Published online Nov 14, 2025. doi: 10.3748/wjg.v31.i42.112196
Revised: August 20, 2025
Accepted: October 11, 2025
Published online: November 14, 2025
Processing time: 116 Days and 0.5 Hours
Artificial intelligence (AI)-augmented contrast-enhanced ultrasonography (CEUS) is emerging as a powerful tool in liver imaging, particularly in enhancing the accuracy of Liver Imaging Reporting and Data System (known as LI-RADS) classification. This review synthesized published data on the integration of machine learning and deep learning techniques into CEUS, revealing that AI algorithms can improve the detection and quantification of contrast enhancement patterns. Such improvements led to more consistent LI-RADS categorization, reduced interoperator variability, and enabled real-time analysis that streamlined work
Core Tip: Artificial intelligence (AI) has shown increasing potential in enhancing liver imaging workflows. This review focused on the integration of AI into contrast-enhanced ultrasound for liver lesion assessment with an emphasis on automating the Liver Imaging Reporting and Data System classification. We summarized recent deep learning architectures, radiomics applications, and clinical impact studies, highlighting both diagnostic performance and technical challenges. The review provided a forward-looking perspective on how contrast-enhanced ultrasound-based AI models may standardize interpretation, support real-time decision-making, and transform hepatocellular carcinoma diagnosis in clinical practice.
- Citation: Ciocalteu A, Urhut CM, Streba CT, Kamal A, Mamuleanu M, Sandulescu LD. Artificial intelligence in contrast enhanced ultrasound: A new era for liver lesion assessment. World J Gastroenterol 2025; 31(42): 112196
- URL: https://www.wjgnet.com/1007-9327/full/v31/i42/112196.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i42.112196
International hepatology society guidelines have established contrast-enhanced computed tomography (CT) and contrast-enhanced magnetic resonance imaging (MRI) as the imaging modalities of choice for diagnosing hepatocellular carcinoma (HCC) lesions larger than 1 cm. MRI remains the gold standard for detecting small HCC nodules in cirrhotic livers due to its superior soft-tissue contrast and functional imaging capabilities. However, early or atypical presentations remain challenging for differential diagnosis, staging, and treatment planning. In these scenarios contrast-enhanced ultrasonography (CEUS) is a valuable second-line tool, offering real-time, radiation-free evaluation and repeatability for follow-up. A recent meta-analysis of head-to-head studies reported comparable diagnostic performance between CEUS and CT/MRI with pooled sensitivities and specificities of 0.67/0.88 for CEUS vs 0.60/0.98 for CT/MRI in non-HCC malignancies, and similar specificities for HCC diagnosis (0.70 for CEUS vs 0.59 for CT; 0.81 for CEUS vs 0.79 for MRI)[1]. Given the limitations of individual imaging modalities, hybrid techniques and multimodal approaches are gaining traction for improving lesion detection, especially in cases where standard methods fall short. Artificial intelligence (AI) has emerged as a powerful tool in medical imaging, enhancing diagnostic accuracy and reliability across platforms. In CEUS liver imaging dynamic enhancement patterns often challenge consistent interpretation across observers. AI holds particular promise for standardizing assessments.
The growing complexity of liver tumor evaluation has also driven interest in approaches that integrate serum bio
In the methodological process of this narrative mini-review, the literature selection was primarily based on targeted PubMed searches. ChatGPT-4o (OpenAI)[4] was employed to assist in refining query parameters and identifying relevant, up-to-date peer-reviewed sources on CEUS-based AI applications.
Recent advancements in AI have improved imaging-based liver disease assessment by enhancing pattern recognition, automating fibrosis and steatosis quantification and aiding in HCC detection[5]. Moreover, AI can identify and categorize liver lesions as benign or malignant using CEUS and complementary imaging modalities[6]. These capabilities support biopsy decision-making, enhance interventional planning, and contribute to greater diagnostic accuracy in liver cancer management.
Deep learning (DL) models, particularly convolutional neural networks (CNNs), have shown strong capabilities in analyzing ultrasound scans and identifying early liver lesions with high sensitivity and pattern recognition potential[7]. Feng et al[8] developed a deep neural network that extracted multiview perfusion patterns from CEUS images, achieving an area under the curve (AUC) of 0.89 in differentiating HCC from other malignant lesions in a multicenter cohort of 1241 patients. Early efforts in quantitative CEUS analysis explored the use of computer-assisted methods to objectively characterize contrast dynamics and vascular patterns. For example, Denis de Senneville et al[9] demonstrated that automated evaluation of microbubble transport patterns could effectively differentiate between focal nodular hyperplasia (FNH) and inflammatory hepatocellular adenoma, outperforming subjective visual assessment. While this approach did not fall within the formal scope of radiomics, it laid important groundwork for the development of more sophisticated image-based feature extraction techniques such as radiomics.
Notably, while Feng et al[8] leveraged multiview perfusion patterns to capture enhancement dynamics from multiple CEUS planes, potentially improving lesion characterization in heterogeneous livers, this approach may be more sensitive to motion artifacts and requires standardized acquisition of multiple views, which can be challenging in routine practice. In contrast, Denis de Senneville et al[9] employed a physics-informed model of microbubble transport, offering robustness in capturing vascular patterns but relying heavily on precise timing and quality of contrast injection. Both strategies show strong diagnostic performance, yet their dependence on specific CEUS phases and acquisition protocols as well as the potential variability in contrast agent administration (e.g., timing, dosage, injection technique) suggests that optimal clinical use may require standardized imaging and injection protocols tailored to local practices and operator expertise. Such variability could still introduce bias even with AI-based automation underscoring the need for rigorous standardization in both clinical practice and multicenter research. Moreover, these results must be viewed in the context of broader challenges for AI in CEUS, such as limited generalizability, reduced interpretability of DL models, and domain shift when applied to data from unfamiliar scanners, protocols, or populations.
Radiomics aims to enhance the diagnosis and prognosis of HCC by leveraging AI techniques to extract clinically relevant information embedded within medical images[10]. It has emerged as a computational approach to identify and validate candidate imaging biomarkers for HCC, supporting a transition toward data-driven precision medicine[2]. The extracted data, known as “radiomic features”, are used in AI-based algorithms to predict clinical endpoints such as histopathological characteristics, tumor aggressiveness, treatment response, and patient outcomes, often surpassing the capabilities of conventional visual assessment[11]. Morphological and hemodynamic alterations remain key diagnostic challenges in HCC and are not fully addressed by current imaging methods. Radiomics is therefore expected to offer a potential solution by quantitatively capturing these subtle features[12].
To date several studies have illustrated how CEUS-based radiomics and DL are transitioning into actionable clinical tools. Liu et al[13] evaluated a DL-based radiomics approach using CEUS to predict progression-free survival after radiofrequency ablation or surgical resection and to optimize treatment selection in very-early and early-stage HCC. Their proposed models and nomograms provided accurate preoperative prediction of progression-free survival and could assist treatment selection between radiofrequency ablation and surgical resection.
Recently, a CEUS-based ultrasomics model was developed to differentiate HCC from intrahepatic cholangiocarcinoma. By integrating CEUS with clinical data, the model achieved an AUC of approximately 0.97, outperforming experienced radiologists although reports remain limited[14]. A key study by Hu et al[15] demonstrated that a DL model trained on CEUS cine loops could reliably distinguish malignant from benign liver lesions. The model achieved an AUC of 0.92 in the validation cohort, significantly outperforming both junior and senior radiologists in accuracy and specificity. This superior performance can be attributed to the CNN-long short-term memory architecture, which captures both spatial and temporal dynamics of CEUS cine loops unlike traditional radiomics approaches limited to static images. These findings highlight the potential of AI to reduce diagnostic variability and improve early lesion characterization.
To address ongoing challenges in differential diagnosis, a recent multicenter study[16] developed a combined radiomics model based on Sonazoid-enhanced CEUS (Kupffer phase) to distinguish well-differentiated HCC from benign focal liver lesions (FLLs). By integrating radiomic features with clinical data, the model achieved an AUC of approximately 0.91, suggesting utility in reducing unnecessary biopsies in indeterminate cases, particularly in cirrhotic livers. However, clinical validation in broader patient cohorts remains limited.
Machine learning (ML), a subfield of AI, focuses on algorithms that learn patterns from data without explicit pro
ML-based radiomics, or “ultrasomics”, has shown promise even in conventional B-mode ultrasound. While radiomics refers broadly to the extraction of quantitative features from medical images, ultrasomics designates the application of this concept specifically to ultrasound data, making it a subset of radiomics (Table 1). For example, Li et al[19] investigated the utility of a ML-based ultrasomics approach to differentiate between FNH and atypical HCC using B-mode ultrasound features. The model significantly outperformed conventional sonographic assessment, achieving an AUC of 0.92 in the validation cohort.
| Term | Definition | CEUS application |
| CNN | Deep learning model using convolutional layers to extract image features | Lesion detection, classification, segmentation |
| Radiomics | Extraction of quantitative handcrafted features from medical images | Supports AI-based lesion characterization |
| Ultrasomics | Radiomics applied specifically to ultrasound images | CEUS feature analysis for HCC risk stratification |
| Transformers | AI models using self-attention to learn feature relationships | Modeling CEUS video sequences for enhancement pattern recognition |
DL, a specialized branch of ML, achieves its performance by structuring information as hierarchical representations in which complex concepts are built upon simpler features[20]. DL uses multilayer neural networks (most notably CNNs) to extract and analyze complex patterns within high-dimensional data. These models are valued for their high sensitivity and strong pattern recognition in early liver lesion detection from ultrasound scans.
Among DL techniques CNNs have proven to be particularly efficient in processing visual data. They are especially effective for medical imaging tasks such as classification (distinguishing tissue types or lesion features), detection (localizing anomalies in images), and segmentation (delineating structures such as lesions or organs)[7].
A CEUS-based DL system distinguished between benign and malignant HCC with high specificity, outperforming two experienced clinicians (100% vs 93.33%), suggesting a strong potential for clinical integration as a supportive diagnostic tool[21]. Zhang and Huo[22] evaluated the use of a CEUS-based DL model to predict early recurrence of HCC preoperatively. The model demonstrated high predictive performance, significantly outperforming the traditional radiomics approach. Also, cross-institutional validation demonstrated that DL outperformed radiomics in terms of robustness for preoperative prediction of microvascular invasion in patients with HCC, offering a promising approach to overcome the variability inherent in non-standardized ultrasound imaging protocols[23]. Yet further improvements in generalizability are still required before clinical use.
In order to enhance clarity and accessibility for AI terminology, we have summarized the main AI concepts, together with their specific applications in CEUS-based liver lesion assessment in Table 1.
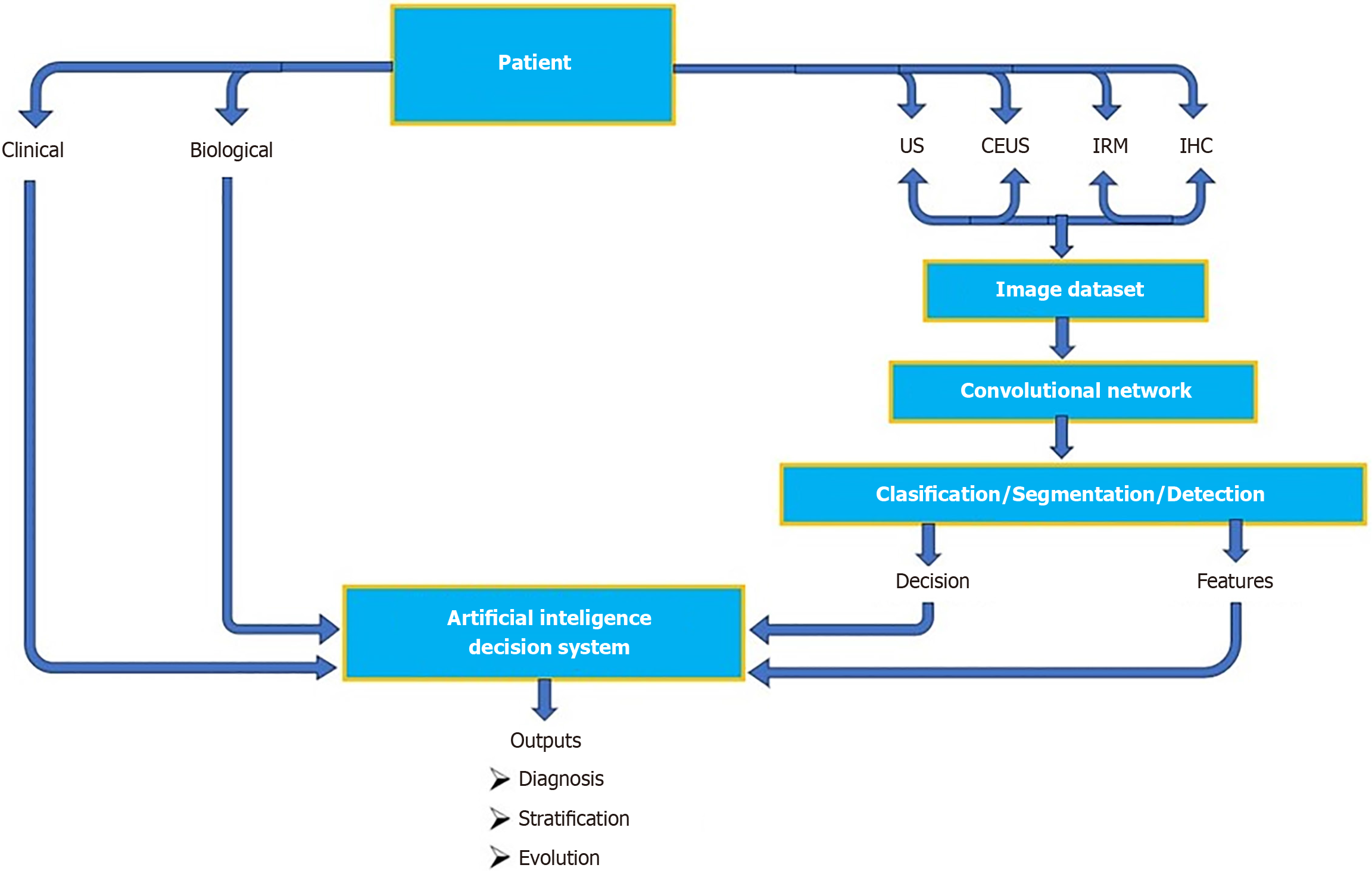
To provide standardization of liver imaging for HCC, LI-RADS[24] was created and supported by the American College of Radiology (commonly referred to as the ACR) (Table 2). The growing complexity of the LI-RADS system has limited its practicality in high-volume clinical settings, hindering widespread adoption. To address this, researchers have turned to AI-based imaging solutions aimed at streamlining workflow through automated lesion detection, classification, and standardized reporting. An example of an AI-driven decision workflow integrating CEUS and multimodal data is pro
| LI-RADS category | Interpretation |
| LR-1 | Definitely benign |
| LR-2 | Probably benign |
| LR-3 | Intermediate probability of malignancy |
| LR-4 | Probably HCC |
| LR-5 | Definitely HCC |
| LR-M | Probably/definite malignancy non-HCC specific |
Foundational studies established the viability of standard CNN models in classifying liver pathology from ultrasound images[25,26]. However, these early models lacked tailored modules for multiphase feature recognition or stepwise interpretability, approaches central to the newer LI-RADS framework. By introducing explicit feature-characterization layers, the latest works represent a meaningful advancement: It goes beyond treating LI-RADS categories as black-box labels and begins to emulate the imagists’ interpretative reasoning, making it the first step toward explainable, clinically aligned DL-assisted LI-RADS scoring.
Thus, building on MRI studies that demonstrated fully automated LI-RADS scoring through DL-based liver and HCC segmentation[27], recent work has extended this approach to CEUS. A U-Net-based model achieved accurate region-of-interest delineation across dynamic CEUS phases, enabling real-time processing and supporting more workflow ef
Xiao et al[29] developed an ML model using quantitative parameters from dynamic CEUS. The model effectively differentiated HCC from non-HCC lesions, including challenging cases categorized as “LI-RADS-M” by capturing contrast-enhancement patterns. Their multicenter results support the use of AI as a valuable adjunct for improving the accuracy and consistency of CEUS-based LI-RADS classification. Extending this application, Yang et al[30] explored correlations between CEUS LI-RADS categories and histopathological differentiation of combined hepatocellular-cholangiocarcinoma. Higher LI-RADS categories were significantly associated with poorer tumor differentiation, suggesting that CEUS-based LI-RADS scoring may offer prognostic as well as diagnostic value in complex liver tumor subtypes.
Recently, Ding et al[31] proposed an AI model based on CEUS for the automated multiclass classification of FLLs, aiming to emulate an LI-RADS-like diagnostic approach. Trained on a multicenter dataset of 1344 Lesions (including HCC, hemangioma, hepatic adenoma, FNH, and metastases), the hybrid CNN-transformer model achieved an external test accuracy of 88.1% with AUCs between 0.93 and 0.97. A notable innovation was the temporal modelling of CEUS enhancement dynamics, arterial, portal, and late phases, replicating the stepwise interpretation characteristic of LI-RADS. The model identified key imaging features such as arterial phase hyperenhancement, washout, and capsule appearance at a level comparable with expert radiologists, underscoring its potential for standardized, CEUS-based lesion classification. Weakly supervised DL approaches applied to CEUS have shown promising performance in automating LI-RADS cate
A recent study[32] analyzed 955938 CEUS images from 370 patients using case-level labels only, achieving impressive diagnostic performance with an AUC of 0.94 along with 80.0% accuracy, 81.8% sensitivity, and 84.6% specificity. This approach markedly reduces the dependence on labor-intensive annotated datasets and streamlines time-consuming steps within the LIRADS workflow, making a compelling case for scalable, real-world deployment in CEUS-based liver lesion evaluation.
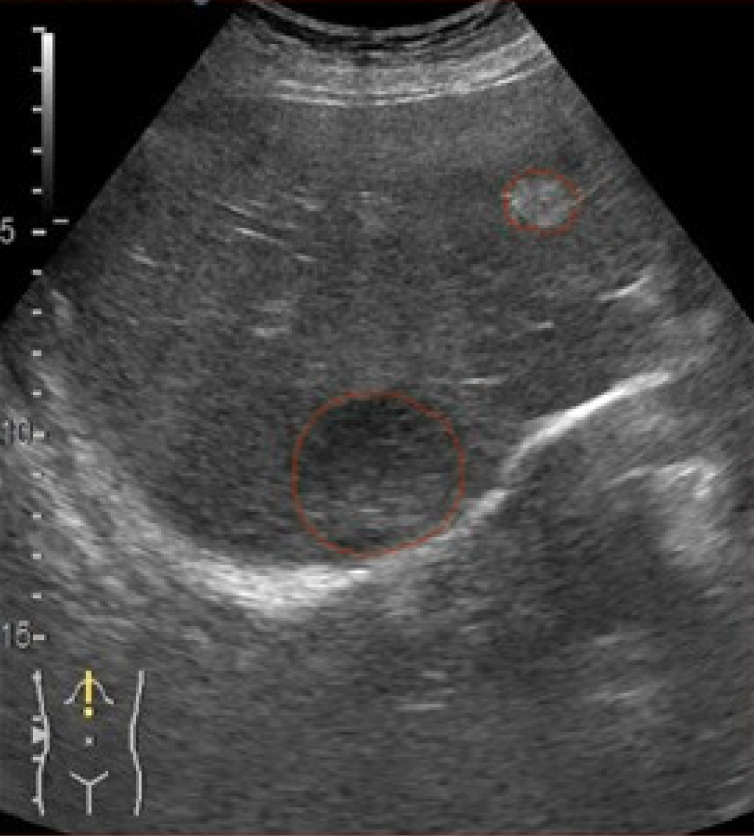
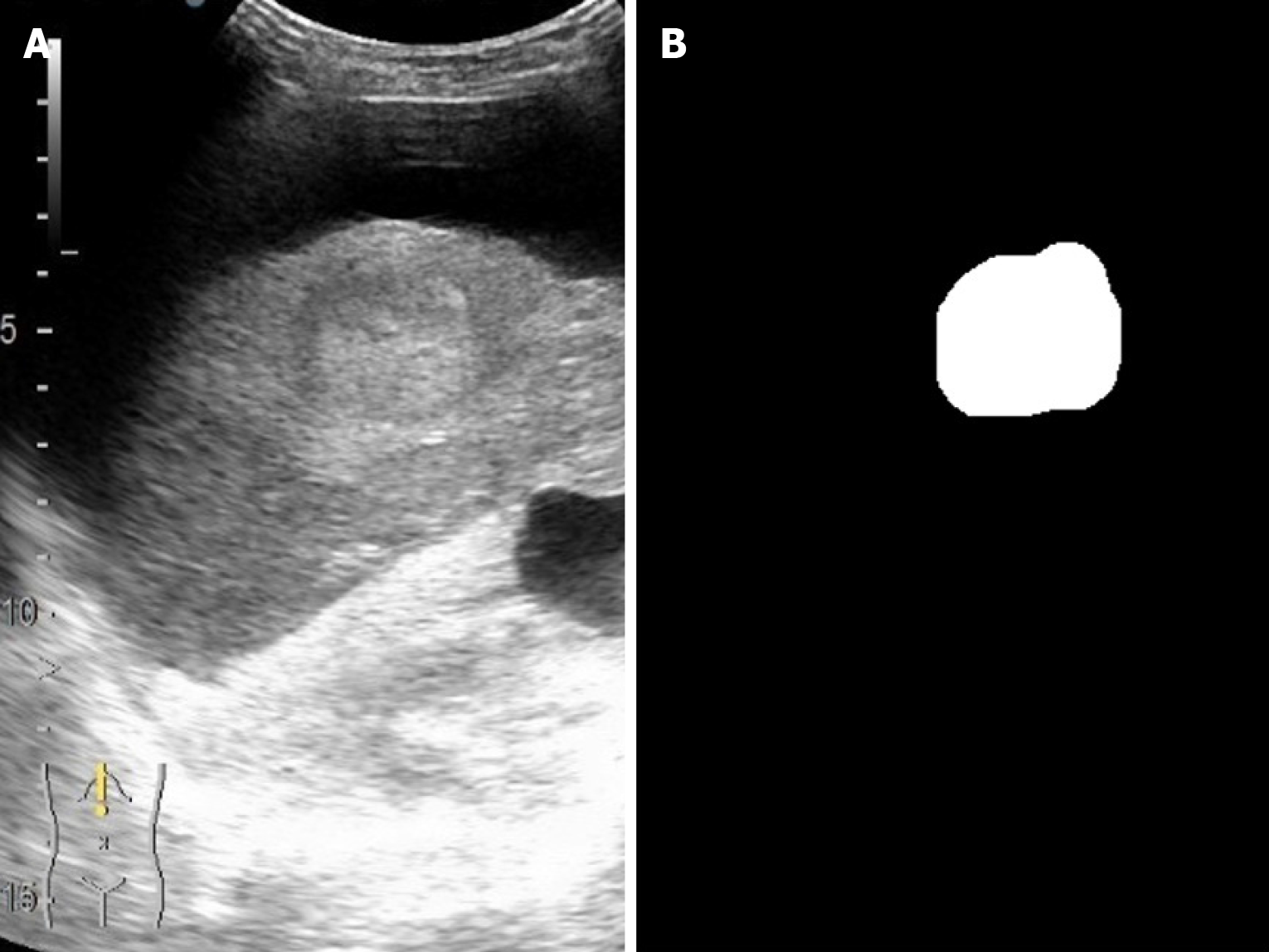
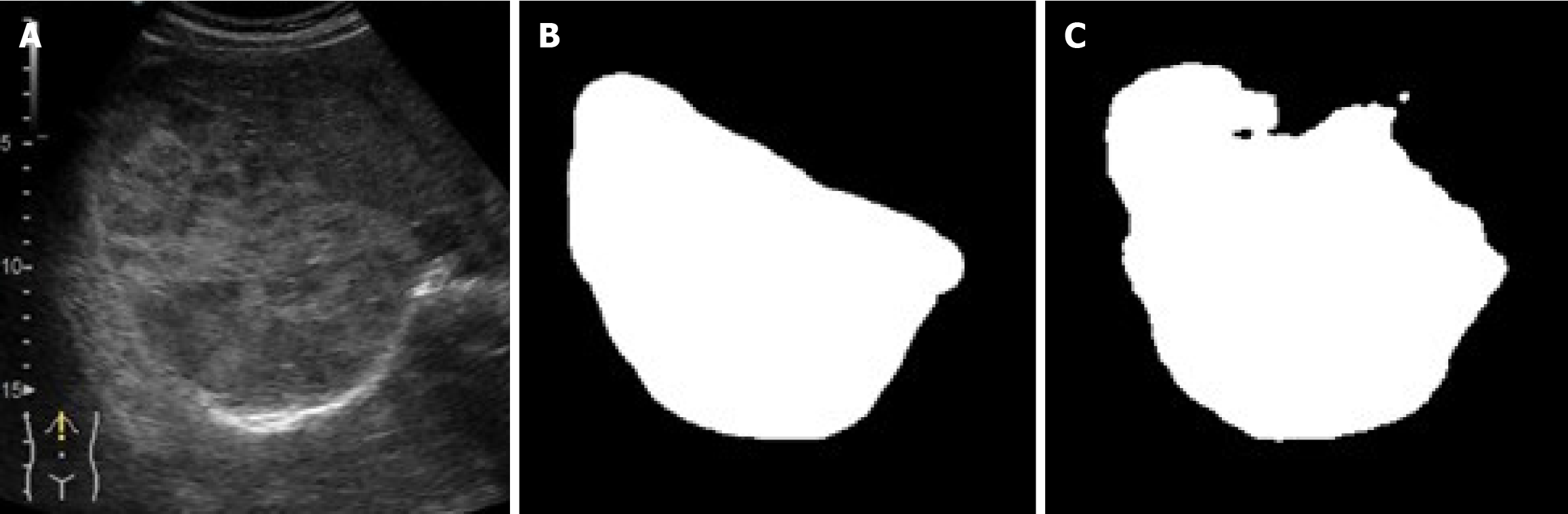
As illustrated in figures below, the U-Net segmentation output closely matches the gastroenterologist’s manual mask, underscoring the ability of the model to reproduce expert-level lesion delineation[21,28]. In Figure 3 the AI output closely matches the manual segmentation with high overlap and accurate lesion boundary delineation, reducing interoperator variability and supporting standardized LI-RADS-based interpretation. Figure 4 demonstrates high intersection over union, indicating strong boundary agreement and high precision, meaning most pixels identified as lesion were correct. Together, these results confirm the robustness and clinical applicability of the model for reproducible CEUS liver lesion analysis, treatment planning, and follow-up (Table 3).
| Ref. | Comparison | Main findings |
| Urhuț et al[21] | Clinicians vs AI model (CEUS) | For differentiating benign from malignant liver tumors, the AI system showed higher specificity than both experienced readers (blinded and unblinded) but lower sensitivity; less accurate for HCC and metastases yet may assist less-experienced clinicians |
| Hu et al[15] | Senior radiologists vs DL model (CEUS) | AI outperformed residents (accuracy 82.9%-84.4%, P = 0.038) and matched experts (87.2%-88.2%, P = 0.438), improving resident performance and reducing CEUS interobserver variability in differentiating benign from malignant |
| Zhou et al[34] | 3D-CNN vs CNN + LSTM (CEUS cine-loops) | High overall AUC (approximately 0.91) for CNN + LSTM, outperforming TIC and 3D-CNN by balancing sensitivity and specificity (3D-CNN: 0.96/0.55), narrowing accuracy gap between less-experienced and more-experienced radiologists (0.82 → 0.87); accuracy for benign vs malignant differentiation (n = 210 Lesions): 0.82 for less-experienced radiologists, 0.87 after AI assistance |
| Oezsoy et al[32] | Weakly supervised DL vs manual LI-RADS scoring | Model matched expert performance using only case-level labels with high accuracy (AUC 0.94) |
AI and DL have shown significant potential to improve efficiency and accuracy in CEUS liver imaging, moving beyond proof-of-concept toward real-world clinical integration. Liu et al[33] proposed a dynamic DL radiomics model (FS3D U) combined with serum markers (alpha-fetoprotein and hepatitis status) that demonstrated superior diagnostic effectiveness in differentiating FLLs. The model achieved AUCs of 0.97 and 0.96 in internal and external cohorts, respectively, and delivered results in under 10 seconds per case, substantially faster than human radiologists whose average inter
Similarly, Zhou et al[34] introduced an end-to-end DL model (CNN + long short-term memory) for CEUS video analysis, achieving an AUC of 0.91 for malignancy prediction and outperforming traditional time-intensity curve analysis and standard three-dimensional CNNs. The end-to-end implementation without manual feature extraction highlights its potential for real-world integration into ultrasound software systems. Moreover, a recent integrative review by Yin et al[5] highlighted the expanding role of AI in liver imaging, noting its capacity to speed pattern recognition, automate dia
Taken together, these studies illustrate the tangible benefits of AI in CEUS: Improved standardization; faster in
| Objective | Clinical impact |
| Reduction in interpretation time | AI-assisted models provide results in approximately 10 seconds, faster than manual reading (23-29 seconds)[33] |
| Improved diagnostic accuracy | Deep learning models achieve AUCs of 0.96-0.97 for benign vs malignant lesions[31,33] |
| Fully automated workflows | End-to-end segmentation and classification eliminate manual intervention[34] |
| Integration into ultrasound systems | Real-time AI implementation feasible within existing CEUS devices[28] |
| Reduction of annotation workload | Weakly supervised learning reduces dependence on manually labeled training data[32] |
| Enhanced LI-RADS standardization | AI models align closely with LI-RADS criteria, improving consistency and objectivity[15,24] |
While several studies compare AI performance directly with that of radiologists[32,34], most do not explicitly account for interobserver variability among human readers when interpreting CEUS images, limiting assessment of true clinical impact. Particularly, Hu et al[15] showed that CEUS-based AI improved resident performance and reduced interobserver heterogeneity.
Although CEUS demonstrates sensitivity for liver metastases comparable to contrast-enhanced CT and superior to conventional ultrasound[36], the role of AI in this context remains underexplored with only preliminary work[37] on highly selected population that restricts generalizability of the findings.
Despite its promise the adoption of AI in HCC surveillance faces challenges, including data heterogeneity, algorithm standardization, and ethical considerations, necessitating further validation in real-world settings[7,37].
The HCC-CDSS may serve as a valuable adjunct for physicians to improve diagnostic precision and guide therapeutic decision-making through personalized algorithms[17]. A CDSS incorporating CEUS data could further enhance its utility by integrating real-time, radiation-free imaging parameters, particularly in resource-limited settings where access to CT or MRI is restricted. Future multicenter validation studies are essential to compare treatment strategies across institutions and to tailor decision models according to institutional expertise and patient-specific variables.
CEUS-based radiomics remains limited by several technical and methodological barriers. Compared with CT or MRI, ultrasound is inherently more operator-dependent, has lower spatial resolution, and exhibits variability in image acquisition, all of which compromise the reproducibility and robustness of radiomic features[12]. Most CEUS-radiomics studies to date are retrospective with potential for selection bias and limited generalizability. Prospective, ideally randomized, trials are necessary to validate its role in clinical decision-making. Still, radiomics holds substantial potential to facilitate early diagnosis, risk stratification, and individualized treatment planning in HCC, supporting the shift toward precision oncology[38].
Technical challenges continue to impact the scalability of DL in CEUS imaging. Transfer learning from DL models trained on data from a single device or institution often fails to generalize well to new datasets, raising concerns about robustness in diverse clinical environments[39]. To address variability ML-guided feedback during real-time image acquisition has been proposed to support sonographers and enhance reproducibility[40]. Furthermore, automated control of image acquisition parameters, real-time quality optimization, and standardized region-of-interest selection are emerging as key strategies to improve consistency and reliability across imaging platforms.
An emerging trend is the integration of DL classification networks with radiomics pipelines to improve predictive accuracy. Such hybrid models have demonstrated improved performance in prognostic tasks, such as early recurrence prediction after resection[41]. However, many of these studies are retrospective in design, and further validation through prospective multicenter studies is needed as well.
Earlier studies often applied generic CNN architectures for liver lesion classification without tailoring models to the LI-RADS framework[23]. More recent efforts have introduced DL architecture specifically designed for LI-RADS categorization[34]. A key technical challenge in automated LI-RADS is that the system was originally developed for CT and MRI, and its direct applicability to CEUS remains under active evaluation. CEUS-specific enhancement kinetics, particularly for the “washout” criterion, differ significantly from cross-sectional imaging. According to the 2017 ACR CEUS LI-RADS guidelines, washout may be classified as early and marked or mild, a distinction that does not exist in CT/MRI-based LI-RADS. Without adaptation AI models risk misclassification if they rigidly apply CT/MRI definitions to CEUS data. Therefore, CEUS-based AI classifiers should be explicitly trained on modality-specific criteria, ensuring that lesion characterization reflects diagnostic standards and mitigates performance bias.
As AI tools are increasingly integrated into clinical workflows, ethical concerns must be addressed, including transparency in algorithm design, data privacy, and the need for human oversight. Clinicians must be trained not only to guide and interpret AI outputs but also to recognize their limitations. Regulatory frameworks and robust validation standards will be critical to ensuring the safe, equitable, and effective deployment of AI-enhanced CEUS tools.
Beyond challenges such as data heterogeneity and algorithm standardization, risks include algorithmic bias in which models trained on limited or non-representative datasets may underperform in certain patient subgroups, potentially leading to inequitable care. In multicenter studies, such as by Ding et al[31], consistent protocols for patient consent and data deidentification are essential. These should specify whether consent covered AI-based secondary analysis and confirm that anonymization methods were applied uniformly across sites.
Explainability remains a key requirement, allowing clinicians to understand the reasoning behind AI-based classifications and to maintain trust in decision-making. Human oversight should be explicitly defined whether AI functions as a “second reader” to validate radiologist findings or as a triage tool to prioritize high-risk cases while ensuring final responsibility for diagnosis and treatment rests with experienced clinicians.
Beyond technical and regulatory hurdles, real-world adoption also depends on clinician acceptance and seamless workflow integration. Early pilot implementations suggest feasibility, but broader validation and training remain essential for routine use.
Therefore, before AI-enhanced CEUS can be fully translated into routine clinical practice for liver lesion assessment, several critical steps must be addressed: (1) Large-scale prospective validation studies are needed to confirm performance across diverse patient populations, imaging devices, and operator skill levels; (2) Real-time deployment frameworks should be developed and tested, ensuring seamless integration with existing ultrasound systems without compromising examination workflow; (3) Regulatory approval pathways must be navigated, requiring compliance with medical device standards, thorough documentation of training data, and demonstration of reproducibility and safety; (4) Multicenter collaborations should focus on creating standardized imaging protocols, harmonized annotation guidelines, and open-access datasets to reduce bias and improve generalizability. This aligns with ongoing efforts by the World Federation for Ultrasound in Medicine and Biology (commonly referred to as the WFUMB) and ACR to develop standardized CEUS acquisition and reporting protocols, which could serve as a foundation for multicenter AI validation studies[24,42]; and (5) Research should prioritize explainability tools that allow clinicians to understand and trust AI outputs, enabling informed decision-making and safe patient care. To our knowledge most current CEUS-AI studies rely on visual in
The fusion of CEUS with AI technologies, particularly DL and radiomics, has the potential to revolutionize liver lesion assessment, including liver cancer diagnostic and care. Addressing technical limitations, ensuring clinical validation, and developing ethically sound implementation strategies will be key to realizing the full clinical impact of these innovations.
| 1. | Wang H, Cao J, Fan H, Huang J, Zhang H, Ling W. Compared with CT/MRI LI-RADS, whether CEUS LI-RADS is worth popularizing in diagnosis of hepatocellular carcinoma?-a direct head-to-head meta-analysis. Quant Imaging Med Surg. 2023;13:4919-4932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 2. | Mansur A, Vrionis A, Charles JP, Hancel K, Panagides JC, Moloudi F, Iqbal S, Daye D. The Role of Artificial Intelligence in the Detection and Implementation of Biomarkers for Hepatocellular Carcinoma: Outlook and Opportunities. Cancers (Basel). 2023;15:2928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 3. | Brooks JA, Kallenbach M, Radu IP, Berzigotti A, Dietrich CF, Kather JN, Luedde T, Seraphin TP. Artificial Intelligence for Contrast-Enhanced Ultrasound of the Liver: A Systematic Review. Digestion. 2025;106:227-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 4. | Motzfeldt Jensen M, Brix Danielsen M, Riis J, Assifuah Kristjansen K, Andersen S, Okubo Y, Jørgensen MG. ChatGPT-4o can serve as the second rater for data extraction in systematic reviews. PLoS One. 2025;20:e0313401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 5. | Yin C, Zhang H, Du J, Zhu Y, Zhu H, Yue H. Artificial intelligence in imaging for liver disease diagnosis. Front Med (Lausanne). 2025;12:1591523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 6. | Chatzipanagiotou OP, Loukas C, Vailas M, Machairas N, Kykalos S, Charalampopoulos G, Filippiadis D, Felekouras E, Schizas D. Artificial intelligence in hepatocellular carcinoma diagnosis: a comprehensive review of current literature. J Gastroenterol Hepatol. 2024;39:1994-2005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 7. | Wongsuwan J, Tubtawee T, Nirattisaikul S, Danpanichkul P, Cheungpasitporn W, Chaichulee S, Kaewdech A. Enhancing ultrasonographic detection of hepatocellular carcinoma with artificial intelligence: current applications, challenges and future directions. BMJ Open Gastroenterol. 2025;12:e001832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 8. | Feng X, Cai W, Zheng R, Tang L, Zhou J, Wang H, Liao J, Luo B, Cheng W, Wei A, Zhao W, Jing X, Liang P, Yu J, Huang Q. Diagnosis of hepatocellular carcinoma using deep network with multi-view enhanced patterns mined in contrast-enhanced ultrasound data. Eng Appl Artif Intell. 2023;118:105635. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Denis de Senneville B, Frulio N, Laumonier H, Salut C, Lafitte L, Trillaud H. Liver contrast-enhanced sonography: computer-assisted differentiation between focal nodular hyperplasia and inflammatory hepatocellular adenoma by reference to microbubble transport patterns. Eur Radiol. 2020;30:2995-3003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Wei T, Wei W, Ma Q, Shen Z, Lu K, Zhu X. Development of a Clinical-Radiomics Nomogram That Used Contrast-Enhanced Ultrasound Images to Anticipate the Occurrence of Preoperative Cervical Lymph Node Metastasis in Papillary Thyroid Carcinoma Patients. Int J Gen Med. 2023;16:3921-3932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 11. | Lewis S, Hectors S, Taouli B. Radiomics of hepatocellular carcinoma. Abdom Radiol (NY). 2021;46:111-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 12. | Maruyama H, Yamaguchi T, Nagamatsu H, Shiina S. AI-Based Radiological Imaging for HCC: Current Status and Future of Ultrasound. Diagnostics (Basel). 2021;11:292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Liu F, Liu D, Wang K, Xie X, Su L, Kuang M, Huang G, Peng B, Wang Y, Lin M, Tian J, Xie X. Deep Learning Radiomics Based on Contrast-Enhanced Ultrasound Might Optimize Curative Treatments for Very-Early or Early-Stage Hepatocellular Carcinoma Patients. Liver Cancer. 2020;9:397-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 111] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 14. | Su LY, Xu M, Chen YL, Lin MX, Xie XY. Ultrasomics in liver cancer: Developing a radiomics model for differentiating intrahepatic cholangiocarcinoma from hepatocellular carcinoma using contrast-enhanced ultrasound. World J Radiol. 2024;16:247-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (5)] |
| 15. | Hu HT, Wang W, Chen LD, Ruan SM, Chen SL, Li X, Lu MD, Xie XY, Kuang M. Artificial intelligence assists identifying malignant versus benign liver lesions using contrast-enhanced ultrasound. J Gastroenterol Hepatol. 2021;36:2875-2883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 16. | Wang Z, Yao J, Jing X, Li K, Lu S, Yang H, Ding H, Li K, Cheng W, He G, Jiang T, Liu F, Yu J, Han Z, Cheng Z, Tan S, Wang Z, Qi E, Wang S, Zhang Y, Li L, Dong X, Liang P, Yu X. A combined model based on radiomics features of Sonazoid contrast-enhanced ultrasound in the Kupffer phase for the diagnosis of well-differentiated hepatocellular carcinoma and atypical focal liver lesions: a prospective, multicenter study. Abdom Radiol (NY). 2024;49:3427-3437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 17. | Choi GH, Yun J, Choi J, Lee D, Shim JH, Lee HC, Chung YH, Lee YS, Park B, Kim N, Kim KM. Development of machine learning-based clinical decision support system for hepatocellular carcinoma. Sci Rep. 2020;10:14855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 18. | Wyner AJ, Olson M, Bleich J, Mease D. Explaining the success of adaboost and random forests as interpolating classifiers. J Mach Learn Res. 2017;18:1-33. |
| 19. | Li W, Lv XZ, Zheng X, Ruan SM, Hu HT, Chen LD, Huang Y, Li X, Zhang CQ, Xie XY, Kuang M, Lu MD, Zhuang BW, Wang W. Machine Learning-Based Ultrasomics Improves the Diagnostic Performance in Differentiating Focal Nodular Hyperplasia and Atypical Hepatocellular Carcinoma. Front Oncol. 2021;11:544979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Goodfellow I, Bengio Y, Courville A. Deep Learning. 1st ed. Cambridge (MA): MIT Press, 2016. |
| 21. | Urhuț MC, Săndulescu LD, Streba CT, Mămuleanu M, Ciocâlteu A, Cazacu SM, Dănoiu S. Diagnostic Performance of an Artificial Intelligence Model Based on Contrast-Enhanced Ultrasound in Patients with Liver Lesions: A Comparative Study with Clinicians. Diagnostics (Basel). 2023;13:3387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 22. | Zhang H, Huo F. Prediction of early recurrence of HCC after hepatectomy by contrast-enhanced ultrasound-based deep learning radiomics. Front Oncol. 2022;12:930458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 23. | Zhang W, Guo Q, Zhu Y, Wang M, Zhang T, Cheng G, Zhang Q, Ding H. Cross-institutional evaluation of deep learning and radiomics models in predicting microvascular invasion in hepatocellular carcinoma: validity, robustness, and ultrasound modality efficacy comparison. Cancer Imaging. 2024;24:142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 24. | American College of Radiology. Liver reporting & data system. [cited 23 September 2025]. Available from: https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS. |
| 25. | Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aubé C, Luciani A, Lassau N, Jégou S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;100:227-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 26. | Brehar R, Mitrea DA, Vancea F, Marita T, Nedevschi S, Lupsor-Platon M, Rotaru M, Badea RI. Comparison of Deep-Learning and Conventional Machine-Learning Methods for the Automatic Recognition of the Hepatocellular Carcinoma Areas from Ultrasound Images. Sensors (Basel). 2020;20:3085. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 27. | Wang K, Liu Y, Chen H, Yu W, Zhou J, Wang X. Fully automating LI-RADS on MRI with deep learning-guided lesion segmentation, feature characterization, and score inference. Front Oncol. 2023;13:1153241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 28. | Mămuleanu M, Urhuț CM, Săndulescu LD, Kamal C, Pătrașcu AM, Ionescu AG, Șerbănescu MS, Streba CT. Deep Learning Algorithms in the Automatic Segmentation of Liver Lesions in Ultrasound Investigations. Life (Basel). 2022;12:1877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Xiao M, Deng Y, Zheng W, Huang L, Wang W, Yang H, Gao D, Guo Z, Wang J, Li C, Li F, Han F. Machine learning model based on dynamic contrast-enhanced ultrasound assisting LI-RADS diagnosis of HCC: A multicenter diagnostic study. Heliyon. 2024;10:e38850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 30. | Yang J, Cao J, Ruan X, Ren Y, Ling W. The correlation between contrast-enhanced ultrasound Liver Imaging Reporting and Data System classification and differentiation grades of combined hepatocellular carcinoma-cholangiocarcinoma. Quant Imaging Med Surg. 2025;15:259-271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 31. | Ding W, Meng Y, Ma J, Pang C, Wu J, Tian J, Yu J, Liang P, Wang K. Contrast-enhanced ultrasound-based AI model for multi-classification of focal liver lesions. J Hepatol. 2025;83:426-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 32. | Oezsoy A, Brooks JA, van Treeck M, Doerffel Y, Morgera U, Berger J, Gustav M, Saldanha OL, Luedde T, Kather JN, Seraphin TP, Kallenbach M. Weakly Supervised Deep Learning Can Analyze Focal Liver Lesions in Contrast-Enhanced Ultrasound. Digestion. 2025;1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Liu L, Tang C, Li L, Chen P, Tan Y, Hu X, Chen K, Shang Y, Liu D, Liu H, Liu H, Nie F, Tian J, Zhao M, He W, Guo Y. Deep learning radiomics for focal liver lesions diagnosis on long-range contrast-enhanced ultrasound and clinical factors. Quant Imaging Med Surg. 2022;12:3213-3226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 34. | Zhou H, Ding J, Zhou Y, Wang Y, Zhao L, Shih CC, Xu J, Wang J, Tong L, Chen Z, Lin Q, Jing X. Malignancy diagnosis of liver lesion in contrast enhanced ultrasound using an end-to-end method based on deep learning. BMC Med Imaging. 2024;24:68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 35. | Li J, Li H, Xiao F, Liu R, Chen Y, Xue M, Yu J, Liang P. Comparison of machine learning models and CEUS LI-RADS in differentiation of hepatic carcinoma and liver metastases in patients at risk of both hepatitis and extrahepatic malignancy. Cancer Imaging. 2023;23:63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 36. | Dănilă M, Popescu A, Sirli R, Sporea I, Martie A, Sendroiu M. Contrast enhanced ultrasound (CEUS) in the evaluation of liver metastases. Med Ultrason. 2010;12:233-237. [PubMed] |
| 37. | Al-Obeidat F, Hafez W, Gador M, Ahmed N, Abdeljawad MM, Yadav A, Rashed A. Diagnostic performance of AI-based models versus physicians among patients with hepatocellular carcinoma: a systematic review and meta-analysis. Front Artif Intell. 2024;7:1398205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 38. | Wei J, Jiang H, Zhou Y, Tian J, Furtado FS, Catalano OA. Radiomics: A radiological evidence-based artificial intelligence technique to facilitate personalized precision medicine in hepatocellular carcinoma. Dig Liver Dis. 2023;55:833-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 39. | Akkus Z, Cai J, Boonrod A, Zeinoddini A, Weston AD, Philbrick KA, Erickson BJ. A Survey of Deep-Learning Applications in Ultrasound: Artificial Intelligence-Powered Ultrasound for Improving Clinical Workflow. J Am Coll Radiol. 2019;16:1318-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 182] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 40. | Brattain LJ, Telfer BA, Dhyani M, Grajo JR, Samir AE. Machine learning for medical ultrasound: status, methods, and future opportunities. Abdom Radiol (NY). 2018;43:786-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 141] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 41. | Gao W, Wang W, Song D, Yang C, Zhu K, Zeng M, Rao SX, Wang M. A predictive model integrating deep and radiomics features based on gadobenate dimeglumine-enhanced MRI for postoperative early recurrence of hepatocellular carcinoma. Radiol Med. 2022;127:259-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 42. | Dietrich CF, Nolsøe CP, Barr RG, Berzigotti A, Burns PN, Cantisani V, Chammas MC, Chaubal N, Choi BI, Clevert DA, Cui X, Dong Y, D'Onofrio M, Fowlkes JB, Gilja OH, Huang P, Ignee A, Jenssen C, Kono Y, Kudo M, Lassau N, Lee WJ, Lee JY, Liang P, Lim A, Lyshchik A, Meloni MF, Correas JM, Minami Y, Moriyasu F, Nicolau C, Piscaglia F, Saftoiu A, Sidhu PS, Sporea I, Torzilli G, Xie X, Zheng R. Guidelines and Good Clinical Practice Recommendations for Contrast Enhanced Ultrasound (CEUS) in the Liver - Update 2020 - WFUMB in Cooperation with EFSUMB, AFSUMB, AIUM, and FLAUS. Ultraschall Med. 2020;41:562-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 167] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/