Published online Nov 28, 2025. doi: 10.3748/wjg.v31.i44.111160
Revised: July 20, 2025
Accepted: October 23, 2025
Published online: November 28, 2025
Processing time: 156 Days and 21.8 Hours
Artificial intelligence (AI) is revolutionizing the field of gastrointestinal (GI) endoscopy, a technology that relies heavily on images and optical data. Precancerous lesions and early cancers of the GI tract can be subtle and easily missed even on high-definition endoscopy and chromoendoscopy. The advancements in machine learning and deep learning led to the development of computer-aided models of high performance in image analysis. The convolutional neural networks of these models are trained to analyze large datasets of endoscopic images th
Core Tip: Artificial intelligence (AI) is revolutionizing healthcare. There is evidence supporting the role of AI in the detection/characterization of premalignant lesions and early cancers of the gastrointestinal (GI) tract, especially in the esophagus, stomach and colon. The utilization of AI may eventually decrease the incidence of interval cancers which are mainly attributed to missed lesions during endoscopy. It benefits mostly novice endoscopists and trainees. AI may be superior to experienced endoscopists in the detection of diminutive lesions. The effect of AI on clinically meaningful outcomes such as incidence of interval GI cancers, morbidity, mortality and cost effectiveness requires further research.
- Citation: El Asmar N, Baydoun M, Mrad J, Barada K. Role of artificial intelligence in the detection and characterization of gastrointestinal premalignant and early malignant lesions. World J Gastroenterol 2025; 31(44): 111160
- URL: https://www.wjgnet.com/1007-9327/full/v31/i44/111160.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i44.111160
The field of artificial intelligence (AI) has witnessed exponential growth during the last few years. It has influenced various aspects of life including business, cybersecurity and education. Its transformative effect on healthcare is not an exception[1].
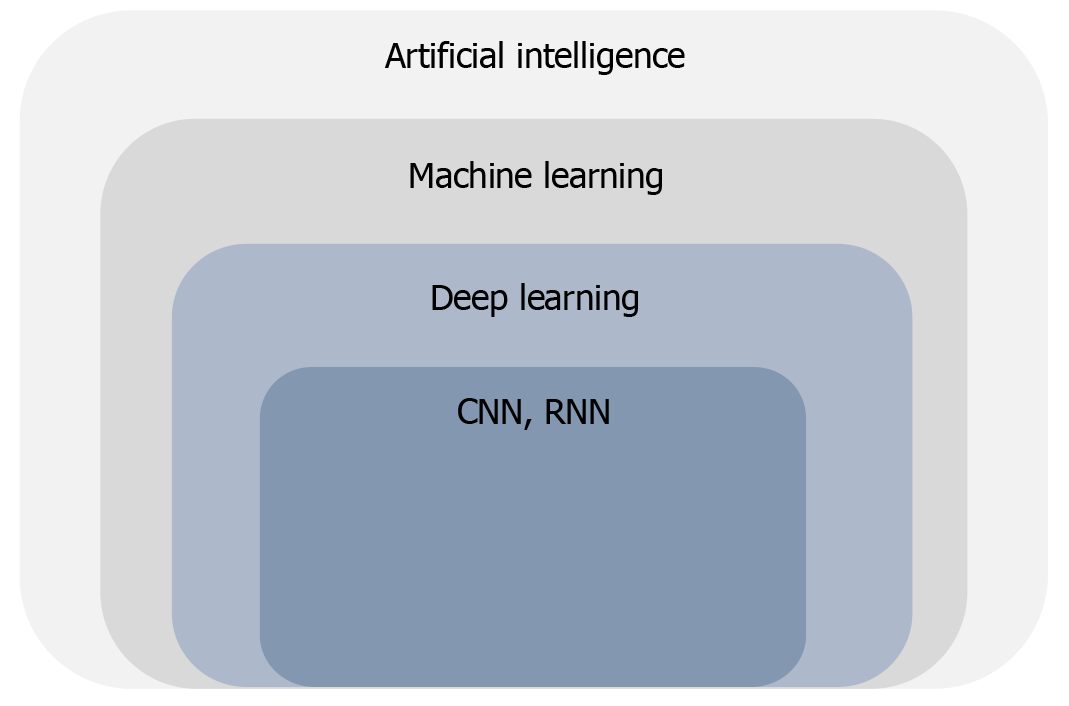
AI evolves around the development and deployment of computer systems capable of performing tasks that require human-level intelligence such as learning, problem solving and decision-making[2]. It is a wide-ranging term that encompasses multiple disciplines including machine learning (ML) and deep learning (DL) (Figure 1).
ML is a subset of AI that enables computer programs to learn from datasets and subsequently improve their per
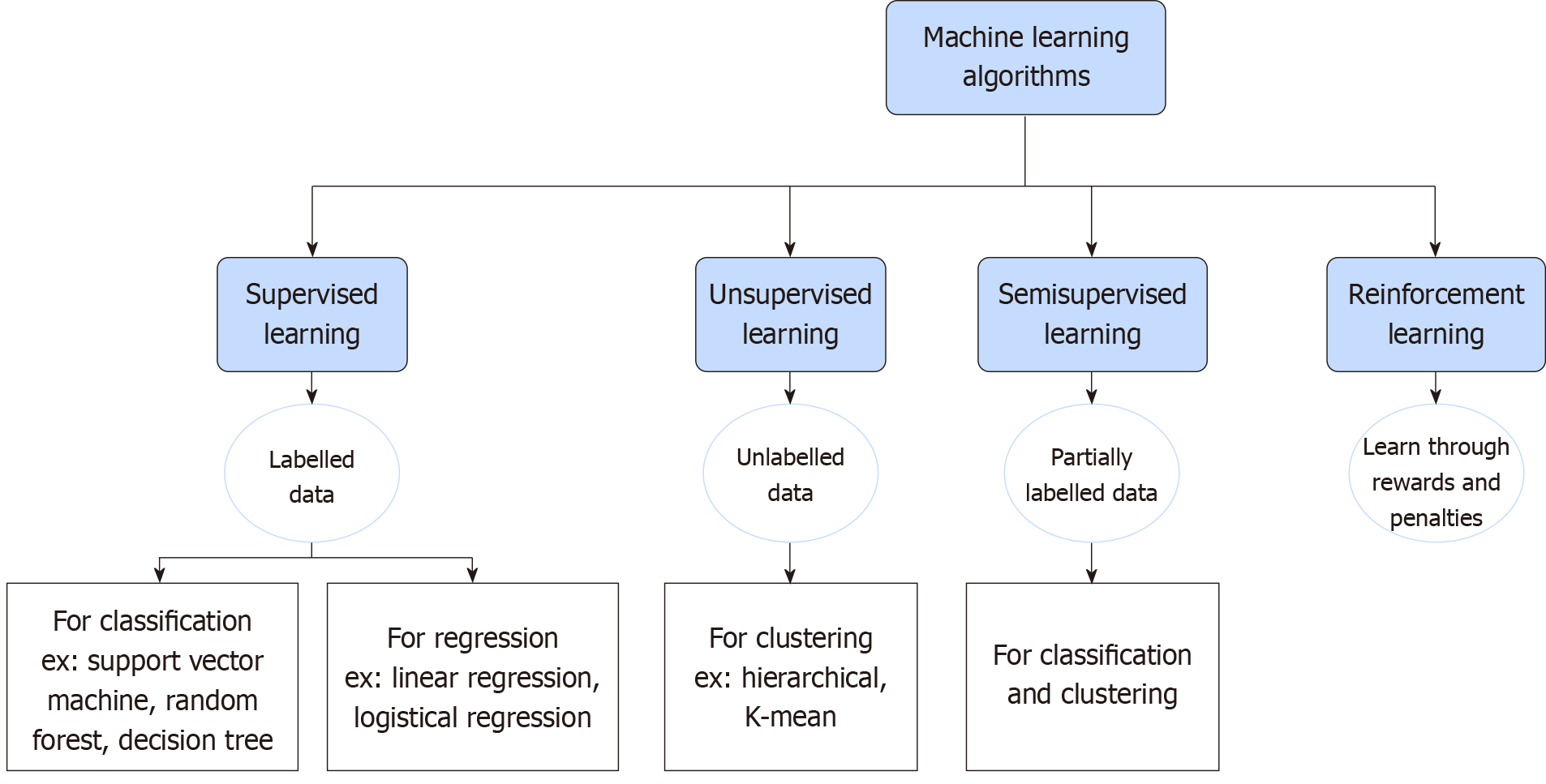
There are four main categories of ML algorithms (Figure 2): Supervised, unsupervised, semi-supervised and reinforcement learning[4]. Supervised learning is the most important methodology in ML. The distinguishing feature of supervised learning is the utilization of labelled training datasets to learn the function of calculating and modifying errors to achieve the desired outcome. This will induce computer models capable of mapping input variables to output variables. It has two main functions: (1) Data classification: Organizing data into categories; and (2) Data regression: Establishing the relationship between changes of an independent variable and its effect on other dependent variables. Examples of supervised learning are face recognition and image tagging feature in social networking sites[4,5]. In unsupervised learning, models learn to discover common hidden patterns among unlabeled datasets without explicit guidance from humans. It is used for data clustering where data is grouped into clusters based on their inherent simi
DL is a subset of ML[7]. A distinctive feature of DL is the outstanding performance with large amounts of data in comparison to traditional ML[8]. The structure and processing function in DL are inspired by the human brain and neuronal interaction. DL is based on the older fundamental concept of artificial neural network (ANN) where it uses multiple layers of algorithms organized in several layers of fully connected ANN, each of which interprets the data it receives differently[9]. Neural networks consist of interconnected nodes called neurons. Each neuron has a specific weight (strength) and threshold. Activation of a neuron and subsequent effect on other neurons occurs only when the output exceeds the threshold value, otherwise no data is propagated to the next layer[10]. A DL neural network com
CNN is among the most used DL networks and a main contributor to its popularity nowadays. They are capable of learning image features extraction from raw datasets without any human intervention, and that makes it superior to traditional ANN. CNNs are specifically designed to process a large array of two-dimensional and three-dimensional figures by utilizing the information across the pixels of an image, and then creating feature maps that are later on pooled together to capture a larger field of the image. Eventually, features from all CNN layers are combined to produce the final intended outcome. This mechanism explains their application in medical imaging analysis, visual recognition and natural language processing[9,12].
RNN is another familiar type of DL networks. It uses sequential or time-series data, i.e., it processes data where a specific order and arrangement of elements are required for a meaningful outcome. Memory is a distinguishing feature of RNN. This memory allows RNN to influence current input by experience learnt from previous inputs. Speech recognition, natural language processing and prediction problems (such as forecasting stock prices and predicting flood levels) are the most common applications of RNN[13].
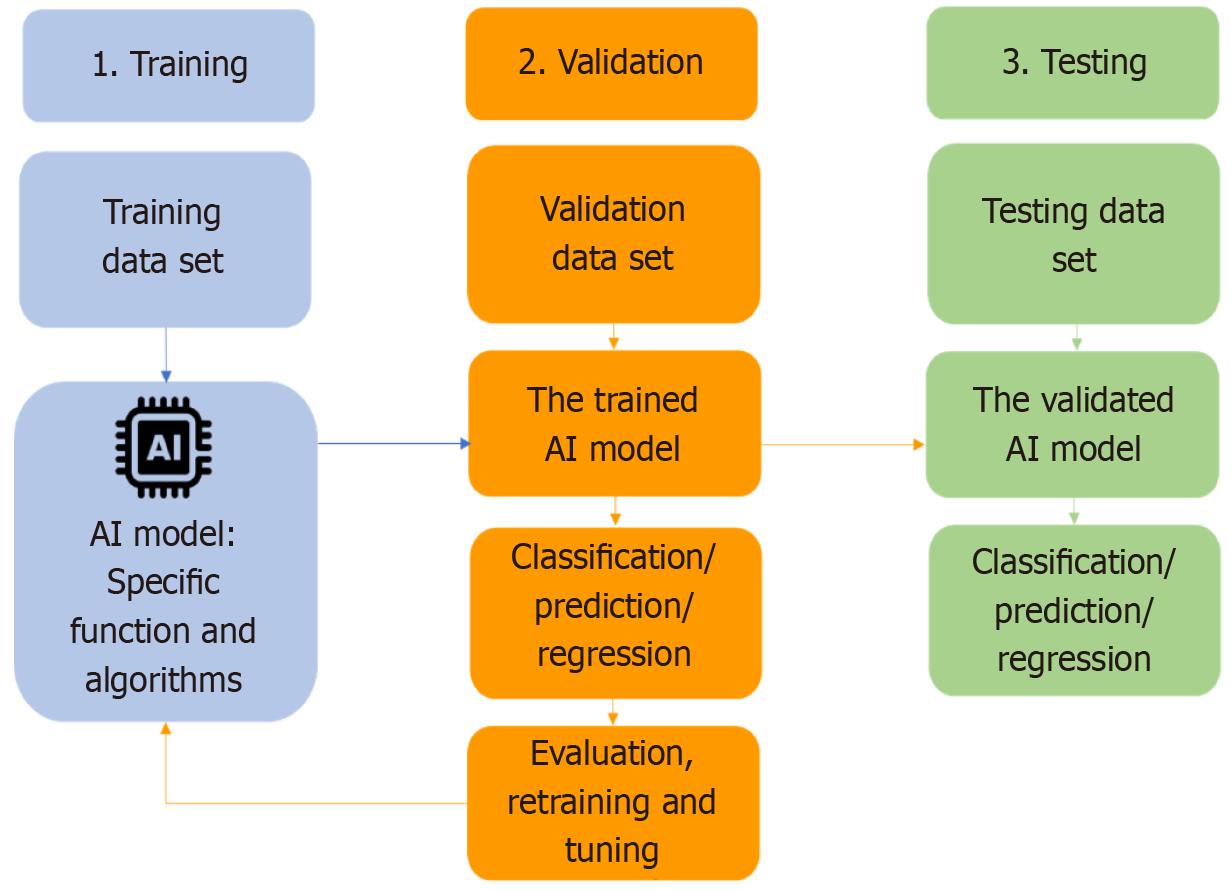
The process of training AI models occurs over several steps. Initially, the targeted raw data goes through the process of data annotation, where the data gets prepared and structured into training data sets to feed the AI models later. Next, and depending on the type and complexity of data, suitable algorithms are deployed. The next step is validating the trained model with new data, the validation dataset. This step is essential to assess if the performance of the model aligns with the expectations. It also allows transfer learning, defined as retraining and fine-tuning of the model with new data exposure. The last step is testing the model’s classification or prediction ability with a separate set of new data, the testing data set (Figure 3).
AI has revolutionized various image-dependent medical fields such as radiology, gastroenterology, dermatology and pathology[14]. However, with the growing bulk of digital images and advances in imaging modalities comes the need for advanced AI models that have the ability to efficiently process and learn from these large optical datasets. The evolution of ML through DL and specifically CNN provided the computer systems with these fundamental features facilitating the implementation of AI in the aforementioned fields.
All disciplines of gastroenterology are subjects for AI research and application. Nevertheless, endoscopy [esophagogastroduodenoscopy, colonoscopy, capsule endoscopy and device assisted enteroscopy (DAE)] remain the cornerstone diagnostic and therapeutic procedures in gastroenterology. They yield enormous digital datasets that represent an ideal substrate to feed and train computer models. As a result, the main focus of AI research is drawn to endoscopic image analysis rather than data analysis as in the fields of hepatology, pancreaticobiliary diseases and inflammatory bowel diseases[15].
Gastrointestinal (GI) malignancies remain a leading cause of mortality worldwide. Despite the availability of well-established screening programs and the decreased mortality associated with their implementation, colorectal, gastric and esophageal cancers are still responsible for more than 3.5 million new cases and around 2 million deaths yearly[16]. Established risk factors contributing to the high percentage of cancers post endoscopy (interval cancers) are high miss rates of early cancers during the procedure, as well as failed recognition and sampling of high-risk premalignant lesions[17,18]. Research has been directed towards the development of AI algorithms to optimize the effectiveness of endoscopic procedures by eliminating inter-endoscopist variability. This led to the development of clinically meaningful computer assisted devices (CAD) for lesion detection [computer aided detection (CADe)], diagnosis [computer-aided diagnosis (CADx)] and quality assessment[19]. CADe helps in the detection of mucosal abnormalities through recognition of changes in shape, color and texture whereas CADx is used for optical diagnosis and prognostication of the detected lesions without the need for histopathological assessment[20].
This review sheds light on the role of AI assisted endoscopy in the detection and characterization of premalignant and early malignant lesions of the upper GI tract, small intestine as well as the colorectum.
A comprehensive literature search was conducted using PubMed databases to identify relevant studies on the application of AI in the detection and characterization of GI premalignant and early malignant lesions. The search covered publications from January 2003 to April 2025. The following keywords were used and combined using Boolean operators: “Artificial intelligence”, “deep learning”, “machine learning”, “convolutional neural networks”, “CADx”, “CADe”, “gastrointestinal”, “colorectal”, “gastric”, “esophagus”, “small bowel”, “Barrett’s esophagus”, “endoscopy”, “colonoscopy”, “gastroscopy”, “upper endoscopy”, “neoplasia”, “precancerous”, “premalignant” and “dysplasia”. In addition, the “related articles” feature in PubMed was used to expand the search and identify further relevant studies. Eligible studies were those involving endoscopic applications of AI in luminal GI neoplasia. Studies focusing on biliary, hepatic, or pancreatic malignancies, and those not involving endoscopic procedures were excluded. Only peer-reviewed articles published in English were considered. Reference lists of relevant reviews and included articles were also screened to identify additional eligible studies.
Precancerous lesions and early cancers of the esophagus and stomach are often subtle, flat, multifocal and easily missed[21,22]. By improving tumor detection rate, AI effectively enhances the diagnostic yield in upper GI cancer screening[23,24].
In the esophagus, the premalignant lesion is Barrett’s esophagus (BE)-associated dysplasia, a precursor of esophageal adenocarcinoma (EAC), while squamous dysplasia is the precursor of esophageal squamous cell carcinoma (ESCC). Furthermore, early-stage or superficial ESCC or EAC is defined histopathologically as high-grade intraepithelial neoplasia or cancer confined to the mucosa and submucosa. Most esophageal cancer cases are still diagnosed at advanced stages, contributing to poor prognosis, higher mortality, and increased economic burden[25,26]. In light of this, AI may improve early detection and characterization of early esophageal cancers and precancerous lesions, enabling timely intervention and better patient outcomes.
BE: Early detection of BE is essential for improving both clinical outcomes and cost-effectiveness. BE affects about 1% of the global population and typically progresses through stages of low-grade and high-grade dysplasia before developing into EAC. Therefore, endoscopic surveillance is essential, as more than 90% of dysplastic lesions are detected either during the initial endoscopy or within six months, suggesting that many lesions are already present but may be initially overlooked[27]. Nevertheless, neoplasia in BE can be missed in up to 33% of cases[26]. The early detection of BE and associated dysplastic changes may reduce cancer-related mortality. Recent advances in AI have significantly improved the endoscopic detection and characterization of BE-associated neoplasia[28]. Table 1 provides a summary of studies evaluating AI models for the detection and characterization of BE and associated neoplasia.
| Ref. | Purpose | Design | Training set | Test set | IEE | AI model | Performance |
| de Groof et al[39] | Real-time Det + Ch of early BE neoplasia | Prospective study | 1247 i | 297 i | WLI | Hybrid: ResNet-UNet | Acc: 89%; Se: 90%; Sp: 88% |
| de Groof et al[40] | Real-time Det + Ch of early BE neoplasia | Prospective pilot study | 1544 i | 144 i | WLI | Hybrid: ResNet-UNet | Acc: 90%; Se 91%; Sp: 89% |
| Hashimoto et al[41] | Distinguish dysplastic from nondysplastic BE | Prospective pilot study | 1374 i | 458 i | WLI; NBI | Inception-ResNet-v2 | Acc: 95%; Se: 96%; Sp: 94% |
| Ali et al[33] | BE risk stratification | Prospective pilot study | 10000 i | 194 v | WLI; NBI | ResNet-50; DeepLabv3 + | Acc: 98% |
| Hussein et al[45] | Det and delineation of BE dysplasia | Retrospective study | 148936 v | 276 v | WLI; i-scan | ResNet-101 | Se: 91%; Sp: 79%; AUC: 0.93 |
| Abdelrahim et al[37] | Det and localization of BE neoplasia | Prospective multicenter trial | 1090171 i + v | 471 i; 75 v | WLI | VGG16; SegNet | I: Acc/Se/Sp: 95%; v: Acc: 92%; Se: 95%; Sp: 91% |
| Tsai et al[30] | Det of BE | Retrospective study | 771 i | 160 i | NBI | EfficientNetV2B2 | Acc: 94%; Se: 94%; Sp: 94% |
| Xin et al[8] | Det of early BE neoplasia | Retrospective multicenter | 14046 i | 400 i; 188 v | EfficientNet-Lite1; MobileNetV2; DeepLabv3 + | I: Se: 88%; Sp: 90%; v: Se: 79%; Sp: 94% | |
| Meinikheim et al[34] | AI impact on nonexp BE neoplasia Det | Multicenter RCT | 51273 i | 96 v | WLI; NBI; Chromo | DeepLabv3; ResNet-50 | Se: 70% to 78% w/AI; Sp: 67% to 73% w/AI |
| Jukema et al[42] | AI vs exp/nonexp in BE neoplasia Det | Prospective | 3468 i | 161 i; 161 v | NBI | EfficientNet-Lite1 | Se: 84% to 96% w/AI; Sp: 90% to 98% w/AI |
| Jong et al[29] | Ensure AI reliability in BE neoplasia Det | Retrospective real word experience | 1102 i; 12011 v | 117 i | WLI; NBI | Hybrid: ResNet-50-vision transformer | Se: 85% (high-quality I); 62% (mod); 47% (low) |
Multiple CADe systems-trained on extensive multicenter image and video datasets have demonstrated high accuracy, sensitivity, and real-time performance[29]. These systems often utilize white light imaging (WLI), and some of them rely on narrow-band imaging (NBI)[30,31] or linked-color imaging (LCI)[32] to enhance visualization and improve lesion detection. Traditional endoscopic detection of BE is challenging, with expert endoscopists demonstrating only 50% sensitivity and low interobserver agreement. AI systems help address these limitations by significantly reducing interobserver variability and improving overall diagnostic consistency. Most of these models were evaluated using static endoscopic images; however, few studies also incorporated video datasets to better reflect real-world clinical settings. Notably, detection performance remained robust across challenging test sets and live settings, including short segment BE[32,33], although image quality and training data diversity influenced outcomes. For instance, some DL models achieved sensitivities exceeding 90% and consistently outperformed nonexpert endoscopists, with some matching and in certain cases surpassing expert endoscopists[30,34-37]. A meta-analysis confirmed diagnostic performance of AI in detecting early BE neoplasia with pooled sensitivity and specificity around 90% and 84%, respectively[38]. In several studies, AI showed higher sensitivity than endoscopists and enabled fast, accurate lesion localization, supporting its integration into routine screening and surveillance[37,39,40].
AI-based CADx systems, particularly those trained on NBI, have demonstrated high standalone sensitivity and specificity in distinguishing dysplastic from non-dysplastic BE[31,41]. Moreover, some AI models have shown the ability to differentiate between low-grade and high-grade dysplasia[39]. In addition to their diagnostic accuracy, CADx systems also boost the confidence and performance of general endoscopists, enabling them to reach expert-level accuracy[42,43]. Other systems using magnifying endoscopy (ME) with i-scan or CNNs trained on NBI zoom videos achieved high area under the curve (AUC) and real-time frame processing speeds, enabling accurate dysplasia classification and optimizing biopsy site selection[41,44,45]. Tools based on WLI were also explored, with some showing lower performance compared to NBI and LCI, reinforcing the value of optical enhancement for AI-based characterization[31,32].
Early ESCC: ESCC is equally challenging to diagnose due to its extremely subtle endoscopic appearance and the absence of specific symptoms in the initial stages[46]. Enhanced imaging techniques, including Lugol dye chromoendoscopy and NBI, are used to improve lesion detection and margin visualization[28]. Population-based screening is typically recommended in high-risk geographic regions and for individuals with known risk factors, including a history of head and neck tumors[47]. Nevertheless, despite advancements in endoscopic imaging, the majority of ESCC cases are still diagnosed at advanced stages, which portends a poor prognosis[28]. DL models have shown the potential to improve early cancer detection and lesion characterization, predict invasion depth, and delineate lesion margins with remarkable accuracy, all contributing to effective treatment planning[28,48]. Table 2 provides a summary of studies evaluating AI models for the detection and characterization of early ESCC and precancerous lesions.
| Ref. | Purpose | Design | Training set | Test set | IEE | AI model | Performance |
| Horie et al[67] | Det of early ESCC | Retrospective | 8428 i | 1118 i | WLI; NBI | SSD | Acc/Se: 98%; NPV: 95%; PPV: 40% |
| Ohmori et al[70] | Det of early ESCC | Retrospective | 11283 non-ME i; 11279 ME i | 523 non-ME i; 204 ME i | WLI; NBI; BLI | SSD | Non-ME Acc: 79%; Se: 95%; Sp: 69%; ME: Acc: 77%; Se: 98%; Sp: 56% |
| Fukuda et al[66] | AI vs exp in early ESCC Det and Ch | Retrospective | 17274 i | 144 v | NBI; BLI | SSD | Det: Acc: 63%; Se: 91%; Sp: 51%; Ch: Acc: 88%; Se: 86%; Sp: 89% |
| Yang et al[52] | AI vs exp in Det of early ESCC | Retrospective | 22994 i | 222 i; 104 v | WLI; NBI; BLI | YOLOv3; ResNet-v2 | Acc: 99%; Se: 100%; Sp: 99% |
| Yuan et al[51] | Prediction of ID in early ESCC | Retrospective multicenter | 7094 i | 1589 i | NBI | HRNet; OCRNet | Acc: 91%; ID prediction: 74% |
| Tani et al[50] | Real-time Det of early ESCC | Prospective single center trial | 25048 i | 237 i | WLI; NBI | ResNet-101 | Acc: 81%; Se: 68%; Sp: 83% |
| Li et al[49] | Det of early ESCC and precancerous lesions | RCT | 26543 i | 3117 i | WLI; NBI | ENDOANGEL-ELD | Acc: 98%; Se: 90%; Sp: 98% |
| Ma et al[54] | Det + optical biopsy for early ESCC | Prospective | 25056 i | 2442 i; 187 v | WLI; pCLE | iCLE inception-ResNet-v2 | Acc: 98%; Se: 95%; Sp: 99% |
| Aoyama et al[53] | AI vs exp/nonexp in early ESCC Det | Prospective | 280 v | 115 v | NBI | YOLOv3 | Acc: 77%; Se: 76%; Sp: 79% |
AI systems have significantly improved the detection of superficial ESCC and precancerous lesions, while also reducing inter-operator variability. Several randomized controlled trials (RCTs) using CNNs[49-51] have demonstrated increased detection rates, reduced miss rates and improved diagnostic confidence among novice and mid-level endoscopists. Further, several models are performing on par with[52] or exceeding experts endoscopists[53-56]. Meta-analyses[57,58] further confirmed the high diagnostic accuracy of AI-assisted endoscopy for early ESCC, with pooled sensitivities and specificities exceeding 90%, consistently outperforming endoscopists in early detection of sub centimetric lesions. Retrospective studies[59-62] utilized you only look once (YOLO)-based architectures trained on large datasets of WLI, NBI, LCI and blue-laser imaging (BLI). CAD systems and other DL models consistently achieved high sensitivity, specificity and accuracy in identifying early ESCC[59,63,64]. Additionally, the integration of hyperspectral imaging with AI further enhanced detection performance, particularly in WLI settings[60,65].
Beyond detection, AI has shown strong potential in the characterization and staging of esophageal neoplasia. DL models can predict invasion depth, classify vascular patterns and enhance optical biopsy interpretation, ultimately reducing the need for unnecessary tissue sampling[54]. For example, they can reliably differentiate between esophageal squamous dysplasia and nondysplastic mucosa, as well as distinguish superficial from advanced cancer with high diagnostic accuracy[60,66,67]. Several modern and sophisticated DL models such as clustering-constrained attention multiple instance learning[68], efficient net-vision transformer hybrids[64], and single shot multi-box detector (SSD) architectures[69], have been trained on histologic slides, ME-NBI images and videos to predict mucosal, submucosal, and deeper invasion. These models can reveal distorted or irregular vascular patterns in invasive ESCC. They consistently achieved high accuracy and outperformed or matched experienced endoscopists[70-72]. For example, the interpretable AI-based invasion depth prediction system significantly improved endoscopists’ accuracy in assessing invasion depth and was validated across multiple centers[73]. Similarly, AI tools trained to classify intrapapillary capillary loops (IPCLs) subtypes using ME enhanced junior endoscopists’ ability to assess invasion depth[74]. The Japanese IPCLs classification is frequently used to guide AI training for depth assessment, helping to differentiate intramucosal from submucosally invasive ESCC and aiding in the selection of lesions suitable for endoscopic resection[75].
Gastric premalignant conditions (GPMC) include atrophic gastritis (AG), gastric intestinal metaplasia (GIM), dysplasia, and certain gastric epithelial polyps such as adenomas, some hyperplastic polyps, as well as type 1 neuroendocrine tumors[47]. Early-stage gastric cancer (EGC) is defined as a carcinoma confined to the mucosa or submucosa, regardless of the presence or absence of lymph node metastasis[47,76]. The risk of progression from GMPC varies based on established risk factors, including age, Helicobacter pylori infection, anatomic extent and histologic severity of AG/GIM, GIM subtype (complete vs incomplete), family history of gastric cancer, smoking and dietary habits[76]. Advanced-stage gastric cancer has a median survival of less than one year and a five-year survival rate below 20%[77]. In contrast, EGC carries a favorable prognosis, with five-year survival exceeding 90% when detected and treated promptly. Hence the critical importance of early detection and surveillance[28,76]. High-quality upper endoscopy is strongly recommended for identifying GPMC and EGC, with an emphasis on adequate mucosal visualization, careful inspection and detailed anatomical mapping[47,78]. AI may enhance the early detection and characterization of GPMC and EGC, facilitating timely intervention, guiding appropriate surveillance strategies, and ultimately improving patient outcomes.
GMPC: Early detection of GPMC is essential for optimizing clinical outcomes. Helicobacter pylori testing, treatment, and eradication confirmation are recommended in all individuals with GPMC[47]. Routine gastric biopsies are not recommended unless there is suspicion of GPMC[47]. Although AG, GIM and dysplasia are detectable using WLI with or without NBI or LCI, they are frequently missed due to limited endoscopist familiarity, endoscopist fatigue, lesion variability and blind spots. AI tools have shown promise in detecting GPMC in a well-visualized stomach, though current evidence remains insufficient to support routine clinical use[76,79]. By providing real-time assistance, DL models can reduce miss rates, improve lesion detection and enhance lesion characterization[80]. Table 3 provides a summary of studies evaluating AI models for the detection and characterization of GPMC.
| Ref. | Purpose | Design | Training set | Test set | IEE | AI model | Performance |
| Zhang et al[92] | Real-time Det of gastric polyps | RCT | 708 i | 50 i | WLI | SSD-GPNet | mAP: 90%; PDR by > 10% |
| Zhang et al[99] | Det of AG | Retrospective | 3829 i | 1641 i | WLI; i-scan | CNN-CAG | Acc: 94%; Se: 95%; Sp: 94% |
| Xu et al[85] | AI vs exp/nonexp in Det of GPMC | Retrospective multicenter | 5198 i | 1052 i; 98 v | ME-NBI; ME-BLI | ENDOANGEL | AG: Acc: 86% (i); 88% (v); GIM: Acc: 86% (i); 90% (v) |
| Lin et al[90] | Det of AG/GIM | Retrospective multicenter | 2193 i | 273 i | WLI | TResNet | AG: Acc: 96%; AUC: 0.98; Se: 96%; Sp: 96%; GIM: Acc: 98%; AUC: 0.99; Se: 98%; Sp: 97% |
| Watanabe et al[100] | Det of gastric indefinite for dysplasia lesions | Retrospective | 2961 i | 248 i | WLI | SSD + miR148a DNA methylation | AUC: 0.93 (exp) > 0.83 (AI + miR148a) > 0.59 (trainees) |
| Kodaka et al[95] | AG Det and OLGA staging in patients w/Helicobacter pylori infection | Retrospective | 11497 i | 7724 i | WLI | ResNet-50 | AUC: 0.75 (AI + Kyoto) > 0.67 (AI + OLGA) > 0.66 (AI alone) |
| Zhao and Chi[98] | Severity classification of AGAI vs endoscopists | Prospective | 2922 i | 268 v | NBI | UNet | (Increase) DR of mod AG (16% vs 8%) and severe AG (7% vs 3%); (decrease) unnecessary Bx |
| Zhao et al[89] | Det of AGAI vs endoscopists | Prospective case-control | 4175 i | 676 v | NBI | UNet | Acc: 91% vs 72%; Se: 84% vs 63%; Sp: 97% vs 82%; AUC: 0.91 vs 0.74 |
| Yang et al[81] | Det of AG/GIM | Retrospective | 21420 i | 5355 i | WLI; LCI | SE-ResNet | AG: Acc: 97%; Se: 99%; Sp: 95%; GIM: Acc: 99%; Se: 99%; Sp: 99% |
| Li et al[96] | Severity classification of GIM | Retrospective | 837 i | 278 i | NBI; LCI | CDCN | Acc: 84% |
| Shi et al[88] | Det of AG | Retrospective | 6216 i | 600 i; 118 v | WLI | GAM-EfficientNet | i: Acc: 94%; Se: 93%; Sp: 94%; v: Acc: 92%; Se: 96%; Sp: 89% |
| Iwaya et al[87] | Det of GIM and OLGIM staging | Retrospective | 5753 i | 1150 i | HE slides | ResNet-50 | Se: 98%; Sp: 95%; OLGIM stage III/IV classified in 18% |
| Fang et al[86] | AI vs pathologists in Det and grading of AG/GIM | Prospective multicenter | 1745 i | 545 i | Pathology slides | GasMIL | (Increase) pathologists’ performance (AUC: 0.95 vs 0.88) |
| Tao et al[97] | AG Det and risk stratification vs exp | Retrospective | 5856 i | 869 i; 119 v | WLI | UNet ++ ResNet-50 ENDOANGEL | (Increase) Se vs exp i: 93% vs 77%; v: 95% vs 86% |
| Niu et al[94] | GIM grading and OLGIM staging | Retrospective multicenter | 470 i | 333 i | ME-NBI | Faster R-CNN | Pred high-risk stage Acc: 84% |
| Zou et al[83] | Det of Helicobacter pylori infection AI vs endoscopists | Multicenter RCT | 7377 i | 2080 i | WLI | EfficientNet | Acc: 93% vs 76%; Se: 92% vs 79%; Sp: 93% vs 75% |
| Xu et al[84] | AI vs exp/nonexp in Det of AG/GIM | Single center RCT | NA | 1968 v | WLI | ENDOANGEL | (Increase) DR of AG: 23% vs 17%; (Increase) DR of GIM: 14% vs 9% |
Numerous studies, RCTs and meta-analyses have demonstrated the effectiveness of AI in detecting GPMC[81-83]. Multiple AI models, particularly CNNs, have shown high accuracy in detecting Helicobacter pylori infection from upper GI endoscopic images, significantly improving diagnostic precision and potentially reducing biopsies requirements and healthcare costs. Several DL models[84-88] were trained on large datasets from diverse imaging modalities (WLI, LCI, BLI, NBI, and histologic images) to detect AG and GIM. They consistently achieved high diagnostic accuracy, sensitivity, and specificity, often surpassing both expert and nonexpert endoscopists[89-91]. Additionally, AI tools like SSD-GPNet[92] showed high speed and accuracy in detecting gastric polyps, particularly small ones. However, another study has shown that the added benefit of AI in polyp detection may be limited compared to the performance of both junior and senior endoscopists[93].
AI is also used to characterize and stage GPMC, particularly in the context of cancer risk stratification. Several DL models were developed to predict standardized histological classifications such as operative link on GIM assessment and operative link on AG assessment, with performance matching or exceeding that of expert pathologists[86,94,95]. They offered highly accurate interpretation of histopathologic and endoscopic features of GIM and AG, enhancing consistency in diagnosis and enabling non-invasive risk stratification and severity grading without the need for biopsy[87,96-99]. Other studies integrated AI with molecular markers such as miR-148a methylation[100]. This combined histologic and molecular approach improves diagnostic accuracy for indefinite for dysplasia gastric lesions, especially among nonexpert endoscopists and pathologists.
EGC: EGC often presents subtly, with a miss rate of around 10%, particularly among less experienced endoscopists or during low-quality exams[78]. Endoscopic resection has become central to EGC staging and treatment, yet 25%-40% of endoscopic submucosal dissections in expert centers target incurable lesions, underscoring limitations in current selection methods[78]. AI has shown strong potential in improving EGC detection, reducing variability across skill levels and enhancing lesion characterization[28,58]. Table 4 summarizes studies evaluating AI models for the detection and characterization of EGC.
| Ref. | Purpose | Design | Training set | Test set | IEE | AI model | Performance |
| Zhu et al[122] | ID prediction of EGC | Retrospective | 790 i | 203 i | WLI | ResNet-50 | Acc: 89%; Se: 76%; Sp: 96%; AUC: 0.94 |
| Horiuchi et al[112] | AI vs exp in EGC Det | Retrospective | 2570 i | 174 v | ME-NBI | GoogLeNet | Acc: 85%; Se: 95%; Sp: 71%; AUC: 0.87 |
| Wu et al[108] | Det of EGC | Multicenter RCT | NA | 1050 v | WLI | ENDOANGEL | Acc: 85%; Se: 100%; Sp: 84% |
| Wu et al[107] | Det of EGC | RCT | NA | 1812 v | WLI | ENDOANGEL-LD | (Decrease) miss rate (AI 6% vs endoscopists 27% RR = 0.22) |
| Wu et al[117] | AI vs exp in ID and DS of EGC | Prospective multicenter | 1131 i | 100 v | ME-NBI | ENDOANGEL | ID: Acc: 79% vs 64%; DS: Acc: 71% vs 64% |
| Wu et al[116] | Real time Det of EGC | Prospective single center trial | 9824 i | 2010 v | WLI | ENDOANGEL-LD | Acc: 92%; Se: 92%; Sp: 92%; PPV: 25%; NPV: 100% |
| Ueyama et al[103] | Det of EGC | Retrospective | 5574 i | 2300 i | ME-NBI | ResNet-50 | Acc: 99%; Se: 99%; Sp: 98% |
| He et al[115] | Det of EGC | Retrospective multicenter | 4667 i | 4702 i; 187 v | ME-NBI | ENDOANGEL-ME | Acc: 90%; Se: 93%; Sp: 94% |
| Li et al[110] | AI vs exp in EGC Det | Retrospective | 1630 i | 267 i; 77 v | ME-NBI | ENDOANGEL-LA | i: Acc: 89%; Se: 86%; Sp: 92%; v: Acc: 87%; Se: 84%; Sp: 88% |
| Tang et al[114] | Det of EGC | Retrospective multicenter | 13151 i | 1577 i; 20 v | NBI | YOLOv3 | Acc: 93%; AUC: 0.95 |
| Jin et al[113] | Det of EGC AI vs exp | Prospective | 5708 i | 1425 i; 10 v | WLI; NBI | Mask R-CNN | Acc: 90%; Se: 91%; Sp: 89% |
| Gong et al[106] | Real-time Det and ID prediction of EGC | RCT | 5017 i | 2524 v | WLI | CDSS | DR: 96%; ID Acc: 86%; lesion class: Acc: 82% |
| Lee et al[101] | EGC pathological Ch | Retrospective | 4336 i; 153 v | 436 i; 89 v | WLI | ENAD CAD-G | Acc: UH: 90%; SMI: 88%; LVI: 88%; LNM: 93% |
| Chang et al[119] | Classification of EGC | Retrospective real-world data | 21918 i | 6785 i; 296 v | WLI | ENAD CAD-G | i: Acc: EGC: 82%; dysplasia: 88%; v: Acc: EGC: 88%; dysplasia: 91% |
| Zhao et al[102] | Det of EGC LCI vs WLI | Retrospective | 9021 i | 116 v | WLI; LCI | CADe | (Increase) Se: LCI: 94% vs WLI: 79%; Sp: 93% in both |
| Lee et al[101] | Det of EGC | Retrospective | 30000 i | 500 i | WLI | CADe (ALPHAON®) | Acc: 88%; Se: 93%; Sp: 87%; AUC: 0.96 |
| Soong et al[105] | Raman spectroscopy for EGC risk strat | RCT | NA | 25 v | WLI | SPECTRA IMDx™ | Acc: 100%; Se: 80%; Sp: 92% |
| Feng et al[104] | AI vs exp/nonexp in EGC Det | Prospective | 12000 i | 1289 i; 130 v | WLI | DCNN | Se: 97%; Sp: 89%; AUC: 0.93 |
AI systems have significantly improved the detection rates of EGC in both retrospective[101-103] and prospective real-time settings[104-106], with RCTs demonstrating reduced miss rates and blind spots, as well as increased inspection time, compared to routine endoscopy[107,108]. Further, endoscopists assisted by CNNs can outperform nonexpert endos
Additionally, CNN-based models have shown strong performance in EGC characterization to support decision-making in endoscopic resection. They classify lesions while assessing key features such as invasion depth, histologic differentiation and lymphovascular involvement[119,120]. For instance, a meta-analysis evaluating invasion depth prediction, reported high pooled sensitivity, specificity and AUC[111]. Moreover, in a prospective real-time human-machine competition, AI outperformed expert endoscopists in predicting the differentiation status and invasion depth of EGC[117]. However, in retrospective studies[121,122], it enabled novice endoscopists to perform at expert levels in differentiating intramucosal from advanced gastric cancer. Similarly, a video-based classifier outperformed static image models in depth prediction[123]. Finally, advancing beyond standard DL models, explainable AI tools like the double-check support system have integrated lesion classification with real-time image quality assessment, improved interobserver agreement, reduced diagnostic time and provided greater consistency in guiding endoscopic resection decisions[124].
Despite advancements in AI for detecting and characterizing early cancers and precancerous lesions of the upper GI tract, widespread clinical integration remains limited by several challenges. These include the reliance on large, high-quality annotated datasets, retrospective study designs, limited generalizability beyond academic centers, and variability in histopathologic reference standards. Real-time implementation also faces technical and logistical barriers, particularly in community settings. While video-based studies suggest AI can outperform endoscopists, especially in diagnostic accuracy and lesion characterization, large-scale, prospective validation in real world settings is essential to improve AI reliability in routine practice. As AI continues to evolve, its potential to enhance detection, characterization, and reduce variability in clinical practice underscores the importance of integrating these tools into routine endoscopic workflows to improve patients’ outcomes.
Small bowel (SB) tumors represent up to 6% and 3% of all GI neoplasms and malignancies, respectively. The incidence of SB tumors has been increasing over the last few years[125]. The majority of SB neoplasms are malignant and fall into four categories: Adenocarcinomas, neuroendocrine tumors, lymphomas and sarcomas[125]. Premalignant lesions in the SB comprise different types of polyps (adenomatous, hamartomatous and serrated) which could be sporadic or part of a hereditary tumor/polyposis syndrome, or associated with immune mediated inflammatory conditions such as Crohn’s or celiac disease[126]. It is worth mentioning that a considerable number of patients with SB tumors have metastatic disease at diagnosis. Late presentations are ascribed to the nonspecific and variable symptoms, the unreachable site by conventional endoscopy, and the subtle changes in appearance in comparison to the surrounding mucosa in visible lesions[127]. There are different staging systems for different SB tumors, and no consensus on the precise definition of an early SB malignancy has been issued in the literature.
Endoscopic evaluation of the SB had formerly been a longstanding challenge for the gastroenterologist, due to the complex anatomy of the SB and the limited direct visualization achieved with traditional esophagogastroduodenoscopy and colonoscopy. The introduction of video capsule endoscopy (VCE) and DAE has rendered the investigation of suspected SB lesions more feasible. VCE allows real-time complete visualization of the entire intestinal mucosal surface yielding thousands of images. DAE serves as a diagnostic as well as endotherapeutic tool[128]. However, this great stride in SB endoscopy carries multiple challenges alongside. Interpreting the large bulk of images in VCE is time consuming and physicians spend up to 120 minutes reading and reporting a full CE recording. On the other hand, DAE is technically demanding and requires high level of experience and skills to overcome the risk of oversight and distraction, and hence reduce lesion miss rate[129,130].
Multiple studies using AI for the development of CAD to help physicians overcome the hurdles associated with the increased utilization of VCE and DAE have been conducted. The main focus of AI implementation is to alleviate the burden on endoscopists through the reduction of reading time, enhanced real time diagnosis and detection of hard-to-detect lesions, and decrease miss rate[131,132]. Table 5 summarizes the studies.
| Ref. | Endoscopy type | Study design | Training set | Test set | AI model | Performance |
| Barbosa et al[135] | VCE | Retrospective | 104 tumors, 100 normal | 700 tumors, 2300 normal | CNN | Se: 94%; Sp: 93% |
| Li and Meng[213] | VCE | Retrospective | 540 tumors, 540 normal | 60 tumors, 60 normal | ML (SVM) | Acc: 84%; Se: 82%; Sp: 8% |
| Constantinescu et al[138] | VCE | Prospective | 54 videos | 90 images (32 polyps, 58 normal) | ANN | Acc: 98%; Se: 94%; Sp: 91% |
| Liu et al[214] | VCE | Retrospective | 1800 (105 patients, 89 videos) | 89 videos (15 tumors, 74 normal) | ML (SVM) | Se: 98%; Sp: 97% |
| Faghih Dinevari et al[215] | VCE | Retrospective | 300 tumors, 300 normal | 100 tumors, 100 normal | ML (SVM) | Acc: 94%; Se: 94%; Sp: 93% |
| Yuan and Meng[216] | VCE | Retrospective | 1000 polyps, 3000 normal | 200 tumors, 600 normal | DL | Acc: 98% |
| Ding et al[217] | VCE | Retrospective | 158, 235 | 113, 268, 334 | CNN | Se: PPA: 99.9%; PLA: 99.9%; Sp: PPA and PLA: 100% |
| Saito et al[218] | VCE | Retrospective | 30, 584 | 17507 (7507 protruding lesions) | CNN | Se: 91%; Sp: 80% |
| Xie et al[219] | VCE | Retrospective | 148, 357, 922 | 146, 956, 145 | CNN | Se: 99% |
| Cardoso et al[139] | Enteroscopy | Retrospective | 6340 | 507 protruding lesions, 1078 normal | CNN | Acc: 97%; Se: 97%; Sp: 97% |
| Inoue et al[220] | EGD | Retrospective | 531 | 1080 | CNN | Se: 95%; Sp: 87% |
| Ding et al[221] | VCE | Retrospective | 280, 426 | 240 videos | CNN | Acc: 81%; Se: 96%; Sp: 98% |
| Zhu et al[140] | DBE | Retrospective | 8222 | 3148 | CNN | Det model: Se: 92%; Sp: 93%; Class model: Se: 80%-93%; Acc: 86% |
Recently, a systematic review and metanalysis[123] including 12 studies on the use of CAD of SB tumors and polyps during VCE showed a pooled sensitivity and specificity of 89% and 91%, respectively. Another study showed that AI assisted VCE reading outperformed expert gastroenterologists in terms of accuracy and sensitivity and decreased the mean reading time by up to 12-fold[133,134]. The majority of the studies detected protruding lesions defined as any elevation above the surface of the mucosa. This includes polyps, tumors and submucosal lesions. On the other hand, data on the role of AI in DAE is scarce and only few studies have been conducted.
Barbosa et al[135] were the first to propose a computer aided system using CNN for the detection of SB tumors called discrete wavelet transform. A novelty in this system was the characterization of texture alteration by statistical modelling of texture descriptors at different scales and colors. It initially showed a sensitivity of 99% and a specificity of 97%. Few years later, the system was tested in real life setting showing a sensitivity of 94% and specificity of 93%[136]. This change in performance metrics can be explained by overfitting training datasets where the models perform well during training but less well when applied to real world clinical settings[137].
Results from other retrospective studies showed comparable sensitivity and specificity (Table 5). AI scores surpassed those of experienced gastroenterologists predominantly in the sensitivity of detecting protruding lesions/polyps and reading time. For trainees, AI assistance increased the overall performance and accuracy to levels on par with experts.
A prospective study was performed[138] using an ANN based system to detect SB polyps during VCE. Feature extraction included primarily shape, but also color and texture. There was no statistically significant difference between AI and experienced physicians’ readings regarding sensitivity and specificity. Experienced physicians had better positive and negative predictive values (NPV).
Regarding DAE, Cardoso et al[139] designed a deep CNN algorithm for the detection of protruding enteric lesions that included epithelial tumors and subepithelial lesions encountered during single or double balloon enteroscopy (DBE). The sensitivity, specificity and overall accuracy of the model reached 97% each. The positive and NPVs were 95% and 99%, respectively.
Zhu et al[140] built a deep CNN system called ENDOANGEL-DBE for SB lesions in DBE. It is composed of detection and classification models. The model classified lesions into 4 categories: Diverticula, erosions/ulcers, angioectasia and protruding lesions. The sensitivity for the detection of protruding lesions was 93%. The total accuracy of ENDOANGEL-DBE was superior to endoscopists who recorded 77% (P < 0.01). The overall performance of ENDOANGEL-DBE was comparable to the experts in lesion classification (85 % and 81%; P = 0.253).
In conclusion, AI assisted SB endoscopy was superior in terms of accuracy and sensitivity for the detection of polyps and protruding lesions (including tumors) even when compared to expert endoscopists. Another significant advantage is VCE reading time reduction, a tedious time-consuming task for the gastroenterologist. Among all the studies investigating the detection of polyps and protruding lesions during VCE, there hasn’t been a clear histopathological classification of these lesions. This significant limitation makes the extent through which AI contributes to the detection of premalignant lesions in the SB unclear. The clinical relevance, effect on morbidity, mortality and cost effectiveness of these advancements are yet to be investigated with long term studies.
Future research might address the role of AI assistance in corelating the anatomical localization during DAE with findings on VCE, paving the way for endoscopic management of these lesions.
Colorectal cancer (CRC) is the third most common cancer worldwide and ranks second in cancer related mortality[141]. Colonoscopy is the gold standard for screening and prevention of CRC as it allows resection of precancerous lesions[142]. Premalignant lesions may be missed during colonoscopy; miss rates for adenomas, serrated lesions and advanced adenomas were 26%, 27% and 9%, respectively in a large meta-analysis[143]. This was shown to account for more than 50% of interval cancers, which represent 10% of all CRC[144,145]. Furthermore, lateral spreading tumors (LSTs) have a high malignant potential and can be elusive and easily missed during colonoscopy. LST non-granular type, including sessile serrated lesions (SSLs), are the most difficult to detect and pose the risk of post-colonoscopy interval cancer[146]. A 1% increase in the detection of right-sided serrated polyps resulted in a substantial 7% reduction in the risk of interval CRC[147]. Adenoma detection rate (ADR) is recognized as the most reliable quality indicator in screening colonoscopy, and a previous study revealed a 3% decrease in the risk of CRC for every 1% increase in ADR[148].
AI can assist in colonoscopy by improving the detection of premalignant lesions, enhancing the characterization of premalignant and early malignant lesions and by decreasing inter-endoscopists’ variability. CADe is the most developed AI system for identifying polyps.
Effect of CADe assisted colonoscopy on ADR: Multiple prospective and retrospective studies evaluated the use of CADe in colonoscopy (Table 6). Maroulis et al[145] described an innovative detection system, colorectal lesions detector, that aids in detecting polyps by combining feature extraction and classification algorithms. The system’s accuracy in lesion detection exceeded 95%. An intelligent model that incorporates color-texture analysis methods and support vector machine’s algorithms into a sound pattern recognition framework could analyze low-quality videos with an accuracy of more than 94%[149].
| Ref. | Study design | Number | Country | Outcomes |
| Park et al[222] | Retrospective ex vivo | 562 images | United States | Se: 86%; Sp: 85% |
| Billah et al[223] | Retrospective ex vivo | 14000 images | Bangladesh | Se: 99%; Sp: 99% |
| Lequan et al[224] | Retrospective ex vivo | 38 videos | China | Se: 71%; PPV: 88% |
| Zhang et al[225] | Retrospective ex vivo | 2442 images | China | Se: 98%; PPV: 99%; Acc: 86% |
| Misawa et al[226] | Retrospective ex vivo | 546 videos | Japan | Per-frame Se: 90%; Sp: 63%; Acc: 76%; Per-polyp Se: 94% |
| Urban et al[150] | Retrospective ex vivo | 63559 images, 40 videos | United States | Se: 90%; Acc: 96% |
| Yamada et al[227] | Retrospective ex vivo | 144823 video images | Japan | Se: 97%; Sp: 99% |
| Klare et al[228] | Prospective in vivo | 55 colonoscopies | Germany | Per-polyp Se: 75%; ADR: 29%; PDR: 51% (31% and 56% in endoscopist) |
| Ozawa et al[229] | Ex vivo | 23495 images | Japan | Se: 92%; PPV: 93%; Acc: 85% |
The first real-time CNNs based model was tested on more than 8000 hand-labeled images with more than 4000 polyps and showed an accuracy of more than 96% for polyp detection. It outperformed expert endoscopists with relatively low false positive rate (around 5%)[150]. Other studies had variable sensitivity and lower accuracy than that study (Table 6).
Multiple RCTs have assessed the performance of CADe compared to standard colonoscopy (Table 7). CADe improved ADR in most studies but had no effect in others when compared to standard colonoscopy. Initial trials showed that CADe improved ADR compared to standard colonoscopy when baseline ADR was rather low[151]. These findings were confirmed by a group with higher baseline ADR who used GI genius, the first Food and Drug Administration-approved CADe system[152]. To note that ADR was significantly higher for diminutive (≤ 5 mm) and small size (6 mm-9 mm) adenomas. The same group showed that AI also improves ADR as well as adenoma per colonoscopy (APC) performed by non-expert endoscopists[153]. More recent RCTs in the East demonstrated high baseline ADR in control arms that showed further increase with CADe systems[154,155].
| Ref. | Study design | Number of patients | Country | Outcomes (AI vs control) |
| Wang et al[151] | Single center RCT | 1058 | China | ADR: 29% vs 20% |
| Wang et al[156] | Single center RCT (double blind) | 962 | China | ADR: 34% vs 28% |
| Gong et al[157] | Single center RCT | 704 | China | ADR: 16% vs 8% |
| Su et al[158] | Single center RCT | 659 | China | ADR: 29% vs 17% |
| Liu et al[159] | RCT | 1026 | China | ADR: 39% vs 23% |
| Wang et al[175] | Tandem RCT | 369 | China | AMR: 14% vs 40% |
| Repici et al[152] | Multicenter RCT | 685 | Italy | ADR: 55% vs 40% |
| Kamba et al[176] | Tandem RCT | 358 | Japan | AMR: 14% vs 37% |
| Glissen Brown et al[177] | Multicenter tandem RCT | 223 | United States | AMR: 20% vs 31% |
| Repici et al[153] | RCT | 660 | Italy, Switzerland | ADR: 53% vs 45% |
| Wallace et al[178] | Tandem RCT | 240 | Italy, United Kingdom, United States | AMR: 16% vs 32% |
| Shaukat et al[166] | RCT | 1359 | United States | APC: 1 vs 0.8 |
| Xu et al[230] | Multicenter RCT | 3059 | China | ADR: 40% vs 32% |
| Gimeno-García et al[231] | Single center RCT | 370 | Spain | ADR: 55% vs 41% |
| Nakashima et al[154] | Single center RCT | 415 | Japan | ADR: 59% vs 48% |
| Karsenti et al[232] | Single center RCT | 2592 | France | ADR: 38% vs 34% |
| Lau et al[155] | Single center RCT | 766 | China | ADR: 58% vs 45% |
| Maas et al[233] | Multicenter RCT | 916 | Germany, Netherlands, United States, Israel | ADR: AI 37% vs 30% |
| Thiruvengadam et al[234] | Single center RCT | 1100 | United States | ADR: 43% vs 34% |
| Park et al[235] | Multicenter RCT | 805 | Korea | ADR: 35% vs 28% |
Interestingly, a double-blind RCT using a sham system (CADe-DB trial) showed that CADe significantly increased ADR[156]. Other RCTs showed significant increase in ADR compared to controls[157-159]. In addition, ENDOANGEL model significantly increased the detection of advanced adenomas (> 10 mm)[157].
An analysis of RCTs assessing deep CNN-based CADe in real-time colonoscopy and including a total of 4962 patients showed a higher pooled ADR in the CADe group, but the pooled rates of relative risk for advanced ADR and sessile serrated ADR were similar to controls[160]. Furthermore, a meta-analysis that studied 10 different CADe systems showed an increase in ADR of 24% but not for advanced adenomas and SSLs. It also showed higher rates of unnecessary removal of non-neoplastic polyps[161]. Another meta-analysis showed an increase in ADR (45% vs 37%) and a mild increase in the detection of advanced colorectal neoplasia (12.7% vs 11.5%)[162].
A network meta-analysis showed that AI increased ADR compared with both mucosal visualization tools and chromoendoscopy, but there was no increase in the detection of SSLs[163]. Another network meta-analysis showed improvement in ADR with AI compared to other endoscopic interventions aimed at increasing ADR (such as distal attachment devices, dye-based/virtual chromoendoscopy, water-based techniques, and balloon-assisted devices)[164]. Finally, a recent RCT comparing LCI colonoscopy to LCA (LCI + AI) colonoscopy found higher ADR and APC in LCA group, particularly for small and diminutive polyps[165].
By contrast, several recent RCTs and a retrospective study failed to demonstrate that AI-assisted colonoscopy improves ADR compared to standard colonoscopy[166-170]. Possible explanations for this include performance of colonoscopies by experienced endoscopists, a higher baseline ADR and a shorter procedure time in the AI group. Moreover, the use of AI in populations with low risk of adenoma can minimize an increase in ADR. Also adding AI in bowel cancer screening programs where expert colonoscopists usually use distal attachment devices may not leave room for improvement in ADR. As such, results may differ between geographic areas due to differences in populations risk and endoscopists behaviors. Another retrospective well-designed pragmatic implementation trial also showed negative results for CADe during a 3-month trial[171]. Quality of mucosal exposure by endoscopists or their behavior regarding CADe could have altered the results in this real-world trial. A recent RCT from Kuwait, one of the few from the Middle East, found a non-significant increase in the ADR (relative risk = 1.26, 0.80-2.00) in the CADe group[172]. The sample size was too small to build solid conclusions. Finally, a recent large United States multicenter RCT revealed a 17% higher APC detection rate in AI-assisted colonoscopy compared with conventional high-definition colonoscopy. Nevertheless, no significant differences were found in adenoma, advanced adenoma and SSL detection rates[173]. The exclusion of trainees and less experienced endoscopists might have contributed to the latter findings.
It’s important to mention the impact of Hawthorne effect which implies that endoscopists might improve their performance when they know they are being monitored, thus reducing the effect of AI. Also, the absence of uniformity in AI systems between different studies may alter ADR results. Lastly, excessive false alarms can desensitize endoscopists who might ignore AI. Further large scale, multicenter and multinational real-world studies are needed to compare the effect of combining CADe with mucosal exposure techniques on ADR in less experienced endoscopists.
In summary, CADe has shown encouraging results in controlled trials, mainly in the detection of diminutive and small polyps. Its role in a real-world setting remains uncertain. The adenoma miss rate (AMR) can be more sensitive than the ADR to assess differences in lesion detectability even among endoscopists with a high ADR[174]. Multiple tandem RCTs comparing CADe with standard colonoscopy showed a significantly lower AMR in the CADe group[175-178], and this was significant only for diminutive and small polyps. Some of these RCTs also showed a significant decrease in SSLs miss rate[176,177]. A systematic review and meta-analysis of 14 RCTs suggested that CADe assisted colonoscopy was associated with a 65% reduction in AMR, a 78% reduction in the SSL miss rate, a 52% increase in ADR and a 93% increase in the number of adenomas > 10 mm[179].
Effect of CADe assisted colonoscopy on the detection of sessile serrated adenomas and LST: Ahmad et al[180] showed that their AI system was able to detect 80% of “subtle lesions”, outperforming experienced and less experienced endoscopists who detected only 37% and 11% of them, respectively. Lin et al[181] recently studied the detection performance of a deep neural network algorithm on LSTs. The accuracy of the model in identifying LSTs exceeded 99%, significantly better than that of novice endoscopists and similar to that of expert ones.
Zhou et al[182] noticed that the per-frame sensitivity for non-granular LSTs and small sessile serrated adenomas/polyps should be further improved. A recent 3-arm RCT showed that AI could increase the SSL and advanced adenoma detection rates when compared with standard high-definition colonoscopy. Further, the combination of endo cuff with AI further improves SSLs detection rate compared to AI alone[183].
In summary, most trials did not show an increase in the detection of advanced adenomas and SSLs. In recent studies, CADe seems promising for the detection of subtle and advanced lesions, but data is still limited, and further studies are needed.
While the real-time prediction of histology was based on virtual chromoendoscopy that enhances mucosal and vascular patterns, there has been growing interest in developing CADx systems to better characterize colonic polyps and differentiate them into neoplastic and non-neoplastic. In this context, the American Society for Gastrointestinal Endoscopy (ASGE) published standards for the performance thresholds required for any endoscopic technology to support a resect and discard (90% or greater agreement with histopathology for post-polypectomy surveillance intervals) and diagnose and leave (90% or greater NPV for adenomatous histology) strategy for diminutive polyps[184]. To note that these thresholds apply only when the endoscopic technology is used with “high confidence”. Modern CADx models utilize CNN with standard NBI or WLI and with high performance (Tables 8 and 9).
| Ref. | Country | AI system | Number | WLI/IEE | Primary outcomes | Results |
| Tischendorf et al[236] | Germany | SVM | 209 polyps | Magnifying NBI | Adenomas vs non adenomas | Se: 90%; Sp: 70%; Acc: 85% |
| Takemura et al[237] | Japan | SVM | 1519 polyps, 371 images | Magnifying NBI | Neoplastic vs non neoplastic | Se: 97%; Sp: 97%; Acc: 97% |
| Misawa et al[195] | Japan | EndoBRAIN | 1179 images | Endocytoscopy, NBI | Prediction of histology | Se: 84%; Sp: 97%; Acc: 90%; PPV: 98%; NPV: 82% |
| Komeda et al[188] | Japan | CNN | 1800 images, 10 videos | WLI | Adenomas vs non adenomas | Acc: 75% |
| Byrne et al[185] | Canada | DCNN | 117000 images, 388 videos | NBI | Adenomas vs non adenomas (DP) | Se: 98%; Sp: 83%; PPV: 90%; NPV: 97%; Acc: 94% |
| Chen et al[238] | China | DCNN | 2441 images | NBI | Neoplastic vs hyperplastic (DP) | Se: 96%; Sp: 78%; PPV: 89%; NPV: 90%; Acc: 90% |
| Sánchez-Montes et al[189] | Spain | ML | 225 polyps | HD WLI | Adenomas vs non adenomas (DP) | Se: 92%; NPV: 91%; Acc: 90%; DP: NPV: 96%; Acc: 87% |
| Song et al[190] | Korea | ENAD | 12480 images, 451 polyps | NBI | Adenomas vs SSLs | Se: 82% vs 84%; Sp: 93% vs 88%; Acc: 81% vs 82% |
| Kudo et al[196] | Japan | EndoBRAIN-EYE | 69142 images | Endocytoscopy, NBI | Neoplastic vs non neoplastic | Se: 96%; Sp: 94%; PPV: 96%; NPV: 94%; Acc: 96% |
| Zachariah et al[192] | United States | CAD-EYE | 5912 images | WLI, NBI | Diminutive adenomas vs SSLs/hyperplastic polyps | Se: 96%; Sp: 90%; PPV: 94%; NPV: 93%; Acc: 94% |
| Jin et al[191] | Korea | DCNN | 2450 polyps | NBI | Adenomas vs non adenomas (DP) | Se: 83%; Sp: 91%; Acc: 87% |
| Ozawa et al[229] | Japan | DCNN (SSD) | 27508 images | WLI, NBI | Adenomas vs non adenomas (DP) | WLI: NPV: 85%; NBI: NPV: 91% |
| Ref. | Country | AI system | Number | WLI/IEE | Primary outcomes | Results |
| Gross et al[239] | Germany | SVM | 434 small polyps (< 10 mm) | Magnifying NBI | Adenomas vs non adenomas (small polyps) | Se: 95%; Sp: 90%; PPV: 93%; NPV: 92%; Acc: 93% |
| Kominami et al[240] | Japan | SVM | 1262 polyps, 118 images | Magnifying NBI | Adenomas vs non adenomas (small polyps) | Se: 93%; Sp: 95%; PPV: 95%; NPV: 93%; Acc: 97% |
| Mori et al[194] | Japan | EndoBRAIN | 61952 images, 466 polyps | Endocytoscopy, NBI | Adenomas vs non adenomas (DP) | Se: 95%; Sp: 92%; PPV: 96%; NPV: 96%; Acc: 98% |
| Barua et al[208] | Norway, United Kingdom, Japan | EndoBRAIN | 892 polyps | WLI, NBI, magnification | Neoplastic vs non neoplastic polyps (DP) | Se: 90%; Sp: 86% |
| Hassan et al[242] | Italy | GI genius | 544 polyps | NBI, BLI | Adenomas vs non adenomas (DP) | NPV: 98%; Se: 82%; Sp: 93%; Acc: 92% |
| Minegishi et al[200] | Japan | EndoBRAIN | 395 polyps | NBI | Adenomas vs non adenomas (DP) | NPV: 94%; Se: 94%; Sp: 63%; Acc: 86% |
| Rondonotti et al[209] | Italy | CAD EYE | 596 polyps | WLI | Adenomas vs non adenomas (DP) | NPV: 91%; Se: 89%; Sp: 88%; Acc: 88% |
| Li et al[241] | Singapore (multicenter) | CAD EYE | 661 polyps | WLI | Neoplastic vs non neoplastic polyps | Se: 62%; Acc: 72% |
| Hassan et al[242] | Italy | CAD EYE; GI genius (head-to-head comparison) | 319 polyps | WLI, BLI | Adenomas vs non adenomas (DP) | NPV: 97% vs 98%; Se: 82% vs 86%; Sp: 92% vs 94% |
| Houwen et al[201] | Netherlands (multicenter) + Spain (1 center) | POLAR | 423 polyps | NBI | Neoplastic (adenomas and SSLs) vs non neoplastic | Se: 89%; Sp: 38%; Acc: 79% |
| Rex et al[243] | United States (multicenter) | GI genius | 2695 polyps | WLI, NBI | Adenomas vs non adenomas | Se: 91%; Sp: 65% |
Effect of CADx on the characterization of polyps into neoplastic vs non-neoplastic: A deep CNN model was evaluated on 106 diminutive polyps. Its accuracy was 94%, the sensitivity for identification of adenomas was 98%, the specificity was 83%, the NPV was 97%, and the positive predictive value (PPV) was 90%. This was the first system to exceed the ASGE threshold requirements[185]. The findings were corroborated by two other studies[186,187]. However, other studies testing CADx with white light failed to demonstrate high accuracy[188,189].
Some studies on CADx assessed its performance depending on the level of experience of endoscopists. A DL CADx model was developed to classify lesions into 3 histological groups: Serrated polyp, benign adenoma/mucosal or superficial submucosal cancer, and deep submucosal cancer. The overall diagnostic accuracy of the model was higher than that of trainees and comparable to experts in NBI. The performance of the trainees improved with CADx assistance[190]. These results were corroborated by another study showing that the overall accuracy of novice endoscopists increased with CADx assistance[191], and by another study that used a CNN-based optical pathology model with both NBI and white light[192].
In another study, a CAD software was developed using densely connected CNNs based on the modified Sano classification. The CAD software was trained with NBI images and tested with separate sets of images using NBI and BLI. The CAD software had an AUC of 94.3% for the internal set and 84.5% and 90.3% for the external sets (NBI and BLI, respectively). This model is unique since it achieved AUCs comparable with experts and similar results with NBI and BLI, although BLI was not part of the training set[193].
In addition, many authors were interested in combining AI with endocytoscopy (EC) for polyp characterization. CAD-EC systems could be helpful for untrained endoscopists. The model achieved high sensitivity and accuracy in diagnosing neoplastic changes, reaching those of expert endoscopists and significantly outperforming those of trainees[194]. Furthermore, EC exceeded the proposed thresholds required for “diagnose and leave’’ strategy and had better outcomes than trainees and experts[186,195,196].
Two recent meta-analyses aimed to explore the benefits and harms of CADx assistance in the optical diagnosis of colorectal polyps[197,198]. The outcome measures for benefit and for harm with use of CADx were the proportion of polyps predicted to be nonneoplastic that could be left in place and the proportion of neoplastic polyps that would be left in situ due to an incorrect diagnosis. In the first meta-analysis, CADx did not show harm or benefit in the “diagnose and leave’’ strategy. The second meta-analysis did not show significant difference for the proportion of polyps that would not be removed, but the proportion of incorrectly predicted neoplastic polyps was lower with the CADx-assisted strategy than with the CADx-unassisted strategy.
It’s worth mentioning that to date there’s one RCT about CADx, comparing autonomous AI to AI-assisted human (AI-H) optical diagnosis of colorectal polyps. Autonomous AI had significantly higher agreement with pathology-based surveillance intervals compared to AI-H (91.5% vs 82.1%, P = 0.016)[199].
Few studies evaluated CADx in the optical diagnosis of SSLs. Minegishi et al[200] prospectively evaluated the performance of an NBI-CAD system in a real-world setting. Its sensitivity for neoplastic lesions, including SSL, was 93%, while the specificity was 61.5% and NPV was 94%. Concordance rates with American, European, and Japanese guidelines were 90%, 93%, and 98%, respectively. A subgroup-analysis including polyps > 10 mm showed sensitivity and specificity for SSLs of 81% and 62%, respectively. A multicenter prospective study in Spain and The Netherlands showed that their CADx system had a low performance in distinguishing neoplastic (adenomas and SSLs) from non-neoplastic lesions (hyperplastic polyps)[201]. In conclusion, most CADx systems classify polyps as either adenomatous or non-adenomatous but still cannot accurately differentiate SSLs from hyperplastic polyps[202].
CADx in the characterization of early CRC: CADx was also investigated in the prediction of early CRC depth, mainly in Japan. Takeda et al[203] developed an EC coupled CADx system that showed an accuracy of 94% in diagnosing invasive cancer, which increased to 99% in high-confidence diagnosis group. A CADx system with magnifying NBI diagnosed invasive cancers with 84% sensitivity, 83% specificity, 53% PPV, 96% NPV, and 83% accuracy[204]. Another model that used white light endoscopy only for the prediction of depth of invasion of CRC had an accuracy of 90%, similar to that in experts, and it assisted trainees to reach experts’ level[205].
In summary, CADe may increase the detection of diminutive polyps and non-advanced adenomas[206,207], and it remains unclear if AI can increase detection of subtle lesions and advanced adenomas such as SSLs and LSTs. Therefore, the exact role of CADe in decreasing the incidence of interval CRC and its related mortality is yet to be demonstrated. Further, CADx systems did not clearly show benefits over endoscopists’ optical diagnosis. For the leave-in situ strategy, CADx did not increase the proportion of non-neoplastic polyps identified for which polypectomy could be avoided. Furthermore, it slightly decreased the misclassification of neoplastic polyps as non-neoplastic. CADx may be beneficial for less experienced endoscopists in optical histology diagnosis. Nevertheless, for expert endoscopists, it increased the proportion of polyps where the diagnosis was made with high confidence[208,209].
Man-AI collaboration is crucial and endoscopists trust their CADe system more when it generates less false-positive results, avoiding unnecessary polypectomies. However, determining a false-positive with CADe model is relatively less challenging than identifying a misdiagnosis with CADx, especially when it comes to novice endoscopists[210,211]. Also, endoscopists must ensure optimal mucosal exposure to the AI system to function effectively. More large-scale RCTs and real-world prospective studies are needed and should focus on patients’ outcomes rather than just polyp detection.
In conclusion, AI is a promising technology that can revolutionize colonoscopy. RCTs and meta-analyses have demonstrated statistically significant benefits with AI regarding ADR, APC, AMR and SSLs miss rate, although these outcomes have been inconsistent. Moreover, AI might be useful in real time optical diagnosis of diminutive colorectal polyps. Clinical practice guidelines on CADe-assisted colonoscopy, published in April 2025 by the American Gastroenterological Association[212] don’t recommend for or against the use of CADe systems in colonoscopy. They state that CADe systems predominantly increase the detection of low-risk polyps, which may result in more frequent and costly follow-up colonoscopies with uncertain benefits in preventing cancer.
AI provides several advantages in the detection and characterization of premalignant and early malignant lesions of the GI tract. Those include improved lesion detection particularly for trainees, enhanced optical diagnosis and disease pattern recognition, improved decision making, reading time-reduction and ability to spend more time with patients. The anticipated benefit of implementing these advancements is to eventually aid in cancer prevention and consequently reduce the cost of healthcare due to decreased treatment expenses. This end result is achieved when increased detection through CADe and surveillance endoscopies are complemented with CADx. This approach allows for less unnecessary biopsies, a resect and discard or a leave in situ strategy when appropriate, or definitive treatment allocation as indicated. Despite these benefits, the growing integration of AI technologies in the medical field is accompanied by multiple challenges including data standardization, overfitting and generalizability of AI models, as well as a multitude of legal and ethical issues. Furthermore, cost effectiveness of AI models, and physicians’ attitudes towards adopting such technology are potential hurdles. When it comes to AI implementation in gastroenterology training programs, the real question is not about using AI or not, but rather about how and when to use it. AI gives trainees an opportunity and a new modality of real-time teaching assistance and an instant feedback system. It has shown promising results so far. On the other hand, overreliance on AI, cognitive overload and distractibility are concerns that should be addressed when AI-assistance is applied in a fellowship program. Some studies proposed a model for AI implementation process and timing during fellowship training. Fellowship programs should ensure that trainees are aware of how AI functions, the ethics of using AI, as well as the potential harm. They should provide feedback on their performance with frequent reassessment. We believe that for AI to augment medical practice sustainably, different parties including medical professionals, policymakers, AI researchers and stakeholders should be involved in the construction and implementation process. AI can reshape the medical landscape when it is positioned along a suitable infrastructure primed for this change with adequate training and resources.
Future research should focus on direct comparisons between CADe and CADx systems on one hand, and expert endoscopists on the other hand, particularly in real-time settings. This could lead to better understanding of their complementary roles and limitations. Outcome-based studies evaluating the long-term clinical impact of AI-assisted endoscopy on cancer detection rates, patient survival, and cost-effectiveness are also essential, although they require extended follow-up and large-scale multicenter collaboration. Additionally, the integration of AI across various imaging modalities, the development of standardized performance benchmarks, and the validation of AI tools in diverse patient populations and clinical environments remain critical areas for future investigation. Exploring AI’s potential in training and decision support may also help bridge the gap between novice and expert performance, enhancing overall endoscopic practice.
The field of gastroenterology and specifically GI endoscopy provided an appealing foundation for AI-assisted image analysis research. The evidence from these studies varies between the upper GI tract, SB and colon. Evidence supporting AI-assisted detection and characterization of premalignant and early cancerous lesions in the esophagus and stomach is the most consistent and promising. For the colon, AI was particularly superior in the detection of diminutive lesions. The long-term outcomes from this increase in sensitivity is yet to be explored. On the other hand, there is insufficient evidence on the role of AI-assisted SB endoscopy and further high-quality studies are required.
| 1. | Yang YC, Islam SU, Noor A, Khan S, Afsar W, Nazir S. Influential Usage of Big Data and Artificial Intelligence in Healthcare. Comput Math Methods Med. 2021;2021:5812499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 2. | Enslin S, Kaul V. Past, Present, and Future: A History Lesson in Artificial Intelligence. Gastrointest Endosc Clin N Am. 2025;35:265-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 3. | Sarker IH. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN Comput Sci. 2021;2:160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 237] [Cited by in RCA: 933] [Article Influence: 186.6] [Reference Citation Analysis (1)] |
| 4. | Mahesh B. Machine Learning Algorithms - A Review. Int J Sci Res. 2020;9:381-386. [DOI] [Full Text] |
| 5. | Pugliese R, Regondi S, Marini R. Machine learning-based approach: global trends, research directions, and regulatory standpoints. Data Sci Manag. 2021;4:19-29. [DOI] [Full Text] |
| 6. | Oyewole GJ, Thopil GA. Data clustering: application and trends. Artif Intell Rev. 2023;56:6439-6475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 7. | Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18:1527-1554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9547] [Cited by in RCA: 3242] [Article Influence: 162.1] [Reference Citation Analysis (0)] |
| 8. | Xin Y, Kong L, Liu Z, Chen Y, Li Y, Zhu H, Gao M, Hou H, Wang C. Machine Learning and Deep Learning Methods for Cybersecurity. IEEE Access. 2018;6:35365-35381. [DOI] [Full Text] |
| 9. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36149] [Cited by in RCA: 21138] [Article Influence: 1921.6] [Reference Citation Analysis (2)] |
| 10. | Krogh A. What are artificial neural networks? Nat Biotechnol. 2008;26:195-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 235] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 11. | Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, Santamaría J, Fadhel MA, Al-Amidie M, Farhan L. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. 2021;8:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3068] [Cited by in RCA: 1318] [Article Influence: 263.6] [Reference Citation Analysis (2)] |
| 12. | Dhillon A, Verma GK. Convolutional neural network: a review of models, methodologies and applications to object detection. Prog Artif Intell. 2020;9:85-112. [RCA] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 222] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 13. | Sarker IH. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput Sci. 2021;2:420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 484] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 14. | Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2376] [Cited by in RCA: 3568] [Article Influence: 509.7] [Reference Citation Analysis (5)] |
| 15. | Koleth G, Emmanue J, Spadaccini M, Mascagni P, Khalaf K, Mori Y, Antonelli G, Maselli R, Carrara S, Galtieri PA, Pellegatta G, Fugazza A, Anderloni A, Selvaggio C, Bretthauer M, Aghemo A, Spinelli A, Savevski V, Sharma P, Hassan C, Repici A. Artificial intelligence in gastroenterology: Where are we heading? Endosc Int Open. 2022;10:E1474-E1480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 16. | Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, Bray F. Cancer statistics for the year 2020: An overview. Int J Cancer. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2411] [Cited by in RCA: 3305] [Article Influence: 661.0] [Reference Citation Analysis (9)] |
| 17. | Ahn SB, Han DS, Bae JH, Byun TJ, Kim JP, Eun CS. The Miss Rate for Colorectal Adenoma Determined by Quality-Adjusted, Back-to-Back Colonoscopies. Gut Liver. 2012;6:64-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 149] [Article Influence: 10.6] [Reference Citation Analysis (1)] |
| 18. | Januszewicz W, Witczak K, Wieszczy P, Socha M, Turkot MH, Wojciechowska U, Didkowska J, Kaminski MF, Regula J. Prevalence and risk factors of upper gastrointestinal cancers missed during endoscopy: a nationwide registry-based study. Endoscopy. 2022;54:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 19. | Byrne MF, Shahidi N, Rex DK. Will Computer-Aided Detection and Diagnosis Revolutionize Colonoscopy? Gastroenterology. 2017;153:1460-1464.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 20. | Roshan A, Byrne MF. Artificial intelligence in colorectal cancer screening. CMAJ. 2022;194:E1481-E1484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 21. | Sharma P, Hassan C. Artificial Intelligence and Deep Learning for Upper Gastrointestinal Neoplasia. Gastroenterology. 2022;162:1056-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 22. | Tokat M, van Tilburg L, Koch AD, Spaander MCW. Artificial Intelligence in Upper Gastrointestinal Endoscopy. Dig Dis. 2022;40:395-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Holt NM, Byrne MF. The Role of Artificial Intelligence and Big Data for Gastrointestinal Disease. Gastrointest Endosc Clin N Am. 2025;35:291-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Adachi K, Ebisutani Y, Matsubara Y, Okimoto E, Ishimura N, Ishihara S. Effectiveness of Artificial Intelligence in Screening Esophagogastroduodenoscopy. Cureus. 2025;17:e79935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 25. | Morgan E, Soerjomataram I, Rumgay H, Coleman HG, Thrift AP, Vignat J, Laversanne M, Ferlay J, Arnold M. The Global Landscape of Esophageal Squamous Cell Carcinoma and Esophageal Adenocarcinoma Incidence and Mortality in 2020 and Projections to 2040: New Estimates From GLOBOCAN 2020. Gastroenterology. 2022;163:649-658.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 796] [Cited by in RCA: 742] [Article Influence: 185.5] [Reference Citation Analysis (1)] |
| 26. | Parlar K, Cakir M, Ozer O, Sharma P. Future of image enhanced endoscopy of esophageal adenocarcinoma. Clin Endosc. 2025;58:503-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 27. | Shaheen NJ, Falk GW, Iyer PG, Souza RF, Yadlapati RH, Sauer BG, Wani S. Diagnosis and Management of Barrett's Esophagus: An Updated ACG Guideline. Am J Gastroenterol. 2022;117:559-587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 405] [Cited by in RCA: 332] [Article Influence: 83.0] [Reference Citation Analysis (0)] |
| 28. | Ebigbo A, Messmann H, Lee SH. Artificial Intelligence Applications in Image-Based Diagnosis of Early Esophageal and Gastric Neoplasms. Gastroenterology. 2025;169:396-415.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 29. | Jong MR, Jaspers TJM, Kusters CHJ, Jukema JB, van Eijck van Heslinga RAH, Fockens KN, Boers TGW, Visser LS, van der Putten JA, van der Sommen F, de With PH, de Groof AJ, Bergman JJ; BONS‐AI consortium. Challenges in Implementing Endoscopic Artificial Intelligence: The Impact of Real-World Imaging Conditions on Barrett's Neoplasia Detection. United European Gastroenterol J. 2025;13:929-937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 30. | Tsai MC, Yen HH, Tsai HY, Huang YK, Luo YS, Kornelius E, Sung WW, Lin CC, Tseng MH, Wang CC. Artificial intelligence system for the detection of Barrett's esophagus. World J Gastroenterol. 2023;29:6198-6207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 31. | Struyvenberg MR, de Groof AJ, van der Putten J, van der Sommen F, Baldaque-Silva F, Omae M, Pouw R, Bisschops R, Vieth M, Schoon EJ, Curvers WL, de With PH, Bergman JJ. A computer-assisted algorithm for narrow-band imaging-based tissue characterization in Barrett's esophagus. Gastrointest Endosc. 2021;93:89-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (2)] |
| 32. | Takeda T, Asaoka D, Ueyama H, Abe D, Suzuki M, Inami Y, Uemura Y, Yamamoto M, Iwano T, Uchida R, Utsunomiya H, Oki S, Suzuki N, Ikeda A, Akazawa Y, Matsumoto K, Ueda K, Hojo M, Nojiri S, Tada T, Nagahara A. Development of an Artificial Intelligence Diagnostic System Using Linked Color Imaging for Barrett's Esophagus. J Clin Med. 2024;13:1990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 33. | Ali S, Bailey A, Ash S, Haghighat M; TGU Investigators, Leedham SJ, Lu X, East JE, Rittscher J, Braden B. A Pilot Study on Automatic Three-Dimensional Quantification of Barrett's Esophagus for Risk Stratification and Therapy Monitoring. Gastroenterology. 2021;161:865-878.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (1)] |
| 34. | Meinikheim M, Mendel R, Palm C, Probst A, Muzalyova A, Scheppach MW, Nagl S, Schnoy E, Römmele C, Schulz DAH, Schlottmann J, Prinz F, Rauber D, Rückert T, Matsumura T, Fernández-Esparrach G, Parsa N, Byrne MF, Messmann H, Ebigbo A. Influence of artificial intelligence on the diagnostic performance of endoscopists in the assessment of Barrett's esophagus: a tandem randomized and video trial. Endoscopy. 2024;56:641-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 35. | Fockens KN, Jong MR, Jukema JB, Boers TGW, Kusters CHJ, van der Putten JA, Pouw RE, Duits LC, Montazeri NSM, van Munster SN, Weusten BLAM, Alvarez Herrero L, Houben MHMG, Nagengast WB, Westerhof J, Alkhalaf A, Mallant-Hent RC, Scholten P, Ragunath K, Seewald S, Elbe P, Baldaque-Silva F, Barret M, Ortiz Fernández-Sordo J, Villarejo GM, Pech O, Beyna T, van der Sommen F, de With PH, de Groof AJ, Bergman JJ; Barrett's Oesophagus Imaging for Artificial Intelligence (BONS-AI) consortium. A deep learning system for detection of early Barrett's neoplasia: a model development and validation study. Lancet Digit Health. 2023;5:e905-e916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 36. | Fockens KN, Jukema JB, Boers T, Jong MR, van der Putten JA, Pouw RE, Weusten BLAM, Alvarez Herrero L, Houben MHMG, Nagengast WB, Westerhof J, Alkhalaf A, Mallant R, Ragunath K, Seewald S, Elbe P, Barret M, Ortiz Fernández-Sordo J, Pech O, Beyna T, van der Sommen F, de With PH, de Groof AJ, Bergman JJ. Towards a robust and compact deep learning system for primary detection of early Barrett's neoplasia: Initial image-based results of training on a multi-center retrospectively collected data set. United European Gastroenterol J. 2023;11:324-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 37. | Abdelrahim M, Saiko M, Maeda N, Hossain E, Alkandari A, Subramaniam S, Parra-Blanco A, Sanchez-Yague A, Coron E, Repici A, Bhandari P. Development and validation of artificial neural networks model for detection of Barrett's neoplasia: a multicenter pragmatic nonrandomized trial (with video). Gastrointest Endosc. 2023;97:422-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 38. | Tan JL, Chinnaratha MA, Woodman R, Martin R, Chen HT, Carneiro G, Singh R. Diagnostic Accuracy of Artificial Intelligence (AI) to Detect Early Neoplasia in Barrett's Esophagus: A Non-comparative Systematic Review and Meta-Analysis. Front Med (Lausanne). 2022;9:890720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 39. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology. 2020;158:915-929.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 239] [Article Influence: 39.8] [Reference Citation Analysis (1)] |
| 40. | de Groof AJ, Struyvenberg MR, Fockens KN, van der Putten J, van der Sommen F, Boers TG, Zinger S, Bisschops R, de With PH, Pouw RE, Curvers WL, Schoon EJ, Bergman JJGHM. Deep learning algorithm detection of Barrett's neoplasia with high accuracy during live endoscopic procedures: a pilot study (with video). Gastrointest Endosc. 2020;91:1242-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (1)] |
| 41. | Hashimoto R, Requa J, Dao T, Ninh A, Tran E, Mai D, Lugo M, El-Hage Chehade N, Chang KJ, Karnes WE, Samarasena JB. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in Barrett's esophagus (with video). Gastrointest Endosc. 2020;91:1264-1271.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 164] [Article Influence: 27.3] [Reference Citation Analysis (1)] |
| 42. | Jukema JB, Kusters CHJ, Jong MR, Fockens KN, Boers T, van der Putten JA, Pouw RE, Duits LC, Weusten BLAM, Herrero LA, Houben MHMG, Nagengast WB, Westerhof J, Alkhalaf A, Mallant-Hent R, Scholten P, Ragunath K, Seewald S, Elbe P, Silva FB, Barret M, Ortiz Fernández-Sordo J, Moral Villarejo G, Pech O, Beyna T, Montazeri NSM, der Sommen FV, de With PH, de Groof AJ, Bergman JJ; BONS-AI consortium. Computer-aided diagnosis improves characterization of Barrett's neoplasia by general endoscopists (with video). Gastrointest Endosc. 2024;100:616-625.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 43. | Iwagami H, Ishihara R, Aoyama K, Fukuda H, Shimamoto Y, Kono M, Nakahira H, Matsuura N, Shichijo S, Kanesaka T, Kanzaki H, Ishii T, Nakatani Y, Tada T. Artificial intelligence for the detection of esophageal and esophagogastric junctional adenocarcinoma. J Gastroenterol Hepatol. 2021;36:131-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (2)] |
| 44. | Hussein M, Lines D, González-Bueno Puyal J, Kader R, Bowman N, Sehgal V, Toth D, Ahmad OF, Everson M, Esteban JM, Bisschops R, Banks M, Haefner M, Mountney P, Stoyanov D, Lovat LB, Haidry R. Computer-aided characterization of early cancer in Barrett's esophagus on i-scan magnification imaging: a multicenter international study. Gastrointest Endosc. 2023;97:646-654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 45. | Hussein M, González-Bueno Puyal J, Lines D, Sehgal V, Toth D, Ahmad OF, Kader R, Everson M, Lipman G, Fernandez-Sordo JO, Ragunath K, Esteban JM, Bisschops R, Banks M, Haefner M, Mountney P, Stoyanov D, Lovat LB, Haidry R. A new artificial intelligence system successfully detects and localises early neoplasia in Barrett's esophagus by using convolutional neural networks. United European Gastroenterol J. 2022;10:528-537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (1)] |
| 46. | Zhou N, Yuan X, Liu W, Luo Q, Liu R, Hu B. Artificial intelligence in endoscopic diagnosis of esophageal squamous cell carcinoma and precancerous lesions. Chin Med J (Engl). 2025;138:1387-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 47. | Morgan DR, Corral JE, Li D, Montgomery EA, Riquelme A, Kim JJ, Sauer B, Shah SC. ACG Clinical Guideline: Diagnosis and Management of Gastric Premalignant Conditions. Am J Gastroenterol. 2025;120:709-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 28] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 48. | Nathani P, Sharma P. Role of Artificial Intelligence in the Detection and Management of Premalignant and Malignant Lesions of the Esophagus and Stomach. Gastrointest Endosc Clin N Am. 2025;35:319-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 49. | Li SW, Zhang LH, Cai Y, Zhou XB, Fu XY, Song YQ, Xu SW, Tang SP, Luo RQ, Huang Q, Yan LL, He SQ, Zhang Y, Wang J, Ge SQ, Gu BB, Peng JB, Wang Y, Fang LN, Wu WD, Ye WG, Zhu M, Luo DH, Jin XX, Yang HD, Zhou JJ, Wang ZZ, Wu JF, Qin QQ, Lu YD, Wang F, Chen YH, Chen X, Xu SJ, Tung TH, Luo CW, Ye LP, Yu HG, Mao XL. Deep learning assists detection of esophageal cancer and precursor lesions in a prospective, randomized controlled study. Sci Transl Med. 2024;16:eadk5395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 11.0] [Reference Citation Analysis (1)] |
| 50. | Tani Y, Ishihara R, Inoue T, Okubo Y, Kawakami Y, Matsueda K, Miyake M, Yoshii S, Shichijo S, Kanesaka T, Yamamoto S, Takeuchi Y, Higashino K, Uedo N, Michida T, Kato Y, Tada T. A single-center prospective study evaluating the usefulness of artificial intelligence for the diagnosis of esophageal squamous cell carcinoma in a real-time setting. BMC Gastroenterol. 2023;23:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 51. | Yuan XL, Liu W, Lin YX, Deng QY, Gao YP, Wan L, Zhang B, Zhang T, Zhang WH, Bi XG, Yang GD, Zhu BH, Zhang F, Qin XB, Pan F, Zeng XH, Chaudhry H, Pang MY, Yang J, Zhang JY, Hu B. Effect of an artificial intelligence-assisted system on endoscopic diagnosis of superficial oesophageal squamous cell carcinoma and precancerous lesions: a multicentre, tandem, double-blind, randomised controlled trial. Lancet Gastroenterol Hepatol. 2024;9:34-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 38] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 52. | Yang XX, Li Z, Shao XJ, Ji R, Qu JY, Zheng MQ, Sun YN, Zhou RC, You H, Li LX, Feng J, Yang XY, Li YQ, Zuo XL. Real-time artificial intelligence for endoscopic diagnosis of early esophageal squamous cell cancer (with video). Dig Endosc. 2021;33:1075-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 53. | Aoyama N, Nakajo K, Sasabe M, Inaba A, Nakanishi Y, Seno H, Yano T. Effects of artificial intelligence assistance on endoscopist performance: Comparison of diagnostic performance in superficial esophageal squamous cell carcinoma detection using video-based models. DEN Open. 2026;6:e70083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (1)] |
| 54. | Ma T, Liu GQ, Guo J, Ji R, Shao XJ, Li YQ, Li Z, Zuo XL. Artificial intelligence-aided optical biopsy improves the diagnosis of esophageal squamous neoplasm. World J Gastroenterol. 2025;31:104370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 55. | Cai SL, Li B, Tan WM, Niu XJ, Yu HH, Yao LQ, Zhou PH, Yan B, Zhong YS. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2019;90:745-753.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 117] [Article Influence: 16.7] [Reference Citation Analysis (4)] |
| 56. | Liu W, Yuan X, Guo L, Pan F, Wu C, Sun Z, Tian F, Yuan C, Zhang W, Bai S, Feng J, Hu Y, Hu B. Artificial Intelligence for Detecting and Delineating Margins of Early ESCC Under WLI Endoscopy. Clin Transl Gastroenterol. 2022;13:e00433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (1)] |
| 57. | Guidozzi N, Menon N, Chidambaram S, Markar SR. The role of artificial intelligence in the endoscopic diagnosis of esophageal cancer: a systematic review and meta-analysis. Dis Esophagus. 2023;36:doad048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 58. | Luo D, Kuang F, Du J, Zhou M, Liu X, Luo X, Tang Y, Li B, Su S. Artificial Intelligence-Assisted Endoscopic Diagnosis of Early Upper Gastrointestinal Cancer: A Systematic Review and Meta-Analysis. Front Oncol. 2022;12:855175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 59. | Baik YS, Lee H, Kim YJ, Chung JW, Kim KG. Early detection of esophageal cancer: Evaluating AI algorithms with multi-institutional narrowband and white-light imaging data. PLoS One. 2025;20:e0321092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 60. | Wang YK, Karmakar R, Mukundan A, Men TC, Tsao YM, Lu SC, Wu IC, Wang HC. Computer-aided endoscopic diagnostic system modified with hyperspectral imaging for the classification of esophageal neoplasms. Front Oncol. 2024;14:1423405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 61. | Meng QQ, Gao Y, Lin H, Wang TJ, Zhang YR, Feng J, Li ZS, Xin L, Wang LW. Application of an artificial intelligence system for endoscopic diagnosis of superficial esophageal squamous cell carcinoma. World J Gastroenterol. 2022;28:5483-5493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (2)] |
| 62. | Wang SX, Ke Y, Liu YM, Liu SY, Song SB, He S, Zhang YM, Dou LZ, Liu Y, Liu XD, Wu HR, Su FX, Zhang FY, Zhang W, Wang GQ. [Establishment and clinical validation of an artificial intelligence YOLOv51 model for the detection of precancerous lesions and superficial esophageal cancer in endoscopic procedure]. Zhonghua Zhong Liu Za Zhi. 2022;44:395-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 63. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (1)] |
| 64. | Chou CK, Nguyen HT, Wang YK, Chen TH, Wu IC, Huang CW, Wang HC. Preparing Well for Esophageal Endoscopic Detection Using a Hybrid Model and Transfer Learning. Cancers (Basel). 2023;15:3783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 65. | Yang PC, Huang CW, Karmakar R, Mukundan A, Chen TH, Chou CK, Yang KY, Wang HC. Precision Imaging for Early Detection of Esophageal Cancer. Bioengineering (Basel). 2025;12:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 66. | Fukuda H, Ishihara R, Kato Y, Matsunaga T, Nishida T, Yamada T, Ogiyama H, Horie M, Kinoshita K, Tada T. Comparison of performances of artificial intelligence versus expert endoscopists for real-time assisted diagnosis of esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2020;92:848-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (1)] |
| 67. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 285] [Article Influence: 40.7] [Reference Citation Analysis (4)] |
| 68. | Urabe A, Adachi M, Sakamoto N, Kojima M, Ishikawa S, Ishii G, Yano T, Sakashita S. Deep learning detected histological differences between invasive and non-invasive areas of early esophageal cancer. Cancer Sci. 2025;116:824-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 69. | Nakagawa K, Ishihara R, Aoyama K, Ohmori M, Nakahira H, Matsuura N, Shichijo S, Nishida T, Yamada T, Yamaguchi S, Ogiyama H, Egawa S, Kishida O, Tada T. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest Endosc. 2019;90:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 113] [Article Influence: 16.1] [Reference Citation Analysis (1)] |
| 70. | Ohmori M, Ishihara R, Aoyama K, Nakagawa K, Iwagami H, Matsuura N, Shichijo S, Yamamoto K, Nagaike K, Nakahara M, Inoue T, Aoi K, Okada H, Tada T. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc. 2020;91:301-309.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (1)] |
| 71. | Tokai Y, Yoshio T, Aoyama K, Horie Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Tsuchida T, Hirasawa T, Sakakibara Y, Yamada T, Yamaguchi S, Fujisaki J, Tada T. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus. 2020;17:250-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (2)] |
| 72. | Yuan XL, Liu W, Liu Y, Zeng XH, Mou Y, Wu CC, Ye LS, Zhang YH, He L, Feng J, Zhang WH, Wang J, Chen X, Hu YX, Zhang KH, Hu B. Artificial intelligence for diagnosing microvessels of precancerous lesions and superficial esophageal squamous cell carcinomas: a multicenter study. Surg Endosc. 2022;36:8651-8662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 73. | Zhang L, Luo R, Tang D, Zhang J, Su Y, Mao X, Ye L, Yao L, Zhou W, Zhou J, Lu Z, Zhang M, Xu Y, Deng Y, Huang X, He C, Xiao Y, Wang J, Wu L, Li J, Zou X, Yu H. Human-Like Artificial Intelligent System for Predicting Invasion Depth of Esophageal Squamous Cell Carcinoma Using Magnifying Narrow-Band Imaging Endoscopy: A Retrospective Multicenter Study. Clin Transl Gastroenterol. 2023;14:e00606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 74. | Wang J, Long Q, Liang Y, Song J, Feng Y, Li P, Sun W, Zhao L. AI-assisted identification of intrapapillary capillary loops in magnification endoscopy for diagnosing early-stage esophageal squamous cell carcinoma: a preliminary study. Med Biol Eng Comput. 2023;61:1631-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 75. | Inoue H, Kaga M, Ikeda H, Sato C, Sato H, Minami H, Santi EG, Hayee B, Eleftheriadis N. Magnification endoscopy in esophageal squamous cell carcinoma: a review of the intrapapillary capillary loop classification. Ann Gastroenterol. 2015;28:41-48. [PubMed] |
| 76. | Shah SC, Wang AY, Wallace MB, Hwang JH. AGA Clinical Practice Update on Screening and Surveillance in Individuals at Increased Risk for Gastric Cancer in the United States: Expert Review. Gastroenterology. 2025;168:405-416.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 50] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 77. | Morgan E, Arnold M, Camargo MC, Gini A, Kunzmann AT, Matsuda T, Meheus F, Verhoeven RHA, Vignat J, Laversanne M, Ferlay J, Soerjomataram I. The current and future incidence and mortality of gastric cancer in 185 countries, 2020-40: A population-based modelling study. EClinicalMedicine. 2022;47:101404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 521] [Article Influence: 130.3] [Reference Citation Analysis (0)] |
| 78. | Lei C, Sun W, Wang K, Weng R, Kan X, Li R. Artificial intelligence-assisted diagnosis of early gastric cancer: present practice and future prospects. Ann Med. 2025;57:2461679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 79. | Messmann H, Bisschops R, Antonelli G, Libânio D, Sinonquel P, Abdelrahim M, Ahmad OF, Areia M, Bergman JJGHM, Bhandari P, Boskoski I, Dekker E, Domagk D, Ebigbo A, Eelbode T, Eliakim R, Häfner M, Haidry RJ, Jover R, Kaminski MF, Kuvaev R, Mori Y, Palazzo M, Repici A, Rondonotti E, Rutter MD, Saito Y, Sharma P, Spada C, Spadaccini M, Veitch A, Gralnek IM, Hassan C, Dinis-Ribeiro M. Expected value of artificial intelligence in gastrointestinal endoscopy: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy. 2022;54:1211-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 105] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 80. | Guimarães P, Keller A, Fehlmann T, Lammert F, Casper M. Deep-learning based detection of gastric precancerous conditions. Gut. 2020;69:4-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 81. | Yang J, Ou Y, Chen Z, Liao J, Sun W, Luo Y, Luo C. A Benchmark Dataset of Endoscopic Images and Novel Deep Learning Method to Detect Intestinal Metaplasia and Gastritis Atrophy. IEEE J Biomed Health Inform. 2023;27:7-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 82. | Parkash O, Lal A, Subash T, Sultan U, Tahir HN, Hoodbhoy Z, Sundar S, Das JK. Use of artificial intelligence for the detection of Helicobacter pylori infection from upper gastrointestinal endoscopy images: an updated systematic review and meta-analysis. Ann Gastroenterol. 2024;37:665-673. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 83. | Zou PY, Zhu JR, Zhao Z, Mei H, Zhao JT, Sun WJ, Wang GH, Chen DF, Fan LL, Lan CH. Development and application of an artificial intelligence-assisted endoscopy system for diagnosis of Helicobacter pylori infection: a multicenter randomized controlled study. BMC Gastroenterol. 2024;24:335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 84. | Xu Z, Li Y, Su P, Zhong Z, Zeng Z, Chen M, Chen D, Lan C. Artificial intelligence system improves the quality of digestive endoscopy: A prospective pretest and post-test single-center clinical trial. Dig Liver Dis. 2025;57:1830-1837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 85. | Xu M, Zhou W, Wu L, Zhang J, Wang J, Mu G, Huang X, Li Y, Yuan J, Zeng Z, Wang Y, Huang L, Liu J, Yu H. Artificial intelligence in the diagnosis of gastric precancerous conditions by image-enhanced endoscopy: a multicenter, diagnostic study (with video). Gastrointest Endosc. 2021;94:540-548.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 86. | Fang S, Liu Z, Qiu Q, Tang Z, Yang Y, Kuang Z, Du X, Xiao S, Liu Y, Luo Y, Gu L, Tian L, Liang X, Fan G, Zhang Y, Zhang P, Zhou W, Liu X, Tian J, Wei W. Diagnosing and grading gastric atrophy and intestinal metaplasia using semi-supervised deep learning on pathological images: development and validation study. Gastric Cancer. 2024;27:343-354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 87. | Iwaya M, Hayashi Y, Sakai Y, Yoshizawa A, Iwaya Y, Uehara T, Kitagawa M, Fukayama M, Mori K, Ota H. Artificial intelligence for evaluating the risk of gastric cancer: reliable detection and scoring of intestinal metaplasia with deep learning algorithms. Gastrointest Endosc. 2023;98:925-933.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 88. | Shi Y, Wei N, Wang K, Wu J, Tao T, Li N, Lv B. Deep learning-assisted diagnosis of chronic atrophic gastritis in endoscopy. Front Oncol. 2023;13:1122247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 89. | Zhao Q, Jia Q, Chi T. Deep learning as a novel method for endoscopic diagnosis of chronic atrophic gastritis: a prospective nested case-control study. BMC Gastroenterol. 2022;22:352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 90. | Lin N, Yu T, Zheng W, Hu H, Xiang L, Ye G, Zhong X, Ye B, Wang R, Deng W, Li J, Wang X, Han F, Zhuang K, Zhang D, Xu H, Ding J, Zhang X, Shen Y, Lin H, Zhang Z, Kim JJ, Liu J, Hu W, Duan H, Si J. Simultaneous Recognition of Atrophic Gastritis and Intestinal Metaplasia on White Light Endoscopic Images Based on Convolutional Neural Networks: A Multicenter Study. Clin Transl Gastroenterol. 2021;12:e00385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 91. | Li N, Yang J, Li X, Shi Y, Wang K. Accuracy of artificial intelligence-assisted endoscopy in the diagnosis of gastric intestinal metaplasia: A systematic review and meta-analysis. PLoS One. 2024;19:e0303421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 92. | Zhang X, Chen F, Yu T, An J, Huang Z, Liu J, Hu W, Wang L, Duan H, Si J. Real-time gastric polyp detection using convolutional neural networks. PLoS One. 2019;14:e0214133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 93. | Yuan XL, Zhou Y, Liu W, Luo Q, Zeng XH, Yi Z, Hu B. Artificial intelligence for diagnosing gastric lesions under white-light endoscopy. Surg Endosc. 2022;36:9444-9453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 94. | Niu W, Liu L, Dong Z, Bu X, Yao F, Wang J, Wu X, Chen C, Mao T, Wu Y, Yuan L, Wan X, Zhou H. A deep learning model based on magnifying endoscopy with narrow-band imaging to evaluate intestinal metaplasia grading and OLGIM staging: A multicenter study. Dig Liver Dis. 2024;56:1565-1571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 95. | Kodaka Y, Futagami S, Watanabe Y, Shichijo S, Uedo N, Aono H, Kirita K, Kato Y, Ueki N, Agawa S, Yamawaki H, Iwakiri K, Tada T. Determination of gastric atrophy with artificial intelligence compared to the assessments of the modified Kyoto and OLGA classifications. JGH Open. 2022;6:704-710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 96. | Li Z, Zheng X, Mu Y, Zhang M, Liu G. Color-guided deformable convolution network for intestinal metaplasia severity classification using endoscopic images. Phys Med Biol. 2023;68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 97. | Tao X, Zhu Y, Dong Z, Huang L, Shang R, Du H, Wang J, Zeng X, Wang W, Wang J, Li Y, Deng Y, Wu L, Yu H. An artificial intelligence system for chronic atrophic gastritis diagnosis and risk stratification under white light endoscopy. Dig Liver Dis. 2024;56:1319-1326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 98. | Zhao Q, Chi T. Deep learning model can improve the diagnosis rate of endoscopic chronic atrophic gastritis: a prospective cohort study. BMC Gastroenterol. 2022;22:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 99. | Zhang Y, Li F, Yuan F, Zhang K, Huo L, Dong Z, Lang Y, Zhang Y, Wang M, Gao Z, Qin Z, Shen L. Diagnosing chronic atrophic gastritis by gastroscopy using artificial intelligence. Dig Liver Dis. 2020;52:566-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 100. | Watanabe Y, Oikawa R, Agawa S, Matsuo Y, Oda I, Futagami S, Yamamoto H, Tada T, Itoh F. Combination of artificial intelligence-based endoscopy and miR148a methylation for gastric indefinite dysplasia diagnosis. J Clin Lab Anal. 2022;36:e24122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 101. | Lee H, Chung JW, Yun SC, Jung SW, Yoon YJ, Kim JH, Cha B, Kayasseh MA, Kim KO. Validation of Artificial Intelligence Computer-Aided Detection on Gastric Neoplasm in Upper Gastrointestinal Endoscopy. Diagnostics (Basel). 2024;14:2706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 102. | Zhao Y, Dohi O, Ishida T, Yoshida N, Ochiai T, Mukai H, Seya M, Yamauchi K, Miyazaki H, Fukui H, Yasuda T, Iwai N, Inoue K, Itoh Y, Liu X, Zhang R, Zhu X. Linked Color Imaging with Artificial Intelligence Improves the Detection of Early Gastric Cancer. Dig Dis. 2024;42:503-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 103. | Ueyama H, Kato Y, Akazawa Y, Yatagai N, Komori H, Takeda T, Matsumoto K, Ueda K, Matsumoto K, Hojo M, Yao T, Nagahara A, Tada T. Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J Gastroenterol Hepatol. 2021;36:482-489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (1)] |
| 104. | Feng J, Zhang Y, Feng Z, Ma H, Gou Y, Wang P, Feng Y, Wang X, Huang X. A prospective and comparative study on improving the diagnostic accuracy of early gastric cancer based on deep convolutional neural network real-time diagnosis system (with video). Surg Endosc. 2025;39:1874-1884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 105. | Soong TK, Kim GW, Chia DKA, So JBY, Lee JWJ, Shabbbir A, Lum JHY, Soon GST, Ho KY. Comparing Raman Spectroscopy-Based Artificial Intelligence to High-Definition White Light Endoscopy for Endoscopic Diagnosis of Gastric Neoplasia: A Feasibility Proof-of-Concept Study. Diagnostics (Basel). 2024;14:2839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 106. | Gong EJ, Bang CS, Lee JJ, Baik GH, Lim H, Jeong JH, Choi SW, Cho J, Kim DY, Lee KB, Shin SI, Sigmund D, Moon BI, Park SC, Lee SH, Bang KB, Son DS. Deep learning-based clinical decision support system for gastric neoplasms in real-time endoscopy: development and validation study. Endoscopy. 2023;55:701-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 107. | Wu L, Shang R, Sharma P, Zhou W, Liu J, Yao L, Dong Z, Yuan J, Zeng Z, Yu Y, He C, Xiong Q, Li Y, Deng Y, Cao Z, Huang C, Zhou R, Li H, Hu G, Chen Y, Wang Y, He X, Zhu Y, Yu H. Effect of a deep learning-based system on the miss rate of gastric neoplasms during upper gastrointestinal endoscopy: a single-centre, tandem, randomised controlled trial. Lancet Gastroenterol Hepatol. 2021;6:700-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 108. | Wu L, He X, Liu M, Xie H, An P, Zhang J, Zhang H, Ai Y, Tong Q, Guo M, Huang M, Ge C, Yang Z, Yuan J, Liu J, Zhou W, Jiang X, Huang X, Mu G, Wan X, Li Y, Wang H, Wang Y, Zhang H, Chen D, Gong D, Wang J, Huang L, Li J, Yao L, Zhu Y, Yu H. Evaluation of the effects of an artificial intelligence system on endoscopy quality and preliminary testing of its performance in detecting early gastric cancer: a randomized controlled trial. Endoscopy. 2021;53:1199-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 119] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 109. | Ishioka M, Osawa H, Hirasawa T, Kawachi H, Nakano K, Fukushima N, Sakaguchi M, Tada T, Kato Y, Shibata J, Ozawa T, Tajiri H, Fujisaki J. Performance of an artificial intelligence-based diagnostic support tool for early gastric cancers: Retrospective study. Dig Endosc. 2023;35:483-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 110. | Li J, Zhu Y, Dong Z, He X, Xu M, Liu J, Zhang M, Tao X, Du H, Chen D, Huang L, Shang R, Zhang L, Luo R, Zhou W, Deng Y, Huang X, Li Y, Chen B, Gong R, Zhang C, Li X, Wu L, Yu H. Development and validation of a feature extraction-based logical anthropomorphic diagnostic system for early gastric cancer: A case-control study. EClinicalMedicine. 2022;46:101366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 111. | Xie F, Zhang K, Li F, Ma G, Ni Y, Zhang W, Wang J, Li Y. Diagnostic accuracy of convolutional neural network-based endoscopic image analysis in diagnosing gastric cancer and predicting its invasion depth: a systematic review and meta-analysis. Gastrointest Endosc. 2022;95:599-609.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 27] [Reference Citation Analysis (1)] |
| 112. | Horiuchi Y, Hirasawa T, Ishizuka N, Tokai Y, Namikawa K, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Performance of a computer-aided diagnosis system in diagnosing early gastric cancer using magnifying endoscopy videos with narrow-band imaging (with videos). Gastrointest Endosc. 2020;92:856-865.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (1)] |
| 113. | Jin J, Zhang Q, Dong B, Ma T, Mei X, Wang X, Song S, Peng J, Wu A, Dong L, Kong D. Automatic detection of early gastric cancer in endoscopy based on Mask region-based convolutional neural networks (Mask R-CNN)(with video). Front Oncol. 2022;12:927868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 114. | Tang D, Ni M, Zheng C, Ding X, Zhang N, Yang T, Zhan Q, Fu Y, Liu W, Zhuang D, Lv Y, Xu G, Wang L, Zou X. A deep learning-based model improves diagnosis of early gastric cancer under narrow band imaging endoscopy. Surg Endosc. 2022;36:7800-7810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 115. | He X, Wu L, Dong Z, Gong D, Jiang X, Zhang H, Ai Y, Tong Q, Lv P, Lu B, Wu Q, Yuan J, Xu M, Yu H. Real-time use of artificial intelligence for diagnosing early gastric cancer by magnifying image-enhanced endoscopy: a multicenter diagnostic study (with videos). Gastrointest Endosc. 2022;95:671-678.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 116. | Wu L, Xu M, Jiang X, He X, Zhang H, Ai Y, Tong Q, Lv P, Lu B, Guo M, Huang M, Ye L, Shen L, Yu H. Real-time artificial intelligence for detecting focal lesions and diagnosing neoplasms of the stomach by white-light endoscopy (with videos). Gastrointest Endosc. 2022;95:269-280.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 117. | Wu L, Wang J, He X, Zhu Y, Jiang X, Chen Y, Wang Y, Huang L, Shang R, Dong Z, Chen B, Tao X, Wu Q, Yu H. Deep learning system compared with expert endoscopists in predicting early gastric cancer and its invasion depth and differentiation status (with videos). Gastrointest Endosc. 2022;95:92-104.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 118. | Chen PC, Lu YR, Kang YN, Chang CC. The Accuracy of Artificial Intelligence in the Endoscopic Diagnosis of Early Gastric Cancer: Pooled Analysis Study. J Med Internet Res. 2022;24:e27694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 119. | Chang YH, Shin CM, Lee HD, Park J, Jeon J, Cho SJ, Kang SJ, Chung JY, Jun YK, Choi Y, Yoon H, Park YS, Kim N, Lee DH. Real-World Application of Artificial Intelligence for Detecting Pathologic Gastric Atypia and Neoplastic Lesions. J Gastric Cancer. 2024;24:327-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 120. | Lee S, Jeon J, Park J, Chang YH, Shin CM, Oh MJ, Kim SH, Kang S, Park SH, Kim SG, Lee HJ, Yang HK, Lee HS, Cho SJ. An artificial intelligence system for comprehensive pathologic outcome prediction in early gastric cancer through endoscopic image analysis (with video). Gastric Cancer. 2024;27:1088-1099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 121. | Tang D, Zhou J, Wang L, Ni M, Chen M, Hassan S, Luo R, Chen X, He X, Zhang L, Ding X, Yu H, Xu G, Zou X. A Novel Model Based on Deep Convolutional Neural Network Improves Diagnostic Accuracy of Intramucosal Gastric Cancer (With Video). Front Oncol. 2021;11:622827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 122. | Zhu Y, Wang QC, Xu MD, Zhang Z, Cheng J, Zhong YS, Zhang YQ, Chen WF, Yao LQ, Zhou PH, Li QL. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc. 2019;89:806-815.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 238] [Article Influence: 34.0] [Reference Citation Analysis (3)] |
| 123. | Kim JH, Oh SI, Han SY, Keum JS, Kim KN, Chun JY, Youn YH, Park H. An Optimal Artificial Intelligence System for Real-Time Endoscopic Prediction of Invasion Depth in Early Gastric Cancer. Cancers (Basel). 2022;14:6000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 124. | Oura H, Matsumura T, Fujie M, Ishikawa T, Nagashima A, Shiratori W, Tokunaga M, Kaneko T, Imai Y, Oike T, Yokoyama Y, Akizue N, Ota Y, Okimoto K, Arai M, Nakagawa Y, Inada M, Yamaguchi K, Kato J, Kato N. Development and evaluation of a double-check support system using artificial intelligence in endoscopic screening for gastric cancer. Gastric Cancer. 2022;25:392-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 125. | Bilimoria KY, Bentrem DJ, Wayne JD, Ko CY, Bennett CL, Talamonti MS. Small bowel cancer in the United States: changes in epidemiology, treatment, and survival over the last 20 years. Ann Surg. 2009;249:63-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 482] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 126. | Vanoli A, Grillo F, Furlan D, Arpa G, Grami O, Guerini C, Riboni R, Mastracci L, Di Sabatino A. Small Bowel Epithelial Precursor Lesions: A Focus on Molecular Alterations. Int J Mol Sci. 2021;22:4388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 127. | Cardoso H, Rodrigues JT, Marques M, Ribeiro A, Vilas-Boas F, Santos-Antunes J, Rodrigues-Pinto E, Silva M, Maia JC, Macedo G. Malignant Small Bowel Tumors: Diagnosis, Management and Prognosis. Acta Med Port. 2015;28:448-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 128. | Pennazio M, Rondonotti E, Despott EJ, Dray X, Keuchel M, Moreels T, Sanders DS, Spada C, Carretero C, Cortegoso Valdivia P, Elli L, Fuccio L, Gonzalez Suarez B, Koulaouzidis A, Kunovsky L, McNamara D, Neumann H, Perez-Cuadrado-Martinez E, Perez-Cuadrado-Robles E, Piccirelli S, Rosa B, Saurin JC, Sidhu R, Tacheci I, Vlachou E, Triantafyllou K. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Guideline - Update 2022. Endoscopy. 2023;55:58-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 195] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 129. | Rondonotti E, Spada C, Adler S, May A, Despott EJ, Koulaouzidis A, Panter S, Domagk D, Fernandez-Urien I, Rahmi G, Riccioni ME, van Hooft JE, Hassan C, Pennazio M. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Technical Review. Endoscopy. 2018;50:423-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 304] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 130. | Hashimoto R, Matsuda T, Nakahori M. False-negative double-balloon enteroscopy in overt small bowel bleeding: long-term follow-up after negative results. Surg Endosc. 2019;33:2635-2641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 131. | Berzin TM, Parasa S, Wallace MB, Gross SA, Repici A, Sharma P. Position statement on priorities for artificial intelligence in GI endoscopy: a report by the ASGE Task Force. Gastrointest Endosc. 2020;92:951-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (1)] |
| 132. | Bang CS, Lim H, Jeong HM, Hwang SH. Use of Endoscopic Images in the Prediction of Submucosal Invasion of Gastric Neoplasms: Automated Deep Learning Model Development and Usability Study. J Med Internet Res. 2021;23:e25167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 133. | Dhali A, Kipkorir V, Maity R, Srichawla BS, Biswas J, Rathna RB, Bharadwaj HR, Ongidi I, Chaudhry T, Morara G, Waithaka M, Rugut C, Lemashon M, Cheruiyot I, Ojuka D, Ray S, Dhali GK. Artificial Intelligence-Assisted Capsule Endoscopy Versus Conventional Capsule Endoscopy for Detection of Small Bowel Lesions: A Systematic Review and Meta-Analysis. J Gastroenterol Hepatol. 2025;40:1105-1118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 134. | Cortegoso Valdivia P, Fantasia S, Kayali S, Deding U, Gualandi N, Manno M, Toth E, Dray X, Yang S, Koulaouzidis A. Conventional small-bowel capsule endoscopy reading vs proprietary artificial intelligence auxiliary systems: Systematic review and meta-analysis. Endosc Int Open. 2025;13:a25442863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 135. | Barbosa DJ, Ramos J, Lima CS. Detection of small bowel tumors in capsule endoscopy frames using texture analysis based on the discrete wavelet transform. Annu Int Conf IEEE Eng Med Biol Soc. 2008;2008:3012-3015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 136. | Barbosa DC, Roupar DB, Ramos JC, Tavares AC, Lima CS. Automatic small bowel tumor diagnosis by using multi-scale wavelet-based analysis in wireless capsule endoscopy images. Biomed Eng Online. 2012;11:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 137. | Aliferis C, Simon G. Overfitting, Underfitting and General Model Overconfidence and Under-Performance Pitfalls and Best Practices in Machine Learning and AI. In: Artificial Intelligence and Machine Learning in Health Care and Medical Sciences: Best Practices and Pitfalls [Internet]. Cham (CH): Springer; 2024–. [PubMed] [DOI] [Full Text] |
| 138. | Constantinescu AF, Ionescu M, Iovănescu VF, Ciurea ME, Ionescu AG, Streba CT, Bunescu MG, Rogoveanu I, Vere CC. A computer-aided diagnostic system for intestinal polyps identified by wireless capsule endoscopy. Rom J Morphol Embryol. 2016;57:979-984. [PubMed] |
| 139. | Cardoso P, Saraiva MM, Afonso J, Ribeiro T, Andrade P, Ferreira J, Cardoso H, Macedo G. Artificial Intelligence and Device-Assisted Enteroscopy: Automatic Detection of Enteric Protruding Lesions Using a Convolutional Neural Network. Clin Transl Gastroenterol. 2022;13:e00514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (2)] |
| 140. | Zhu Y, Lyu X, Tao X, Wu L, Yin A, Liao F, Hu S, Wang Y, Zhang M, Huang L, Wang J, Zhang C, Gong D, Jiang X, Zhao L, Yu H. A newly developed deep learning-based system for automatic detection and classification of small bowel lesions during double-balloon enteroscopy examination. BMC Gastroenterol. 2024;24:10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 141. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56665] [Article Influence: 7083.1] [Reference Citation Analysis (135)] |
| 142. | Brenner H, Stock C, Hoffmeister M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ. 2014;348:g2467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 663] [Cited by in RCA: 634] [Article Influence: 52.8] [Reference Citation Analysis (2)] |
| 143. | Robertson DJ, Lieberman DA, Winawer SJ, Ahnen DJ, Baron JA, Schatzkin A, Cross AJ, Zauber AG, Church TR, Lance P, Greenberg ER, Martínez ME. Colorectal cancers soon after colonoscopy: a pooled multicohort analysis. Gut. 2014;63:949-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 353] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 144. | Aslanian HR, Shieh FK, Chan FW, Ciarleglio MM, Deng Y, Rogart JN, Jamidar PA, Siddiqui UD. Nurse observation during colonoscopy increases polyp detection: a randomized prospective study. Am J Gastroenterol. 2013;108:166-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 145. | Maroulis DE, Iakovidis DK, Karkanis SA, Karras DA. CoLD: a versatile detection system for colorectal lesions in endoscopy video-frames. Comput Methods Programs Biomed. 2003;70:151-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 62] [Article Influence: 2.7] [Reference Citation Analysis (1)] |
| 146. | Zhao S, Wang S, Pan P, Xia T, Chang X, Yang X, Guo L, Meng Q, Yang F, Qian W, Xu Z, Wang Y, Wang Z, Gu L, Wang R, Jia F, Yao J, Li Z, Bai Y. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-analysis. Gastroenterology. 2019;156:1661-1674.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 433] [Article Influence: 61.9] [Reference Citation Analysis (1)] |
| 147. | le Clercq CM, Bouwens MW, Rondagh EJ, Bakker CM, Keulen ET, de Ridder RJ, Winkens B, Masclee AA, Sanduleanu S. Postcolonoscopy colorectal cancers are preventable: a population-based study. Gut. 2014;63:957-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 281] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 148. | Corley DA, Jensen CD, Marks AR, Zhao WK, Lee JK, Doubeni CA, Zauber AG, de Boer J, Fireman BH, Schottinger JE, Quinn VP, Ghai NR, Levin TR, Quesenberry CP. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:1298-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1251] [Cited by in RCA: 1666] [Article Influence: 138.8] [Reference Citation Analysis (1)] |
| 149. | Iakovidis DK, Maroulis DE, Karkanis SA. An intelligent system for automatic detection of gastrointestinal adenomas in video endoscopy. Comput Biol Med. 2006;36:1084-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 150. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069-1078.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 453] [Article Influence: 56.6] [Reference Citation Analysis (1)] |
| 151. | Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 591] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 152. | Repici A, Badalamenti M, Maselli R, Correale L, Radaelli F, Rondonotti E, Ferrara E, Spadaccini M, Alkandari A, Fugazza A, Anderloni A, Galtieri PA, Pellegatta G, Carrara S, Di Leo M, Craviotto V, Lamonaca L, Lorenzetti R, Andrealli A, Antonelli G, Wallace M, Sharma P, Rosch T, Hassan C. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology. 2020;159:512-520.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 445] [Article Influence: 74.2] [Reference Citation Analysis (2)] |
| 153. | Repici A, Spadaccini M, Antonelli G, Correale L, Maselli R, Galtieri PA, Pellegatta G, Capogreco A, Milluzzo SM, Lollo G, Di Paolo D, Badalamenti M, Ferrara E, Fugazza A, Carrara S, Anderloni A, Rondonotti E, Amato A, De Gottardi A, Spada C, Radaelli F, Savevski V, Wallace MB, Sharma P, Rösch T, Hassan C. Artificial intelligence and colonoscopy experience: lessons from two randomised trials. Gut. 2022;71:757-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 164] [Article Influence: 41.0] [Reference Citation Analysis (3)] |
| 154. | Nakashima H, Kitazawa N, Fukuyama C, Kawachi H, Kawahira H, Momma K, Sakaki N. Clinical Evaluation of Computer-Aided Colorectal Neoplasia Detection Using a Novel Endoscopic Artificial Intelligence: A Single-Center Randomized Controlled Trial. Digestion. 2023;104:193-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 155. | Lau LHS, Ho JCL, Lai JCT, Ho AHY, Wu CWK, Lo VWH, Lai CMS, Scheppach MW, Sia F, Ho KHK, Xiao X, Yip TCF, Lam TYT, Kwok HYH, Chan HCH, Lui RN, Chan TT, Wong MTL, Ho MF, Ko RCW, Hon SF, Chu S, Futaba K, Ng SSM, Yip HC, Tang RSY, Wong VWS, Chan FKL, Chiu PWY; ENDOAID-TRAIN study group. Effect of Real-Time Computer-Aided Polyp Detection System (ENDO-AID) on Adenoma Detection in Endoscopists-in-Training: A Randomized Trial. Clin Gastroenterol Hepatol. 2024;22:630-641.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 156. | Wang P, Liu X, Berzin TM, Glissen Brown JR, Liu P, Zhou C, Lei L, Li L, Guo Z, Lei S, Xiong F, Wang H, Song Y, Pan Y, Zhou G. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol. 2020;5:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 315] [Article Influence: 52.5] [Reference Citation Analysis (1)] |
| 157. | Gong D, Wu L, Zhang J, Mu G, Shen L, Liu J, Wang Z, Zhou W, An P, Huang X, Jiang X, Li Y, Wan X, Hu S, Chen Y, Hu X, Xu Y, Zhu X, Li S, Yao L, He X, Chen D, Huang L, Wei X, Wang X, Yu H. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol. 2020;5:352-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 284] [Article Influence: 47.3] [Reference Citation Analysis (1)] |
| 158. | Su JR, Li Z, Shao XJ, Ji CR, Ji R, Zhou RC, Li GC, Liu GQ, He YS, Zuo XL, Li YQ. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastrointest Endosc. 2020;91:415-424.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 223] [Article Influence: 37.2] [Reference Citation Analysis (3)] |
| 159. | Liu WN, Zhang YY, Bian XQ, Wang LJ, Yang Q, Zhang XD, Huang J. Study on detection rate of polyps and adenomas in artificial-intelligence-aided colonoscopy. Saudi J Gastroenterol. 2020;26:13-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 135] [Article Influence: 19.3] [Reference Citation Analysis (1)] |
| 160. | Mohan BP, Facciorusso A, Khan SR, Chandan S, Kassab LL, Gkolfakis P, Tziatzios G, Triantafyllou K, Adler DG. Real-time computer aided colonoscopy versus standard colonoscopy for improving adenoma detection rate: A meta-analysis of randomized-controlled trials. EClinicalMedicine. 2020;29-30:100622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 161. | Hassan C, Spadaccini M, Mori Y, Foroutan F, Facciorusso A, Gkolfakis P, Tziatzios G, Triantafyllou K, Antonelli G, Khalaf K, Rizkala T, Vandvik PO, Fugazza A, Rondonotti E, Glissen-Brown JR, Kamba S, Maida M, Correale L, Bhandari P, Jover R, Sharma P, Rex DK, Repici A. Real-Time Computer-Aided Detection of Colorectal Neoplasia During Colonoscopy : A Systematic Review and Meta-analysis. Ann Intern Med. 2023;176:1209-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 140] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 162. | Soleymanjahi S, Huebner J, Elmansy L, Rajashekar N, Lüdtke N, Paracha R, Thompson R, Grimshaw AA, Foroutan F, Sultan S, Shung DL. Artificial Intelligence-Assisted Colonoscopy for Polyp Detection : A Systematic Review and Meta-analysis. Ann Intern Med. 2024;177:1652-1663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 44] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 163. | Spadaccini M, Iannone A, Maselli R, Badalamenti M, Desai M, Chandrasekar VT, Patel HK, Fugazza A, Pellegatta G, Galtieri PA, Lollo G, Carrara S, Anderloni A, Rex DK, Savevski V, Wallace MB, Bhandari P, Roesch T, Gralnek IM, Sharma P, Hassan C, Repici A. Computer-aided detection versus advanced imaging for detection of colorectal neoplasia: a systematic review and network meta-analysis. Lancet Gastroenterol Hepatol. 2021;6:793-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 164. | Aziz M, Haghbin H, Sayeh W, Alfatlawi H, Gangwani MK, Sohail AH, Zahdeh T, Weissman S, Kamal F, Lee-Smith W, Nawras A, Sharma P, Shaukat A. Comparison of Artificial Intelligence With Other Interventions to Improve Adenoma Detection Rate for Colonoscopy: A Network Meta-analysis. J Clin Gastroenterol. 2024;58:143-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 165. | Miyaguchi K, Tsuzuki Y, Hirooka N, Matsumoto H, Ohgo H, Nakamoto H, Imaeda H. Linked-color imaging with or without artificial intelligence for adenoma detection: a randomized trial. Endoscopy. 2024;56:376-383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 166. | Shaukat A, Lichtenstein DR, Somers SC, Chung DC, Perdue DG, Gopal M, Colucci DR, Phillips SA, Marka NA, Church TR, Brugge WR; SKOUT™ Registration Study Team. Computer-Aided Detection Improves Adenomas per Colonoscopy for Screening and Surveillance Colonoscopy: A Randomized Trial. Gastroenterology. 2022;163:732-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 100] [Article Influence: 25.0] [Reference Citation Analysis (1)] |
| 167. | Ahmad A, Wilson A, Haycock A, Humphries A, Monahan K, Suzuki N, Thomas-Gibson S, Vance M, Bassett P, Thiruvilangam K, Dhillon A, Saunders BP. Evaluation of a real-time computer-aided polyp detection system during screening colonoscopy: AI-DETECT study. Endoscopy. 2023;55:313-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 168. | Mangas-Sanjuan C, de-Castro L, Cubiella J, Díez-Redondo P, Suárez A, Pellisé M, Fernández N, Zarraquiños S, Núñez-Rodríguez H, Álvarez-García V, Ortiz O, Sala-Miquel N, Zapater P, Jover R; CADILLAC study investigators. Role of Artificial Intelligence in Colonoscopy Detection of Advanced Neoplasias : A Randomized Trial. Ann Intern Med. 2023;176:1145-1152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 52] [Article Influence: 17.3] [Reference Citation Analysis (3)] |
| 169. | Levy I, Bruckmayer L, Klang E, Ben-Horin S, Kopylov U. Artificial Intelligence-Aided Colonoscopy Does Not Increase Adenoma Detection Rate in Routine Clinical Practice. Am J Gastroenterol. 2022;117:1871-1873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 170. | Zimmermann-Fraedrich K, Sehner S, Rösch T, Aschenbeck J, Schubert S, Liceni T, Moog G, Neumann H, Berndt R, Weigt J, Kaczmarek DJ, May A, Hoffmeister A, Möschler O, Wiessner C, Schachschal G. No Effect of Computer-Aided Diagnosis on Colonoscopic Adenoma Detection in a Large Pragmatic Multicenter Randomized Study. Am J Gastroenterol. 2025;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 171. | Ladabaum U, Shepard J, Weng Y, Desai M, Singer SJ, Mannalithara A. Computer-aided Detection of Polyps Does Not Improve Colonoscopist Performance in a Pragmatic Implementation Trial. Gastroenterology. 2023;164:481-483.e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 66] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 172. | Alali AA, Alhashmi A, Alotaibi N, Ali N, Alali M, Alfadhli A. Artificial Intelligence for Adenoma and Polyp Detection During Screening and Surveillance Colonoscopy: A Randomized-Controlled Trial. J Clin Med. 2025;14:581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 173. | Desai M, Ausk K, Brannan D, Chhabra R, Chan W, Chiorean M, Gross SA, Girotra M, Haber G, Hogan RB, Jacob B, Jonnalagadda S, Iles-Shih L, Kumar N, Law J, Lee L, Lin O, Mizrahi M, Pacheco P, Parasa S, Phan J, Reeves V, Sethi A, Snell D, Underwood J, Venu N, Visrodia K, Wong A, Winn J, Wright CH, Sharma P. Use of a Novel Artificial Intelligence System Leads to the Detection of Significantly Higher Number of Adenomas During Screening and Surveillance Colonoscopy: Results From a Large, Prospective, US Multicenter, Randomized Clinical Trial. Am J Gastroenterol. 2024;119:1383-1391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (1)] |
| 174. | Aniwan S, Orkoonsawat P, Viriyautsahakul V, Angsuwatcharakon P, Pittayanon R, Wisedopas N, Sumdin S, Ponuthai Y, Wiangngoen S, Kullavanijaya P, Rerknimitr R. The Secondary Quality Indicator to Improve Prediction of Adenoma Miss Rate Apart from Adenoma Detection Rate. Am J Gastroenterol. 2016;111:723-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 175. | Wang P, Liu P, Glissen Brown JR, Berzin TM, Zhou G, Lei S, Liu X, Li L, Xiao X. Lower Adenoma Miss Rate of Computer-Aided Detection-Assisted Colonoscopy vs Routine White-Light Colonoscopy in a Prospective Tandem Study. Gastroenterology. 2020;159:1252-1261.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (3)] |
| 176. | Kamba S, Tamai N, Saitoh I, Matsui H, Horiuchi H, Kobayashi M, Sakamoto T, Ego M, Fukuda A, Tonouchi A, Shimahara Y, Nishikawa M, Nishino H, Saito Y, Sumiyama K. Reducing adenoma miss rate of colonoscopy assisted by artificial intelligence: a multicenter randomized controlled trial. J Gastroenterol. 2021;56:746-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (1)] |
| 177. | Glissen Brown JR, Mansour NM, Wang P, Chuchuca MA, Minchenberg SB, Chandnani M, Liu L, Gross SA, Sengupta N, Berzin TM. Deep Learning Computer-aided Polyp Detection Reduces Adenoma Miss Rate: A United States Multi-center Randomized Tandem Colonoscopy Study (CADeT-CS Trial). Clin Gastroenterol Hepatol. 2022;20:1499-1507.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 147] [Article Influence: 36.8] [Reference Citation Analysis (1)] |
| 178. | Wallace MB, Sharma P, Bhandari P, East J, Antonelli G, Lorenzetti R, Vieth M, Speranza I, Spadaccini M, Desai M, Lukens FJ, Babameto G, Batista D, Singh D, Palmer W, Ramirez F, Palmer R, Lunsford T, Ruff K, Bird-Liebermann E, Ciofoaia V, Arndtz S, Cangemi D, Puddick K, Derfus G, Johal AS, Barawi M, Longo L, Moro L, Repici A, Hassan C. Impact of Artificial Intelligence on Miss Rate of Colorectal Neoplasia. Gastroenterology. 2022;163:295-304.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 169] [Article Influence: 42.3] [Reference Citation Analysis (1)] |
| 179. | Shah S, Park N, Chehade NEH, Chahine A, Monachese M, Tiritilli A, Moosvi Z, Ortizo R, Samarasena J. Effect of computer-aided colonoscopy on adenoma miss rates and polyp detection: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2023;38:162-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 180. | Ahmad OF, González-Bueno Puyal J, Brandao P, Kader R, Abbasi F, Hussein M, Haidry RJ, Toth D, Mountney P, Seward E, Vega R, Stoyanov D, Lovat LB. Performance of artificial intelligence for detection of subtle and advanced colorectal neoplasia. Dig Endosc. 2022;34:862-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 181. | Lin Y, Zhang X, Li F, Zhang R, Jiang H, Lai C, Yi L, Li Z, Wu W, Qiu L, Yang H, Guan Q, Wang Z, Deng L, Zhao Z, Lu W, Lun W, Dai J, He S, Bai Y. A deep neural network improves endoscopic detection of laterally spreading tumors. Surg Endosc. 2025;39:776-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 182. | Zhou G, Xiao X, Tu M, Liu P, Yang D, Liu X, Zhang R, Li L, Lei S, Wang H, Song Y, Wang P. Computer aided detection for laterally spreading tumors and sessile serrated adenomas during colonoscopy. PLoS One. 2020;15:e0231880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 183. | Lui TK, Lam CP, To EW, Ko MK, Tsui VWM, Liu KS, Hui CK, Cheung MK, Mak LL, Hui RW, Wong SY, Seto WK, Leung WK. Endocuff With or Without Artificial Intelligence-Assisted Colonoscopy in Detection of Colorectal Adenoma: A Randomized Colonoscopy Trial. Am J Gastroenterol. 2024;119:1318-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 29] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 184. | Rex DK, Kahi C, O'Brien M, Levin TR, Pohl H, Rastogi A, Burgart L, Imperiale T, Ladabaum U, Cohen J, Lieberman DA. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2011;73:419-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 483] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 185. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 433] [Article Influence: 61.9] [Reference Citation Analysis (0)] |
| 186. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 376] [Article Influence: 47.0] [Reference Citation Analysis (4)] |
| 187. | Hassan C, Balsamo G, Lorenzetti R, Zullo A, Antonelli G. Artificial Intelligence Allows Leaving-In-Situ Colorectal Polyps. Clin Gastroenterol Hepatol. 2022;20:2505-2513.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 188. | Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T, Okamoto A, Minami T, Kono M, Arizumi T, Takenaka M, Hagiwara S, Matsui S, Nishida N, Kashida H, Kudo M. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology. 2017;93 Suppl 1:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 130] [Article Influence: 14.4] [Reference Citation Analysis (1)] |
| 189. | Sánchez-Montes C, Sánchez FJ, Bernal J, Córdova H, López-Cerón M, Cuatrecasas M, Rodríguez de Miguel C, García-Rodríguez A, Garcés-Durán R, Pellisé M, Llach J, Fernández-Esparrach G. Computer-aided prediction of polyp histology on white light colonoscopy using surface pattern analysis. Endoscopy. 2019;51:261-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 190. | Song EM, Park B, Ha CA, Hwang SW, Park SH, Yang DH, Ye BD, Myung SJ, Yang SK, Kim N, Byeon JS. Endoscopic diagnosis and treatment planning for colorectal polyps using a deep-learning model. Sci Rep. 2020;10:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (1)] |
| 191. | Jin EH, Lee D, Bae JH, Kang HY, Kwak MS, Seo JY, Yang JI, Yang SY, Lim SH, Yim JY, Lim JH, Chung GE, Chung SJ, Choi JM, Han YM, Kang SJ, Lee J, Chan Kim H, Kim JS. Improved Accuracy in Optical Diagnosis of Colorectal Polyps Using Convolutional Neural Networks with Visual Explanations. Gastroenterology. 2020;158:2169-2179.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 192. | Zachariah R, Samarasena J, Luba D, Duh E, Dao T, Requa J, Ninh A, Karnes W. Prediction of Polyp Pathology Using Convolutional Neural Networks Achieves "Resect and Discard" Thresholds. Am J Gastroenterol. 2020;115:138-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 193. | Zorron Cheng Tao Pu L, Maicas G, Tian Y, Yamamura T, Nakamura M, Suzuki H, Singh G, Rana K, Hirooka Y, Burt AD, Fujishiro M, Carneiro G, Singh R. Computer-aided diagnosis for characterization of colorectal lesions: comprehensive software that includes differentiation of serrated lesions. Gastrointest Endosc. 2020;92:891-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 194. | Mori Y, Kudo SE, Wakamura K, Misawa M, Ogawa Y, Kutsukawa M, Kudo T, Hayashi T, Miyachi H, Ishida F, Inoue H. Novel computer-aided diagnostic system for colorectal lesions by using endocytoscopy (with videos). Gastrointest Endosc. 2015;81:621-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (1)] |
| 195. | Misawa M, Kudo SE, Mori Y, Nakamura H, Kataoka S, Maeda Y, Kudo T, Hayashi T, Wakamura K, Miyachi H, Katagiri A, Baba T, Ishida F, Inoue H, Nimura Y, Mori K. Characterization of Colorectal Lesions Using a Computer-Aided Diagnostic System for Narrow-Band Imaging Endocytoscopy. Gastroenterology. 2016;150:1531-1532.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 127] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 196. | Kudo SE, Misawa M, Mori Y, Hotta K, Ohtsuka K, Ikematsu H, Saito Y, Takeda K, Nakamura H, Ichimasa K, Ishigaki T, Toyoshima N, Kudo T, Hayashi T, Wakamura K, Baba T, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Artificial Intelligence-assisted System Improves Endoscopic Identification of Colorectal Neoplasms. Clin Gastroenterol Hepatol. 2020;18:1874-1881.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 162] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 197. | Hassan C, Rizkala T, Mori Y, Spadaccini M, Misawa M, Antonelli G, Rondonotti E, Dekker E, Houwen BBSL, Pech O, Baumer S, Li JW, von Renteln D, Haumesser C, Maselli R, Facciorusso A, Correale L, Menini M, Schilirò A, Khalaf K, Patel H, Radadiya DK, Bhandari P, Kudo SE, Sultan S, Vandvik PO, Sharma P, Rex DK, Foroutan F, Repici A; CADx analysis study group. Computer-aided diagnosis for the resect-and-discard strategy for colorectal polyps: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2024;9:1010-1019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 198. | Hassan C, Misawa M, Rizkala T, Mori Y, Sultan S, Facciorusso A, Antonelli G, Spadaccini M, Houwen BBSL, Rondonotti E, Patel H, Khalaf K, Li JW, Fernandez GM, Bhandari P, Dekker E, Gross S, Berzin T, Vandvik PO, Correale L, Kudo SE, Sharma P, Rex DK, Repici A, Foroutan F; CADx Analysis Study Group. Computer-Aided Diagnosis for Leaving Colorectal Polyps In Situ : A Systematic Review and Meta-analysis. Ann Intern Med. 2024;177:919-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 199. | Djinbachian R, Haumesser C, Taghiakbari M, Pohl H, Barkun A, Sidani S, Liu Chen Kiow J, Panzini B, Bouchard S, Deslandres E, Alj A, von Renteln D. Autonomous Artificial Intelligence vs Artificial Intelligence-Assisted Human Optical Diagnosis of Colorectal Polyps: A Randomized Controlled Trial. Gastroenterology. 2024;167:392-399.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 44] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 200. | Minegishi Y, Kudo SE, Miyata Y, Nemoto T, Mori K, Misawa M; Showa University and Nagoya University Ai Research Group. Comprehensive Diagnostic Performance of Real-Time Characterization of Colorectal Lesions Using an Artificial Intelligence-Assisted System: A Prospective Study. Gastroenterology. 2022;163:323-325.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 201. | Houwen BBSL, Hazewinkel Y, Giotis I, Vleugels JLA, Mostafavi NS, van Putten P, Fockens P, Dekker E; POLAR Study Group. Computer-aided diagnosis for optical diagnosis of diminutive colorectal polyps including sessile serrated lesions: a real-time comparison with screening endoscopists. Endoscopy. 2023;55:756-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 202. | Kamitani Y, Nonaka K, Isomoto H. Current Status and Future Perspectives of Artificial Intelligence in Colonoscopy. J Clin Med. 2022;11:2923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 203. | Takeda K, Kudo SE, Mori Y, Misawa M, Kudo T, Wakamura K, Katagiri A, Baba T, Hidaka E, Ishida F, Inoue H, Oda M, Mori K. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy. 2017;49:798-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 106] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 204. | Tamai N, Saito Y, Sakamoto T, Nakajima T, Matsuda T, Sumiyama K, Tajiri H, Koyama R, Kido S. Effectiveness of computer-aided diagnosis of colorectal lesions using novel software for magnifying narrow-band imaging: a pilot study. Endosc Int Open. 2017;5:E690-E694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 205. | Tokunaga M, Matsumura T, Nankinzan R, Suzuki T, Oura H, Kaneko T, Fujie M, Hirai S, Saiki R, Akizue N, Okimoto K, Arai M, Kato J, Kato N. Computer-aided diagnosis system using only white-light endoscopy for the prediction of invasion depth in colorectal cancer. Gastrointest Endosc. 2021;93:647-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 206. | Barua I, Vinsard DG, Jodal HC, Løberg M, Kalager M, Holme Ø, Misawa M, Bretthauer M, Mori Y. Artificial intelligence for polyp detection during colonoscopy: a systematic review and meta-analysis. Endoscopy. 2021;53:277-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 178] [Article Influence: 35.6] [Reference Citation Analysis (1)] |
| 207. | Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, Antonelli G, Yu H, Areia M, Dinis-Ribeiro M, Bhandari P, Sharma P, Rex DK, Rösch T, Wallace M, Repici A. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc. 2021;93:77-85.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 351] [Article Influence: 70.2] [Reference Citation Analysis (5)] |
| 208. | Barua I, Wieszczy P, Kudo SE, Misawa M, Holme Ø, Gulati S, Williams S, Mori K, Itoh H, Takishima K, Mochizuki K, Miyata Y, Mochida K, Akimoto Y, Kuroki T, Morita Y, Shiina O, Kato S, Nemoto T, Hayee B, Patel M, Gunasingam N, Kent A, Emmanuel A, Munck C, Nilsen JA, Hvattum SA, Frigstad SO, Tandberg P, Løberg M, Kalager M, Haji A, Bretthauer M, Mori Y. Real-Time Artificial Intelligence-Based Optical Diagnosis of Neoplastic Polyps during Colonoscopy. NEJM Evid. 2022;1:EVIDoa2200003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 209. | Rondonotti E, Hassan C, Tamanini G, Antonelli G, Andrisani G, Leonetti G, Paggi S, Amato A, Scardino G, Di Paolo D, Mandelli G, Lenoci N, Terreni N, Andrealli A, Maselli R, Spadaccini M, Galtieri PA, Correale L, Repici A, Di Matteo FM, Ambrosiani L, Filippi E, Sharma P, Radaelli F. Artificial intelligence-assisted optical diagnosis for the resect-and-discard strategy in clinical practice: the Artificial intelligence BLI Characterization (ABC) study. Endoscopy. 2023;55:14-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 88] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 210. | Zhang C, Yao L, Jiang R, Wang J, Wu H, Li X, Wu Z, Luo R, Luo C, Tan X, Wang W, Xiao B, Hu H, Yu H. Assessment of the role of false-positive alerts in computer-aided polyp detection for assistance capabilities. J Gastroenterol Hepatol. 2024;39:1623-1635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 211. | Okumura T, Imai K, Misawa M, Kudo SE, Hotta K, Ito S, Kishida Y, Takada K, Kawata N, Maeda Y, Yoshida M, Yamamoto Y, Minamide T, Ishiwatari H, Sato J, Matsubayashi H, Ono H. Evaluating false-positive detection in a computer-aided detection system for colonoscopy. J Gastroenterol Hepatol. 2024;39:927-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 212. | Sultan S, Shung DL, Kolb JM, Foroutan F, Hassan C, Kahi CJ, Liang PS, Levin TR, Siddique SM, Lebwohl B. AGA Living Clinical Practice Guideline on Computer-Aided Detection-Assisted Colonoscopy. Gastroenterology. 2025;168:691-700. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 25] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 213. | Li BP, Meng MQ. Comparison of several texture features for tumor detection in CE images. J Med Syst. 2012;36:2463-2469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 214. | Liu G, Yan G, Kuang S, Wang Y. Detection of small bowel tumor based on multi-scale curvelet analysis and fractal technology in capsule endoscopy. Comput Biol Med. 2016;70:131-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 215. | Faghih Dinevari V, Karimian Khosroshahi G, Zolfy Lighvan M. Singular Value Decomposition Based Features for Automatic Tumor Detection in Wireless Capsule Endoscopy Images. Appl Bionics Biomech. 2016;2016:3678913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 216. | Yuan Y, Meng MQ. Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys. 2017;44:1379-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (1)] |
| 217. | Ding Z, Shi H, Zhang H, Meng L, Fan M, Han C, Zhang K, Ming F, Xie X, Liu H, Liu J, Lin R, Hou X. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology. 2019;157:1044-1054.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 221] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 218. | Saito H, Aoki T, Aoyama K, Kato Y, Tsuboi A, Yamada A, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. 2020;92:144-151.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (1)] |
| 219. | Xie X, Xiao YF, Zhao XY, Li JJ, Yang QQ, Peng X, Nie XB, Zhou JY, Zhao YB, Yang H, Liu X, Liu E, Chen YY, Zhou YY, Fan CQ, Bai JY, Lin H, Koulaouzidis A, Yang SM. Development and Validation of an Artificial Intelligence Model for Small Bowel Capsule Endoscopy Video Review. JAMA Netw Open. 2022;5:e2221992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 220. | Inoue S, Shichijo S, Aoyama K, Kono M, Fukuda H, Shimamoto Y, Nakagawa K, Ohmori M, Iwagami H, Matsuno K, Iwatsubo T, Nakahira H, Matsuura N, Maekawa A, Kanesaka T, Yamamoto S, Takeuchi Y, Higashino K, Uedo N, Ishihara R, Tada T. Application of Convolutional Neural Networks for Detection of Superficial Nonampullary Duodenal Epithelial Tumors in Esophagogastroduodenoscopic Images. Clin Transl Gastroenterol. 2020;11:e00154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 221. | Ding Z, Shi H, Zhang H, Zhang H, Tian S, Zhang K, Cai S, Ming F, Xie X, Liu J, Lin R. Artificial intelligence-based diagnosis of abnormalities in small-bowel capsule endoscopy. Endoscopy. 2023;55:44-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 222. | Park SY, Sargent D. Colonoscopic polyp detection using convolutional neural networks. SPIE Proceedings. 2016;9785:978528. [DOI] [Full Text] |
| 223. | Billah M, Waheed S, Rahman MM. An Automatic Gastrointestinal Polyp Detection System in Video Endoscopy Using Fusion of Color Wavelet and Convolutional Neural Network Features. Int J Biomed Imaging. 2017;2017:9545920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 224. | Lequan Yu, Hao Chen, Qi Dou, Jing Qin, Pheng Ann Heng. Integrating Online and Offline Three-Dimensional Deep Learning for Automated Polyp Detection in Colonoscopy Videos. IEEE J Biomed Health Inform. 2017;21:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 225. | Zhang R, Zheng Y, Mak TW, Yu R, Wong SH, Lau JY, Poon CC. Automatic Detection and Classification of Colorectal Polyps by Transferring Low-Level CNN Features From Nonmedical Domain. IEEE J Biomed Health Inform. 2017;21:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 172] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 226. | Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, Nakamura H, Yagawa Y, Toyoshima N, Ogata N, Kudo T, Hisayuki T, Hayashi T, Wakamura K, Baba T, Ishida F, Itoh H, Roth H, Oda M, Mori K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology. 2018;154:2027-2029.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 279] [Article Influence: 34.9] [Reference Citation Analysis (1)] |
| 227. | Yamada M, Saito Y, Imaoka H, Saiko M, Yamada S, Kondo H, Takamaru H, Sakamoto T, Sese J, Kuchiba A, Shibata T, Hamamoto R. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci Rep. 2019;9:14465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 170] [Article Influence: 24.3] [Reference Citation Analysis (2)] |
| 228. | Klare P, Sander C, Prinzen M, Haller B, Nowack S, Abdelhafez M, Poszler A, Brown H, Wilhelm D, Schmid RM, von Delius S, Wittenberg T. Automated polyp detection in the colorectum: a prospective study (with videos). Gastrointest Endosc. 2019;89:576-582.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 229. | Ozawa T, Ishihara S, Fujishiro M, Kumagai Y, Shichijo S, Tada T. Automated endoscopic detection and classification of colorectal polyps using convolutional neural networks. Therap Adv Gastroenterol. 2020;13:1756284820910659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 230. | Xu H, Tang RSY, Lam TYT, Zhao G, Lau JYW, Liu Y, Wu Q, Rong L, Xu W, Li X, Wong SH, Cai S, Wang J, Liu G, Ma T, Liang X, Mak JWY, Xu H, Yuan P, Cao T, Li F, Ye Z, Shutian Z, Sung JJY. Artificial Intelligence-Assisted Colonoscopy for Colorectal Cancer Screening: A Multicenter Randomized Controlled Trial. Clin Gastroenterol Hepatol. 2023;21:337-346.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 132] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 231. | Gimeno-García AZ, Hernández Negrin D, Hernández A, Nicolás-Pérez D, Rodríguez E, Montesdeoca C, Alarcon O, Romero R, Baute Dorta JL, Cedrés Y, Castillo RD, Jiménez A, Felipe V, Morales D, Ortega J, Reygosa C, Quintero E, Hernández-Guerra M. Usefulness of a novel computer-aided detection system for colorectal neoplasia: a randomized controlled trial. Gastrointest Endosc. 2023;97:528-536.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 46] [Reference Citation Analysis (0)] |
| 232. | Karsenti D, Tharsis G, Perrot B, Cattan P, Percie du Sert A, Venezia F, Zrihen E, Gillet A, Lab JP, Tordjman G, Cavicchi M. Effect of real-time computer-aided detection of colorectal adenoma in routine colonoscopy (COLO-GENIUS): a single-centre randomised controlled trial. Lancet Gastroenterol Hepatol. 2023;8:726-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 233. | Maas MHJ, Neumann H, Shirin H, Katz LH, Benson AA, Kahloon A, Soons E, Hazzan R, Landsman MJ, Lebwohl B, Lewis SK, Sivanathan V, Ngamruengphong S, Jacob H, Siersema PD. A computer-aided polyp detection system in screening and surveillance colonoscopy: an international, multicentre, randomised, tandem trial. Lancet Digit Health. 2024;6:e157-e165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 234. | Thiruvengadam NR, Solaimani P, Shrestha M, Buller S, Carson R, Reyes-Garcia B, Gnass RD, Wang B, Albasha N, Leonor P, Saumoy M, Coimbra R, Tabuenca A, Srikureja W, Serrao S. The Efficacy of Real-time Computer-aided Detection of Colonic Neoplasia in Community Practice: A Pragmatic Randomized Controlled Trial. Clin Gastroenterol Hepatol. 2024;22:2221-2230.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 235. | Park DK, Kim EJ, Im JP, Lim H, Lim YJ, Byeon JS, Kim KO, Chung JW, Kim YJ. A prospective multicenter randomized controlled trial on artificial intelligence assisted colonoscopy for enhanced polyp detection. Sci Rep. 2024;14:25453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 236. | Tischendorf JJ, Gross S, Winograd R, Hecker H, Auer R, Behrens A, Trautwein C, Aach T, Stehle T. Computer-aided classification of colorectal polyps based on vascular patterns: a pilot study. Endoscopy. 2010;42:203-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 99] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 237. | Takemura Y, Yoshida S, Tanaka S, Onji K, Oka S, Tamaki T, Kaneda K, Yoshihara M, Chayama K. Quantitative analysis and development of a computer-aided system for identification of regular pit patterns of colorectal lesions. Gastrointest Endosc. 2010;72:1047-1051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (2)] |
| 238. | Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 284] [Article Influence: 35.5] [Reference Citation Analysis (1)] |
| 239. | Gross S, Trautwein C, Behrens A, Winograd R, Palm S, Lutz HH, Schirin-Sokhan R, Hecker H, Aach T, Tischendorf JJ. Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest Endosc. 2011;74:1354-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 99] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 240. | Kominami Y, Yoshida S, Tanaka S, Sanomura Y, Hirakawa T, Raytchev B, Tamaki T, Koide T, Kaneda K, Chayama K. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc. 2016;83:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 163] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 241. | Li JW, Wu CCH, Lee JWJ, Liang R, Soon GST, Wang LM, Koh XH, Koh CJ, Chew WD, Lin KW, Thian MY, Matthew R, Kim G, Khor CJL, Fock KM, Ang TL, So JBY; Artificial Intelligence in Gastrointestinal Endoscopy Singapore (AIGES) Study Group. Real-World Validation of a Computer-Aided Diagnosis System for Prediction of Polyp Histology in Colonoscopy: A Prospective Multicenter Study. Am J Gastroenterol. 2023;118:1353-1364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 36] [Reference Citation Analysis (0)] |
| 242. | Hassan C, Sharma P, Mori Y, Bretthauer M, Rex DK; Combo Study Group, Repici A. Comparative Performance of Artificial Intelligence Optical Diagnosis Systems for Leaving in Situ Colorectal Polyps. Gastroenterology. 2023;164:467-469.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (1)] |
| 243. | Rex DK, Bhavsar-Burke I, Buckles D, Burton J, Cartee A, Comar K, Edwards A, Fennimore B, Fischer M, Gerich M, Gilmore A, Hamdeh S, Hoffman J, Ibach M, Jackson M, James-Stevenson T, Kaltenbach T, Kaplan J, Kapur S, Kohm D, Kriss M, Kundumadam S, Kyanam Kabir Baig KR, Menard-Katcher P, Kraft C, Langworthy J, Misra B, Molloy E, Munoz JC, Norvell J, Nowak T, Obaitan I, Patel S, Patel M, Peter S, Reid BM, Rogers N, Ross J, Ryan J, Sagi S, Saito A, Samo S, Sarkis F, Scott FI, Siwiec R, Sullivan S, Wieland A, Zhang J, Repici A, Hassan C, Byrne MF, Rastogi A. Artificial Intelligence for Real-Time Prediction of the Histology of Colorectal Polyps by General Endoscopists. Ann Intern Med. 2024;177:911-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 35] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/