Published online Oct 28, 2025. doi: 10.3748/wjg.v31.i40.111120
Revised: August 11, 2025
Accepted: September 23, 2025
Published online: October 28, 2025
Processing time: 125 Days and 6.4 Hours
Computer-aided diagnosis (CAD) may assist endoscopists in identifying and classifying polyps during colonoscopy for detecting colorectal cancer.
To build a system using CAD to detect and classify polyps based on the Yamada classification.
A total of 24045 polyp and 72367 nonpolyp images were obtained. We established a computer-aided detection and Yamada classification model based on the YOLOv7 neural network algorithm. Frame-based and image-based evaluation metrics were employed to assess the performance.
Computer-aided detection and Yamada classification screened polyps with a precision of 96.7%, a recall of 95.8%, and an F1-score of 96.2%, outperforming those of all groups of endoscopists. In regard to the Yamada classification of polyps, the CAD system displayed a precision of 82.3%, a recall of 78.5%, and an F1-score of 80.2%, outperforming all levels of endoscopists. In addition, according to the image-based method, the CAD had an accuracy of 99.2%, a specificity of 99.5%, a sensitivity of 98.5%, a positive predictive value of 99.0%, a negative predictive value of 99.2% for polyp detection and an accuracy of 97.2%, a specificity of 98.4%, a sensitivity of 79.2%, a positive predictive value of 83.0%, and a negative predictive value of 98.4% for poly Yamada classification.
We developed a novel CAD system based on a deep neural network for polyp detection, and the Yamada classification outperformed that of nonexpert endoscopists. This CAD system could help community-based hospitals enhance their effectiveness in polyp detection and classification.
Core Tip: This study developed a novel deep learning (YOLOv7-based) computer-aided detection and classification system that significantly outperformed endoscopists in both detecting colorectal polyps (96.7% precision, 95.8% recall) and classifying them morphologically via the Yamada classification (80.2% F1-score). Achieving high image-based accuracy (detection: 99.2%; classification: 97.2%), this computer-aided detection and classification system offers a powerful tool to enhance polyp identification and characterization, particularly benefiting community hospital settings.
- Citation: Qiu L, Ding J, Lai CX, Yang H, Li F, Li ZJ, Wu W, Liu GM, Guan QS, Zhang XG, Zhang RY, Yi LZ, Zhao ZF, Deng L, Lun WJ, Wang ZY, Lu WM, Qiao WG, Wang SL, Chen SM, Shen WQ, Cheng LM, Zhu BG, He SH, Dai J, Bai Y. Bridging the gap: Computer-aided detection and Yamada classification system matches expert performance. World J Gastroenterol 2025; 31(40): 111120
- URL: https://www.wjgnet.com/1007-9327/full/v31/i40/111120.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i40.111120
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide and ranks third in cancer mortality rate[1]. Most CRC patients are diagnosed at late stages due to latent and nonspecific signs and symptoms. In the early stages, the 5-year survival rate of CRC patients is estimated to be 90%. However, the 5-year survival rate decreases dramatically to less than 20% in the metastatic stage[1,2]. Therefore, early detection of CRC is particularly important for improving patient survival. Approximately 90% of CRCs progress from adenomas, and a high adenoma misdiagnosis rate (25.7%) poses a challenge for the early diagnosis of CRC[3]. Colonoscopy plays a crucial role in screening for precancerous CRC lesions and reducing the incidence and mortality of CRC[4,5].
In recent years, several groundbreaking studies have emerged, exploring the potential of computer-aided diagnosis (CAD) in enhancing the accuracy and efficiency of polyp detection during colonoscopies[6-8]. These studies have primarily focused on leveraging advanced machine learning algorithms, particularly deep learning models like convolutional neural networks, to analyze endoscopic images and identify polyps with remarkable precision[6-8]. For instance, a CAD based on YOLOv3 has been developed for real-time polyp detection in CRC screening, showing high sensitivity and specificity, and is suggested to be effectively integrated into a CAD system[9]. Despite these promising advancements, a critical limitation lies in the current artificial intelligence (AI) systems’ inability to perform detailed subclassification of polyps. The lack of subclassification in AI-based polyp detection systems leads to a significant gap in clinical decision-making. Without precise differentiation, physicians may struggle to determine the appropriate follow-up or management plan for patients, potentially resulting in over- or under-treatment. Besides, their research possesses several limitations (i.e., single-center studies, limited numbers of samples, complicated imaging methods).
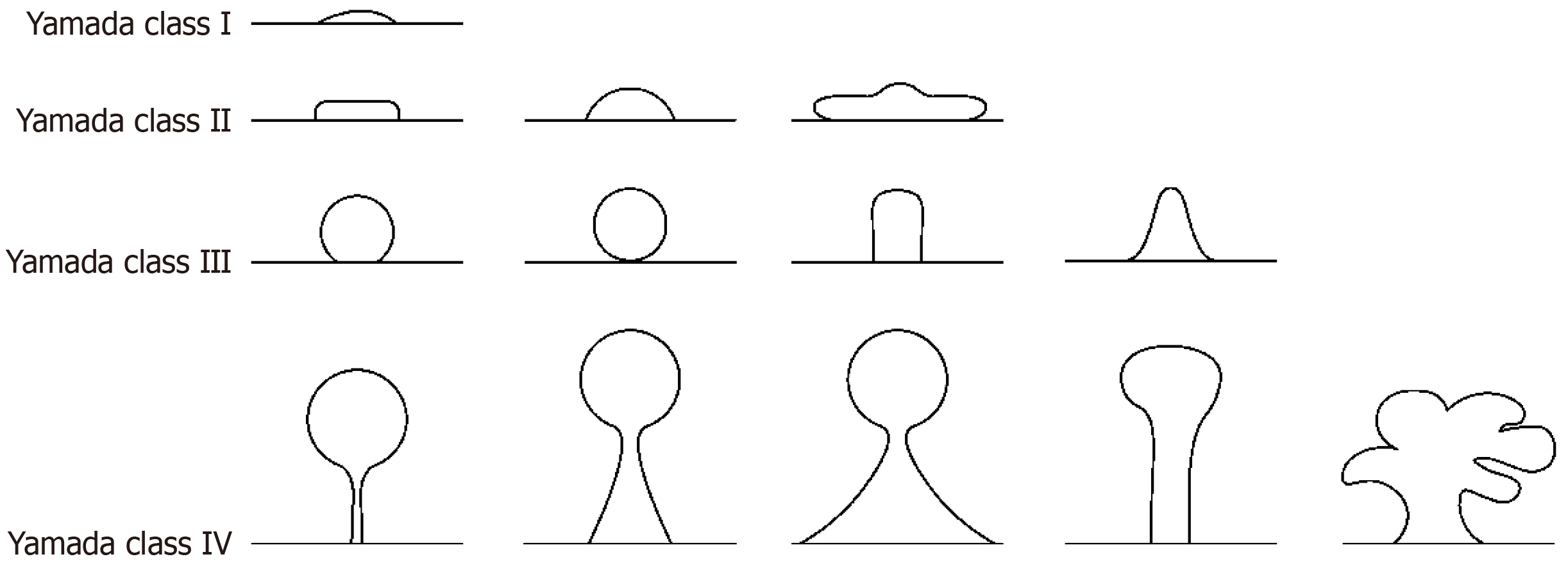
The Yamada classification system is a widely recognized method for the endoscopic assessment of polyps, providing valuable insights into their clinical significance. The Yamada classification categorizes polyps into four types based on their gross morphological features observed during endoscopy[10]. The clinical significance of the Yamada classification lies in its ability to predict the potential for malignancy, with type I and II polyps typically having a lower risk than the higher-risk type III and IV polyps, which exhibit villous or adenomatous features[11]. The system provides a stan
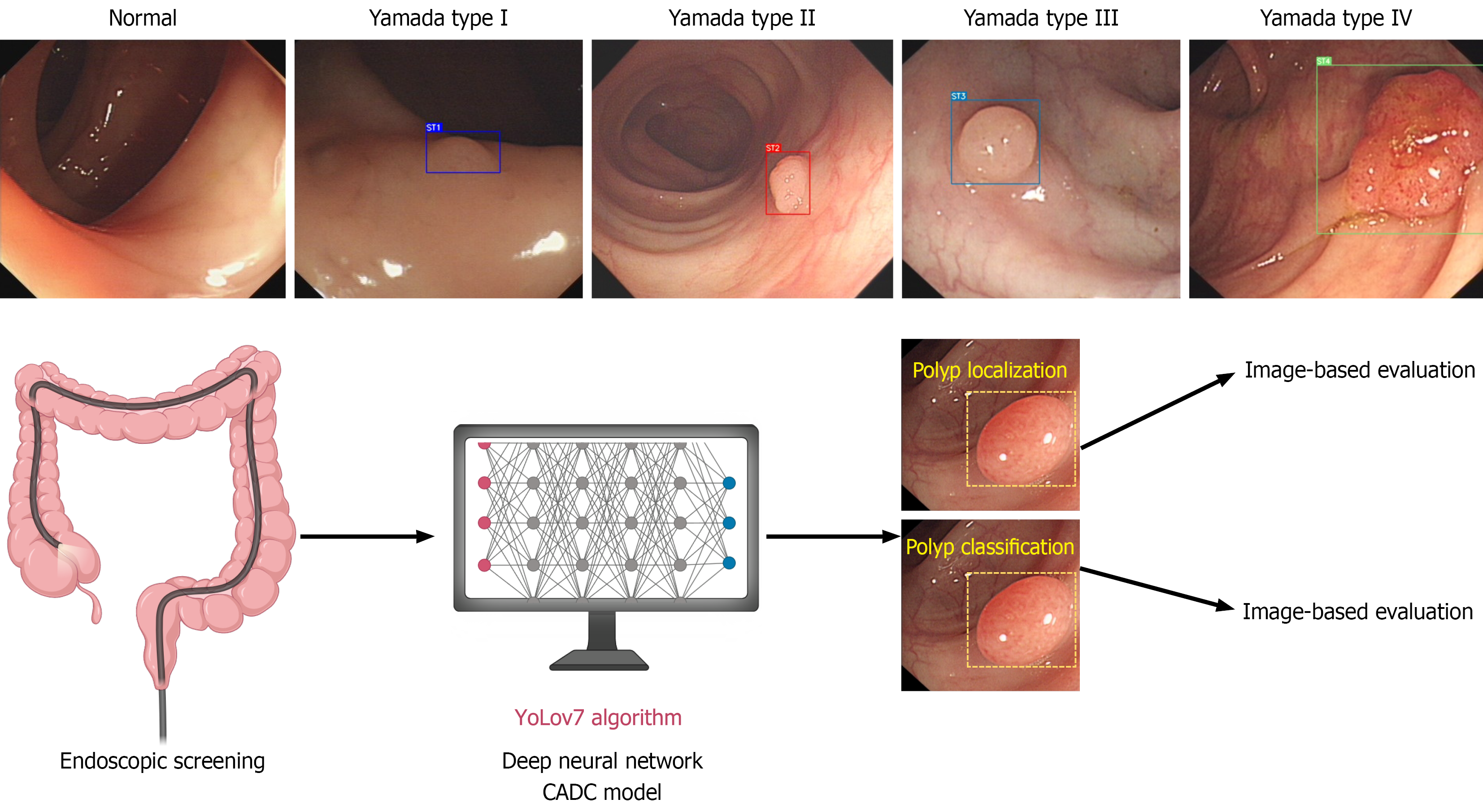
YOLOv7 is the latest iteration of the YOLO object detection model, which focuses on improving the accuracy and speed of bounding box-based object detection[13]. YOLOv7 can effectively detect and classify different types of colorectal polyps, including those with weak contrast, small size, and even multiple polyps, outperforming previous models such as Faster R-convolutional neural network and YOLOv5[14,15]. Hence, we first developed a novel computer-aided detection and Yamada classification (CADC) platform based on the YOLOv7 neural network to detect and classify polyps according to the Yamada classification using real-world endoscopic images from three hospitals for the endoscopic screening of polyps.
We retrospectively collected endoscopic images for the establishment and validation of the CADC system from our data collection spans from 2006 to 2021 and includes images from the following hospitals: (1) Nanfang Hospital, Southern Medical University; (2) Baiyun Branch of Nanfang Hospital, Southern Medical University; (3) Shenzhen Second People’s Hospital; (4) Shunde Hospital of Southern Medical University; (5) The People’s Hospital of Leshan; (6) Traditional Chinese Medicine Hospital of Shayang County; (7) Shanxi Academy of Traditional Chinese Medicine; and (8) Rizhao Hospital of Traditional Chinese Medicine.
We excluded patients with a history of cancer, familial polyposis syndromes or nonpolyposis cancer. This study was approved by the institutional review boards of the participating hospitals. All patients were fully informed and provided written consent. We randomly categorized the images into training and validation datasets for establishing and validating our CADC system and a test dataset for testing our CADC system (Supplementary Figure 1). A detailed summary of the patients’ clinical characteristics for the training, validation and test datasets is shown in Supplementary Table 1.
All images were taken at high resolution but with different endoscopes (Fujifilm-4450, SONOSCAPE-550, and OLYMPUS-290, Tokyo, Japan). These endoscopes undergo regular calibration and quality control checks to minimize resolution differences and avoid impacts on polyp classification. Strict standard operating procedures for pictures, covering lighting, shooting angles, and image processing, ensure the reliability of pictures for accurate polyp classification. Preprocessing was subsequently performed to prepare good-quality colonoscopy images, and the procedures are described in the Supplementary material. Dye-stained images, narrow-band images, and poor-quality images with halation, blurs, defocus, mucus, and poor air insufflation were excluded. Four experienced endoscopists were responsible for quality control, labeling, and classification. Two trained endoscopists independently selected, labeled and classified all the images. Then, two senior endoscopists cooperated in supervising the annotations and reannotated the images with inconsistent annotations. The image selection, labeling, and classification were finalized only when the two senior endoscopists reached a consensus.
To simplify differentiation, enhance labeling accuracy, and minimize errors, a refined definition was devised (Figure 1): (1) Yamada type I: Height < 1/2 radius sessile polyps; (2) Yamada type II: 1/2 radius ≤ height ≤ 3/4 diameter sessile polyps; a sub pedunculated polyp with height < radius; (3) Yamada type III: Sub pedunculated polyp with a prominent waistline; spherical, with the spherical notch attached to the mucosa but not leaving the mucosal plane; 3/4 diameter < height sessile polyps; and (4) Yamada type IV: Pedunculated polyp.
The CADC was based on the YOLOv7 model, an improved YOLO model with increased accuracy and faster processing times[13]. The YOLOv7 architecture comprises multiple scales to detect objects and determine the size of the predicted bounding boxes. In addition, the model can also classify objects. The labeling and classification annotations from the four endoscopists were considered the gold standard for training our CADC system. Additional details of the deep learning algorithm are described in the Supplementary material.
We first validated the performance of the CADC system in identifying and classifying polyps in patients using a validation dataset. We then tested the performance of CADC using test datasets. A subset of patient images from the test set was randomly selected. Three endoscopists of different levels (experienced, competent, and trainee) were required to independently annotate the same test pictures, and their annotations were compared with those of CADC. Three endoscopists were excluded from the previous quality control and labeling of images. The trainee, the competent and the experienced endoscopists had 2 years, more than 5 years and 10 years of experience in endoscopic procedures, respectively.
The intersection over union (IoU), which is a commonly used evaluation metric in the field of computer vision for object detection tasks, was used in the experiment to assess the effectiveness of polyp localization. The IoU threshold utilized in this study is set at 0.5. Additionally, two sets of classification metrics were applied to evaluate the detection and classification performance based on the threshold of IoU. Three classification evaluation metrics, precision, recall and F1-score, were used to measure the polyp detection performance and classification performance for a given category based on labeled frames, as defined in Equations 1-3. For a polyp whose predicted category is the given category, when the predicted category is correct, the result is considered a true positive (TP). In contrast, it is a false positive (FP). Fora polyp whose ground-truth category is the given category, when either the polyp is not detected or the ground-truth category is not correctly predicted, the result is considered a false negative (FN).
Precision = TP/(TP + FP) (1)
Recall = TP/(TP + FN) (2)
F1 = 2 × (precision × recall)/(precision + recall) (3)
Accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) defined in Equations 4-8 were applied for assessing the performance of the detection and Yamada classification of polyps based on images. For a polyp whose predicted category is the given category, the result is considered TP when the predicted category is correct. In contrast, it is FP. For a polyp whose ground-truth category is the given category, the result is considered to be an FN. For a polyp whose ground-truth category is not the given category, when either the polyp is not detected or the ground-truth category is not correctly predicted, the result is true negative (TN).
Accuracy = (TP + TN)/(TP + TN + FP + FN) (4)
Sensitivity = TP/(TP + FN) (5)
Specificity = TN/(TN + FP) (6)
PPV = TP/(TP + FP) (7)
NPV = TN/(TP + FN) (8)
Student’s t-test with Holm-Bonferroni correction was used to analyze differences in the diagnostic precision, recall, F1-score, accuracy, sensitivity, specificity, PPV, and NPV for the detection and Yamada classification of polyps. SPSS was used to conduct all the statistical analyses.
Figure 2 depicts the scheme of our study. Table 1 displays the performance of the CADCs and endoscopists for screening out polyps. The CADC correctly detected polyps with a precision of 96.7%, a recall of 95.80%, and an F1-score of 96.2% among 1192 randomly selected endoscopic images, with or without polyps. Four experts, four seniors, and four novices achieved a precision of 93.1% (SD 4.2%), 91.6% (SD 3.1%), and 89.6% (SD 4.3%) for the tested images, respectively. The precision of the CADC system was significantly greater than that of senior and junior endoscopists and on par with that of experienced endoscopists. In addition, we used image-based methods to evaluate the polyp detection performance. Supplementary Table 2 shows the image-based performance of the CADC system and endoscopists for identifying polyps. The CADC system identified polyps with an accuracy of 99.2%, specificity of 99.5%, sensitivity of 98.5%, PPV of 99.0%, and NPV of 99.2%, which were significantly better than those of all groups of endoscopists.
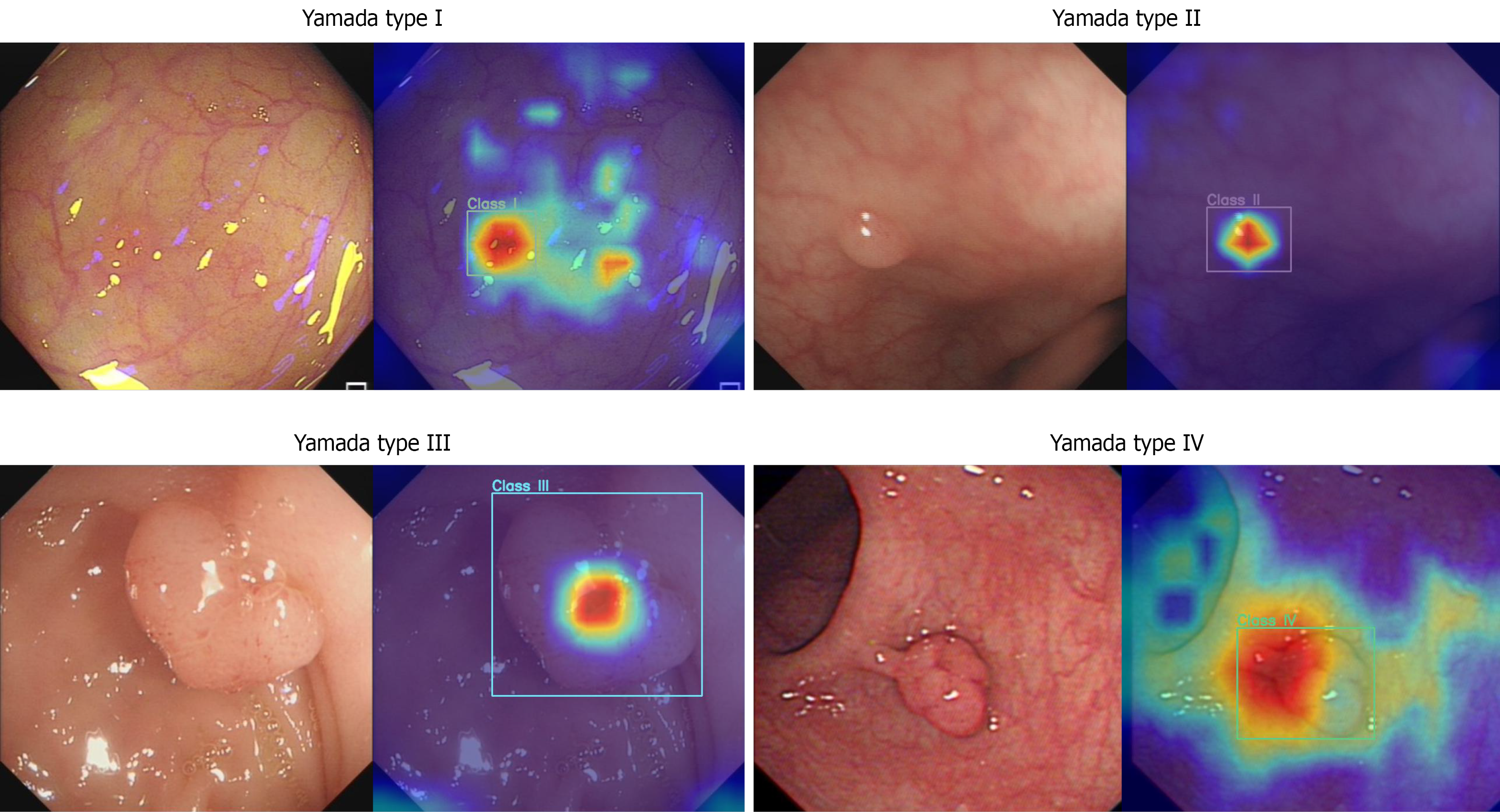
As shown in Table 2, four experts, four seniors, and four juniors classified endoscopic images based on the Yamada classification with a precision of 75.9% (SD 2.5%), 69.0% (1.8%), and 61.7% (4.9%), respectively. The CADC system correctly classified the images into four types with a precision of 82.3%, which was significantly greater than that of any of the endoscopists. Figure 2 displays representative test endoscopic images identified by the CADC system for classifying polyps into four types. Regarding model interpretability, as shown in Figure 3, heatmap visualizations were applied to illustrate how the model focuses on morphological features for classification, with relative density represented by a color scale (e.g., red-yellow-blue, where brighter colors indicate higher attention from the AI). For categories 1-3, high-density values are primarily concentrated in the central area of the polyp, gradually decreasing from the center to the periphery. Specifically, category 1 shows the smallest decrease in density values, with relatively uniform high density across the entire polyp region; category 2 shows a moderate decrease; category 3 exhibits the most significant decrease, with high-density values confined to a small area around the central point and rapid density drop in other regions. For category 4, high-density points are mainly focused on the junction between the polyp body and the stalk, indicating that the AI has learned the key feature of pedunculated polyps. These heatmaps effectively demonstrate the model’s attention patterns, enhancing the transparency of its classification logic. Moreover, we employed image-based methods to assess the performance of the Yamada classification of polyps. As shown in Supplementary Table 3, the CADC classified polyps with an accuracy of 97.2%, specificity of 98.4%, sensitivity of 79.2%, PPV of 83.0%, and NPV of 98.4%, significantly surpassing the other groups.
To further compare the performance of our model with that of endoscopists, we analyzed the precision, recall and F1-score for every Yamada class. The results for Yamada class I are shown in Table 3. The precision of four experts, four seniors, and four juniors reached 74.4% (SD 6.3%), 72.2% (SD 7.8%), and 62.1% (SD 3.1%), respectively. The CADC correctly classified polyps into Yamada class I with a precision of 84.0%, a recall of 79.3%, and an F1-score of 81.6%, outperforming all levels of endoscopists. Additionally, an image-based evaluation method was employed to measure the performance for every Yamada class. Our CADC model classified Yamada class I polyps with an accuracy of 96.6%, a specificity of 98.4%, a sensitivity of 79.1%, a PPV of 83.7%, and a NPV of 97.9%, which significantly exceeded the values reported by all endoscopists (Supplementary Table 4).
For Yamada class II, the precision of CADC was 82.1%, which was significantly different from that of seniors, but the F1-score of CADC (84.9%) was significantly greater than that of the other groups. The experienced, senior and junior doctors had F1-scores of 78.2% (SD 3.4%), 74.9% (SD 4.3%), and 64.7% (SD 9.9%), respectively (Table 4). Besides, Supplementary Table 5 displayed that the CADC model classified Yamada class II polyps with an accuracy of 95.6%, specificity of 96.7%, sensitivity of 90.6%, PPV of 84.7%, and NPV of 98.1%, showing significantly better accuracy than all the other groups of endoscopists.
In regard to Yamada class III, the experienced, senior and junior endoscopists displayed a precision of 62.3% (SD 6.4%), 52.1% (SD 12.1%), and 37.4% (SD 8.9%), respectively. The precision of the CADC system was significantly greater (76.1%) than that of all endoscopists (Table 5). In addition, the CADC model classified Yamada class II polyps with an accuracy of 97.4%, specificity of 99.0%, sensitivity of 63.6%, PPV of 76.1%, NPV 98.3%, displaying significantly higher accuracy than that of the seniors and the juniors and are on par with that of experts (Supplementary Table 6).
For Yamada class IV, the precision of CADC (87.1%) was not significantly different from that of the other groups. However, the F1-score of the CADC group (85.2%) was significantly higher, compared with that of the other groups. The experienced, senior and junior had F1-scores of 78.3% (SD 1.9%), 72.2% (SD 5.5%), and 72.6% (SD 4.8%), respectively (Table 6). Moreover, Supplementary Table 7 shows that the CADC model classified Yamada class IV polyps with an accuracy of 99.0%, specificity of 99.6%, sensitivity of 83.3%, PPV of 87.5%, and NPV of 99.4%, showing significantly greater accuracy than all the other groups of endoscopists.
The precisions of CADC systems on Yamada class I, II, III, and IV were 84.0%, 82.1%, 76.1%, and 87.1%, respectively, outperforming all levels of endoscopists on Yamada class I, III and CADC best classified class I and IV polyps. In addition, the F1-scores of CADC on Yamada class I, II, III, and IV were 81.6%, 84.9%, 69.3%, and 85.2%, respectively, exceeding those of all groups of endoscopists on Yamada class I, II, IV and CADC best classified polyps of classes II and IV polyps based on the F1-score. For the image-based method, the accuracies of CADC for Yamada classes I, II, III, and IV were 96.6%, 95.6%, 97.4%, and 99.0%, respectively, surpassing those of the other groups for Yamada classes I, II, and IV, and our model best classified classes I and IV.
Endoscopy performs a crucial role in diagnosing CRC, the third lethal cancer globally[1]. However, it is difficult to detect early CRC by endoscopy, which demands experienced endoscopists and advanced equipment[16]. It is time- and cost-consuming to train experienced endoscopists. In countries with large populations, such as India and Africa, endoscopy experts are scarce, causing misdiagnosis and missed diagnosis of CRC[17,18]. A deep neural network is considered to be the most valuable deep learning method for object identification and classification[19]. It was reported that CAD classified polyps as neoplastic or hyperplastic type with 98% accuracy using deep neural learning[8]. In addition, CAD could differentiate adenomatous and hyperplastic diminutive polyps on real-time colonoscopy videos[20]. However, the simplicity of these basic classifications is insufficient to meet the complex clinical demands. The Yamada and Paris classifications are among the prevalent endoscopic polyp classification methodologies. The Yamada classification categorizes polyps into four types based on their morphological characteristics[11]. This categorization is more straightforward for computer-based AI systems to analyze and interpret, enabling algorithms to quickly process and distinguish polyp types. In contrast, the Paris classification considers a broader range of aspects such as elevation, depression, lesion size, and surface features[21], making it more complex for AI algorithms to process and apply. Regarding treatment guidance, the Yamada classification is closely linked to treatment options and directly offers preliminary strategies for different polyp types[12]. However, the Paris classification is mainly designed for the detailed evaluation of early-stage gastrointestinal cancer and precancerous lesions, so its guidance for polyp treatment is less explicit[21]. Classifications like Kudo, narrow-band imaging international colorectal endoscopic, and Japanese narrow-band imaging expert team focus on detailed histological features and require advanced imaging techniques or specialized training, making them less practical for routine polyp management[22]. Conversely, as it relies solely on white-light images, the Yamada classification is more cost-effective and user-friendly in routine examination.
Krenzer et al[23] fabricated an AI model to classify polyps based on the Paris classification, but the precision and F1-score of semipedunculated sessile polyp (type Isp) (68.84% and 52.49%) and type IIa (78.90% and 77.56%) were unsatisfactory, and comparisons between the CAD model and endoscopists were lacking. In the narrow-band imaging international colorectal endoscopic classification with scarce data, the few-shot learning algorithm based on deep metric learning obtained an accuracy rate of 81.13%[23]. In comparison, we developed a CADC system to identify polyps based on 124165 white-light view images from eight hospitals, tested the system on 1192 images, with an accuracy of 99.2%, and classified polyps based on the Yamada classification, with an accuracy of 97.2%, precision of 82.3% and F1-score of 80.2%, which are even better than the levels of experienced endoscopists.
In this study, we applied image-based and object-based detection and classification with different calculation metrics due to their unique focuses and methodologies. Object-based classification focuses on accurately classifying individual polyps, treating each polyp as a separate object. IoU is a common evaluation metric used in object detection tasks. It measures the overlap between the predicted bounding box and the ground truth bounding box for an object. Metrics such as precision, recall, and F1-score evaluate the model’s performance in correctly categorizing each polyp, which is crucial for clinical decision-making[24]. Image-based classification evaluates the overall performance across entire images. Metrics such as sensitivity, specificity, PPV, and NPV assess the model’s ability to identify images containing polyps or not, which is valuable for broad screening purposes[25]. Object-based classification prioritizes detailed analysis of individual polyps, while image-based classification focuses on overall screening efficiency at the population level. These approaches complement each other in a comprehensive polyp detection and classification system.
The Yamada classification plays a crucial role in the early detection and accurate classification of polyps, which is vital for preventing CRC[10]. However, despite its utility, the system can be subject to interobserver variability for several reasons: Subjectivity, experience and imaging quality. Interobserver disagreement can lead to inconsistencies in clinical decision-making and research outcomes. Therefore, the development and application of CAD systems can help standardize the classification process and reduce interobserver variability. Hence, our CADC model was better at standardizing the interpretation of endoscopic images.
Additionally, our CADC performance is limited by the quality of retrospectively collected endoscopic images and videos used for training. In real-life clinical settings, the model’s effectiveness may vary due to differences in imaging techniques, equipment, and endoscopist expertise, which can impact the clarity and diagnostic value of the images. To improve the clinical application of the algorithm, it is essential to ensure consistent image quality and consider prospective studies for further validation. Furthermore, our study faces significant ethical challenges when it comes to deploying clinical AI. To address these challenges, potential bias could be mitigated by expanding training data diversity (encompassing varied ethnicities, ages, disease severities) and involving third parties in fairness testing. Secondly, to address over-reliance, clarifying the AI system’s auxiliary role in guidelines and strengthening physician training could ensure rational use. For medical error risks, establishing robust prevention mechanisms (real-time monitoring, manual reviews, emergency plans) might tackle such issues.
We developed a CADC based on a deep neural network algorithm that provides automated, accurate, and stable diagnoses for detecting and classifying polyps. The CADC is a reliable and powerful tool to help endoscopists identify and classify polyps according to the Yamada classification. Moreover, this system should be further improved and validated by conducting more research.
| 1. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1746] [Article Influence: 349.2] [Reference Citation Analysis (1)] |
| 2. | Aguiar Junior S, Oliveira MM, Silva DRME, Mello CAL, Calsavara VF, Curado MP. Survival of patients with colorectal cancer in a cancer center. Arq Gastroenterol. 2020;57:172-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 3. | Park JH, Moon HS, Kwon IS, Kim JS, Kang SH, Lee ES, Kim SH, Sung JK, Lee BS, Jeong HY. Quality of Preoperative Colonoscopy Affects Missed Postoperative Adenoma Detection in Colorectal Cancer Patients. Dig Dis Sci. 2020;65:2063-2070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Young GP, Senore C, Mandel JS, Allison JE, Atkin WS, Benamouzig R, Bossuyt PM, Silva MD, Guittet L, Halloran SP, Haug U, Hoff G, Itzkowitz SH, Leja M, Levin B, Meijer GA, O'Morain CA, Parry S, Rabeneck L, Rozen P, Saito H, Schoen RE, Seaman HE, Steele RJ, Sung JJ, Winawer SJ. Recommendations for a step-wise comparative approach to the evaluation of new screening tests for colorectal cancer. Cancer. 2016;122:826-839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Gupta S. Screening for Colorectal Cancer. Hematol Oncol Clin North Am. 2022;36:393-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 6. | Sharma P, Balabantaray BK, Bora K, Mallik S, Kasugai K, Zhao Z. An Ensemble-Based Deep Convolutional Neural Network for Computer-Aided Polyps Identification From Colonoscopy. Front Genet. 2022;13:844391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 7. | Itoh H, Misawa M, Mori Y, Kudo SE, Oda M, Mori K. Positive-gradient-weighted object activation mapping: visual explanation of object detector towards precise colorectal-polyp localisation. Int J Comput Assist Radiol Surg. 2022;17:2051-2063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 8. | Kudo SE, Misawa M, Mori Y, Hotta K, Ohtsuka K, Ikematsu H, Saito Y, Takeda K, Nakamura H, Ichimasa K, Ishigaki T, Toyoshima N, Kudo T, Hayashi T, Wakamura K, Baba T, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Artificial Intelligence-assisted System Improves Endoscopic Identification of Colorectal Neoplasms. Clin Gastroenterol Hepatol. 2020;18:1874-1881.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 162] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 9. | Nogueira-Rodríguez A, Domínguez-Carbajales R, Campos-Tato F, Herrero J, Puga M, Remedios D, Rivas L, Sánchez E, Iglesias Á, Cubiella J, Fdez-Riverola F, López-Fernández H, Reboiro-Jato M, Glez-Peña D. Real-time polyp detection model using convolutional neural networks. Neural Comput Applic. 2022;34:10375-10396. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 10. | Yamada T, Ichikawa H. X-ray diagnosis of elevated lesions of the stomach. Radiology. 1974;110:79-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 11. | Zhang DX, Niu ZY, Wang Y, Zu M, Wu YH, Shi YY, Zhang HJ, Zhang J, Ding SG. Endoscopic and pathological features of neoplastic transformation of gastric hyperplastic polyps: Retrospective study of 4010 cases. World J Gastrointest Oncol. 2024;16:4424-4435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 12. | Yu L, Li N, Zhang XM, Wang T, Chen W. Analysis of 234 cases of colorectal polyps treated by endoscopic mucosal resection. World J Clin Cases. 2020;8:5180-5187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Wang CY, Bochkovskiy A, Liao HYM. YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2023 Jun 17-24; Vancouver, Canada. NJ, United States: IEEE, 2023: 7464-7475. [DOI] [Full Text] |
| 14. | Zhu PC, Wan JJ, Shao W, Meng XC, Chen BL. Colorectal image analysis for polyp diagnosis. Front Comput Neurosci. 2024;18:1356447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 15. | Hong LTH, Nhuong LH, Tu DQ, Huy NS, Hanh ND, Luong TT, Do NG, Dung LA. Real-time detection of colon polyps during colonoscopy using YOLOv7. JMST. 2023;122-134. [DOI] [Full Text] |
| 16. | Winawer SJ; National Polyp Study Workgroup. Colonoscopy Screening and Colorectal Cancer Incidence and Mortality. N Engl J Med. 2023;388:377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Zacharias P, Mathew S, Mathews J, Somu A, Peethambaran M, Prashanth M, Philip M. Sedation practices in gastrointestinal endoscopy-A survey from southern India. Indian J Gastroenterol. 2018;37:164-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Parker RK, Mwachiro MM, Topazian HM, Davis R, Nyanga AF, O'Connor Z, Burgert SL, Topazian MD. Gastrointestinal endoscopy experience of surgical trainees throughout rural Africa. Surg Endosc. 2021;35:6708-6716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Wang J, Li Y, Chen B, Cheng D, Liao F, Tan T, Xu Q, Liu Z, Huang Y, Zhu C, Cao W, Yao L, Wu Z, Wu L, Zhang C, Xiao B, Xu M, Liu J, Li S, Yu H. A real-time deep learning-based system for colorectal polyp size estimation by white-light endoscopy: development and multicenter prospective validation. Endoscopy. 2024;56:260-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 20. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 433] [Article Influence: 61.9] [Reference Citation Analysis (0)] |
| 21. | Endoscopic Classification Review Group. Update on the paris classification of superficial neoplastic lesions in the digestive tract. Endoscopy. 2005;37:570-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 559] [Cited by in RCA: 677] [Article Influence: 32.2] [Reference Citation Analysis (2)] |
| 22. | Johnson GGRJ, Helewa R, Moffatt DC, Coneys JG, Park J, Hyun E. Colorectal polyp classification and management of complex polyps for surgeon endoscopists. Can J Surg. 2023;66:E491-E498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 23. | Krenzer A, Heil S, Fitting D, Matti S, Zoller WG, Hann A, Puppe F. Automated classification of polyps using deep learning architectures and few-shot learning. BMC Med Imaging. 2023;23:59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 26] [Reference Citation Analysis (1)] |
| 24. | Lalinia M, Sahafi A. Colorectal polyp detection in colonoscopy images using YOLO-V8 network. Signal Image Video Process. 2024;18:2047-2058. [DOI] [Full Text] |
| 25. | Shen MH, Huang CC, Chen YT, Tsai YJ, Liou FM, Chang SC, Phan NN. Deep Learning Empowers Endoscopic Detection and Polyps Classification: A Multiple-Hospital Study. Diagnostics (Basel). 2023;13:1473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/