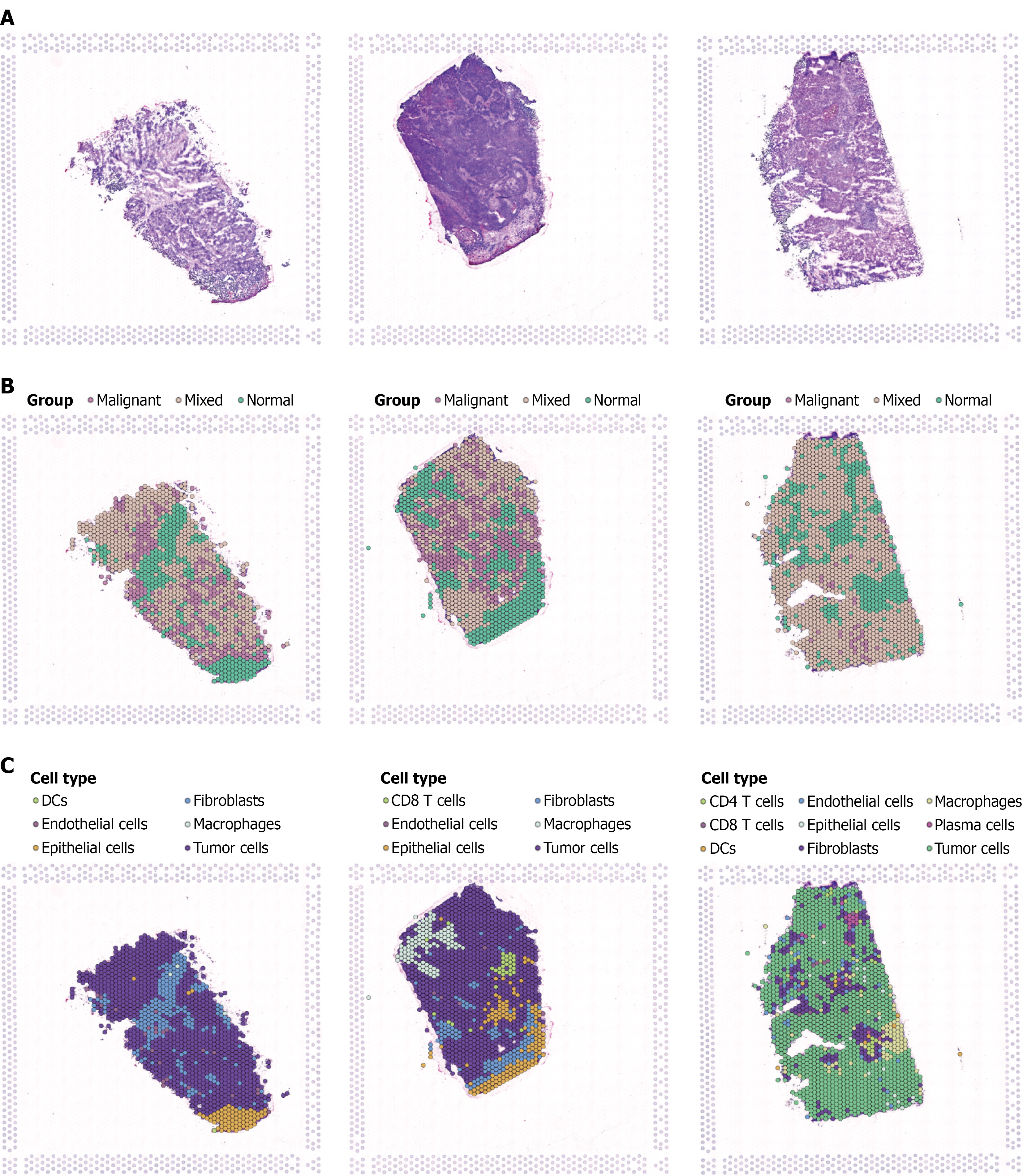
Figure 6 Single-cell spatial transcriptomic analysis of breast cancer tissues.
A: Tissue morphology of three breast cancer samples showing typical tumor architecture with cancer cell aggregation and stromal distribution; B: Spatial distribution of risk scores based on stem cell-related genes, with high-risk regions (red/orange) and low-risk regions (green/blue) displaying distinct clustering patterns; C: Cell type annotation identifying major cellular components including cancer cells, immune cells, fibroblasts, and endothelial cells with specific spatial distribution patterns throughout the tumor microenvironment.
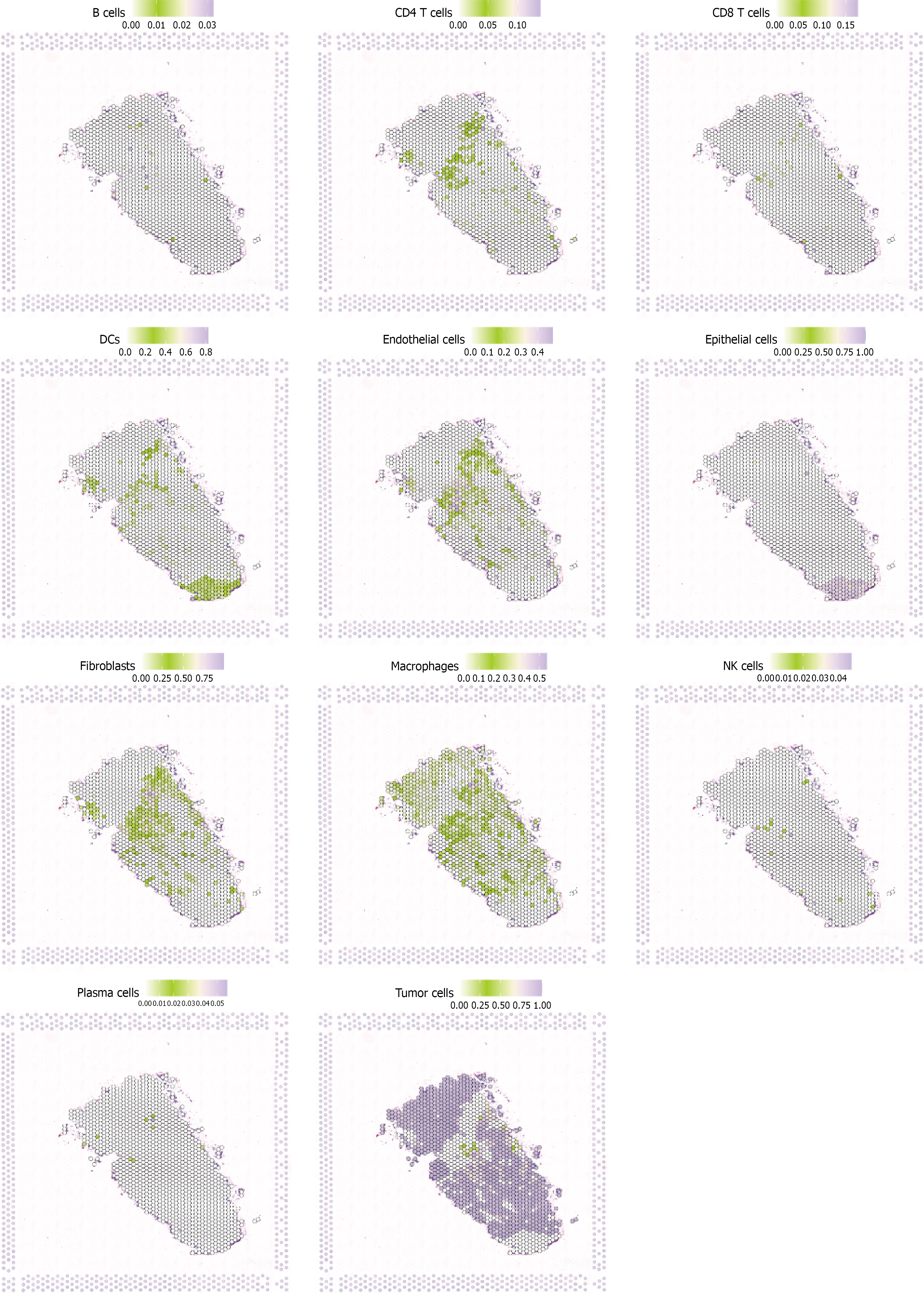
Figure 7 Spatial expression patterns of key stem cell-related genes in breast cancer tissues.
Spatial transcriptomic analysis showing the expression distribution of 11 core genes selected by least absolute shrinkage and selection operator regression, including BCL6, COL7A2, COL15A1, BCL6, CosmicPairs, EphrinMulti, macrophages, MXRA8, fibronectin, macrophage, and TumorCells. Expression levels are indicated by color intensity (green to purple scale), with gray areas representing low or no expression. The analysis reveals heterogeneous spatial distribution patterns, with some genes showing localized expression in specific tumor regions while others display broader tissue distribution or gradient patterns from tumor center to periphery.
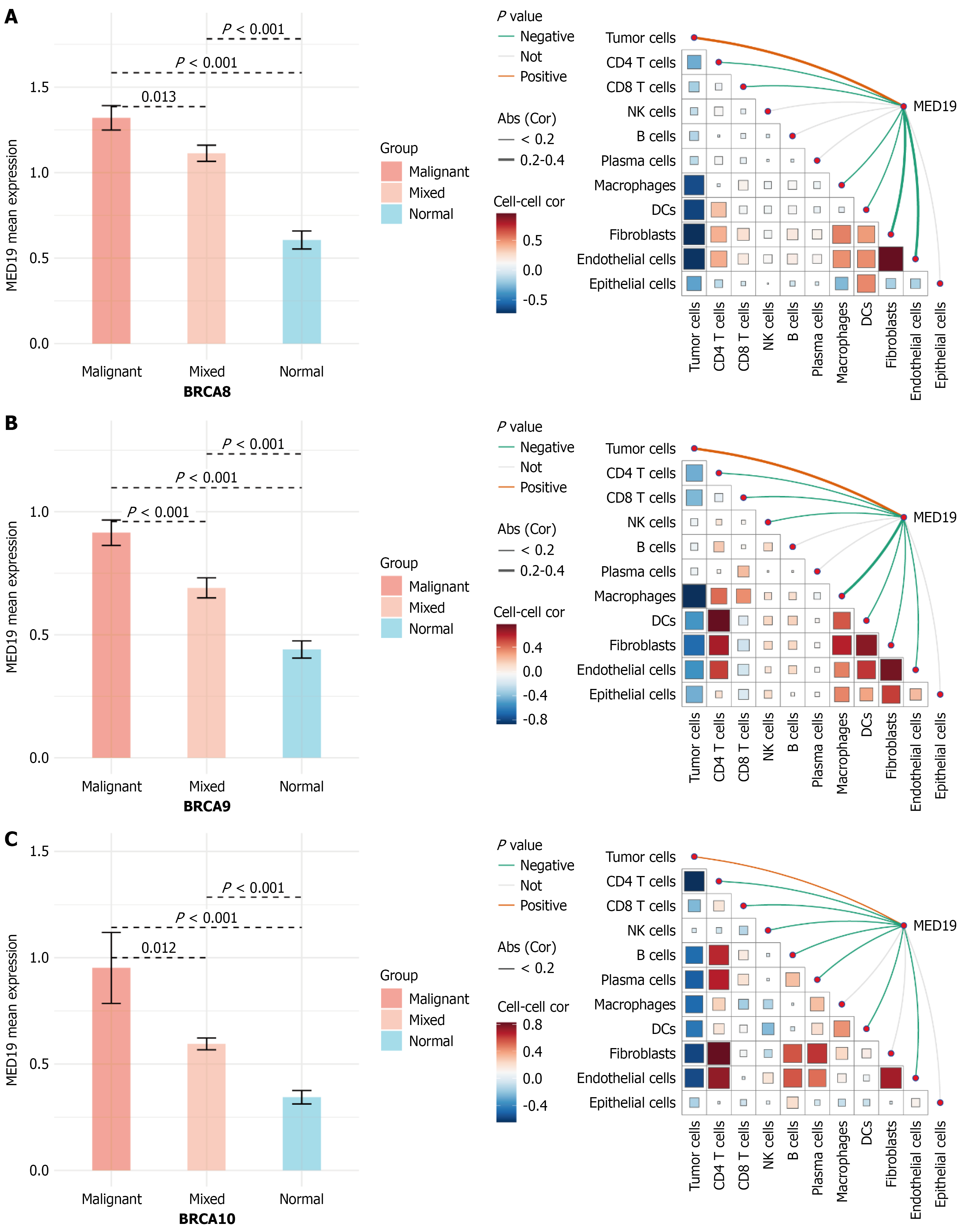
Figure 8 Expression validation and cell-cell communication analysis of MED18 in breast cancer.
A-C: Expression analysis of MED18 across three independent breast cancer samples showing significantly higher expression in malignant tissues compared to normal controls and upregulation in breast cancer subtypes (P < 0.001). Right panels display correlation heatmaps and network diagrams illustrating relationships between MED18 expression and various cell types including tumor cells, macrophages, fibroblasts, and endothelial cells. Network analysis reveals MED18’s central position in cell-cell communication networks, with correlation strength indicated by color intensity (red: Positive correlation, blue: Negative correlation).
- Citation: Guo DY, Liu ZY, Yi QC. Breast cancer stem cell activity driven by ME18D gene expression in the tumor microenvironment. World J Stem Cells 2026; 18(1): 111348
- URL: https://www.wjgnet.com/1948-0210/full/v18/i1/111348.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v18.i1.111348