Published online Sep 21, 2025. doi: 10.3748/wjg.v31.i35.112220
Revised: August 24, 2025
Accepted: September 3, 2025
Published online: September 21, 2025
Processing time: 59 Days and 13.9 Hours
The study by Dos Santos et al marks a significant advancement in understanding the genetics of colorectal polyposis, particularly within the underrepresented Brazilian population. Utilizing whole-exome sequencing in 27 patients with unexplained polyposis, the researchers identified 16 candidate genes in 44.4% of cases-an impressive outcome given strict exclusion criteria. Many identified variants were linked to the Wnt/β-catenin signaling pathway, reinforcing their biological relevance. However, the study underscores key challenges in genomic medicine, especially the gap between gene discovery and clinical application. A substantial proportion of variants (60.1%) were classified as of uncertain sig
Core Tip: This study applies whole-exome sequencing to a Brazilian cohort with unexplained colorectal polyposis, ide
- Citation: Dhali A, Maity R, Biswas J. Expanding the genetic landscape of colorectal polyposis: Progress and challenges. World J Gastroenterol 2025; 31(35): 112220
- URL: https://www.wjgnet.com/1007-9327/full/v31/i35/112220.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i35.112220
The colorectal polyposis landscape has undergone significant transformation over the past two decades, evolving from a relatively simple genetic paradigm dominated by APC and MUTYH mutations to a complex heterogeneous spectrum involving multiple genes and pathways. The manuscript by Dos Santos et al[1] exemplifies both the promise and challenges inherent in contemporary genetic research for polyposis syndromes, offering valuable insights into the expanding genetic architecture while highlighting persistent diagnostic gaps.
The traditional understanding of polyposis genetics has been fundamentally challenged by advances in whole-exome sequencing (WES) and multi-gene panel testing. While APC mutations account for the majority of classic familial adenomatous polyposis (FAP) cases, with detection rates of 70%-80% in patients with ≥ 1000 adenoma, and biallelic MUTYH mutations represent the second most common cause at approximately 7% of adenomatous polyposis cases, a substantial proportion of patients remain genetically unexplained[2,3].
Recent studies have expanded the genetic spectrum to include genes such as POLE, POLD1, NTHL1, MBD4, MSH3, and MLH3[4]. The manuscript by Dos Santos et al[1] extends this paradigm further, identifying pathogenic or likely pathogenic variants in 16 novel candidate genes, including ST7L, A1CF, DKK4, NTHL1, PNKP, PMS2, and FRK, in 44.4% of their APC- and MUTYH-negative polyposis cohort. This represents a significant diagnostic yield that challenges the conventional notion of “unexplained” polyposis.
The identification of variants in Wnt/β-catenin signaling pathway genes such as DKK4 and A1CF is particularly noteworthy, as dysregulation of this pathway is central to colorectal cancer development[5]. The authors’ pathway enrichment analysis, revealing alterations in “negative regulation of protein adenosine diphosphate-ribosylation”, provides mechanistic insight, as this process is involved in Wnt signaling pathway activation. These findings align with recent comprehensive genomic studies that have identified over 250 putative colorectal cancer driver genes, many previously unassociated with polyposis syndromes[6].
The current study’s methodological approach represents current best practices in polyposis genetics research. The use of WES covering 203058 target regions across 19682 genes with a mean coverage of 150 × ensures comprehensive variant detection. The filtering strategy, requiring ≥ 30 reads, ≥ 25% variant allele fraction, and < 1% population frequency, helps distinguish genuine pathogenic variants from technical artifacts and common polymorphisms. However, there were a few limitations that warrant consideration. The lack of functional validation for most identified variants represents a significant constraint. While the authors applied American College of Medical Genetics and Genomics criteria for variant classification, many variants remain classified as variants of uncertain significance (VUS), comprising 60.1% of all identified variants. This high VUS rate reflects broader challenges in variant interpretation that plague the field of clinical genetics[7]. The absence of tumor analysis represents another limitation. Loss of heterozygosity analysis could have provided crucial evidence supporting the pathogenic role of identified germline variants. Additionally, the study’s relatively small sample size (n = 27) limits statistical power for detecting rare variants and establishing genotype-phenotype correlations.
The diagnostic yield reported in this study (44.4% carrying at least one pathogenic/Likely pathogenic variant) is remarkably high compared to other recent studies. For context, a large United Kingdom (UK) study of 259 patients with 10-99 adenomas reported a 25.5% diagnostic yield for pathogenic variants, while studies focusing on patients with ≥ 10 adenomas typically report yields of 10%-30%[8]. This disparity may reflect differences in patient selection, with the Brazilian cohort potentially representing a more enriched population given the context of the specialized referral center. The finding that participants with pathogenic/Likely pathogenic variants tended to be younger at diagnosis (mean age 47 vs 55 years) aligns with established patterns in hereditary cancer syndromes. However, the difference did not reach statistical significance in this small cohort.
Notably, the lack of correlation between genetic findings and family history is intriguing and potentially concerning. This observation challenges traditional risk assessment models that heavily weight family history in determining genetic testing eligibility. Current National Comprehensive Cancer Network (NCCN) guidelines recommend genetic testing for patients with ≥ 10-20 cumulative adenomas, but the optimal threshold remains debated[9]. Table 1 compares diagnostic yields and pathway findings across ethnic cohorts of colorectal polyposis.
| Ref. | Country | Sample size (n) | Diagnostic yield (% with P/LP variants) | Major mutations identified | Key pathway findings |
| Dos Santos et al[1] | Brazil | 27 | 44.4 | ST7L, A1CF, DKK4, NTHL1, PNKP, PMS2, FRK | Wnt/β-catenin dysregulation; oligogenicity |
| Yang et al[20] | China | 120 | 74.2 | APC, MUTYH, POLE, POLD1, PTEN, MLH3, SMAD4 | Structural variants, APC promoter mutations |
| Mak et al[8] | United Kingdom | 259 | 25.5 | APC, biallelic MUTYH, POLD1, GREM1, MLH1, MSH2 | DNA repair, base excision repair |
Despite technological advances, significant challenges persist in polyposis genetics. First, the “missing heritability” problem remains substantial, with approximately 20%-35% of patients meeting clinical criteria for polyposis syndromes lacking identifiable genetic causes[10,11]. While studies like Dos Santos et al[1] make important contributions to addressing this gap, the clinical actionability of many newly identified genes remains uncertain. Second, the inter
Professional guidelines have evolved to recommend multi-gene panel testing for polyposis evaluation, typically including APC, MUTYH, POLE, POLD1, and NTHL1[3]. The European Society for Medical Oncology guidelines specifically recommend panel testing, including these five genes, for patients with > 10 adenomas, while the NCCN guidelines suggest testing for patients with ≥ 20 cumulative adenomas[9,14].
However, significant variation exists in testing thresholds globally. UK guidelines recommend an individualized approach for patients with multiple colorectal adenomas (≥ 10 metachronous adenomas), considering factors such as age at diagnosis, family history, and total polyp burden[15]. Japanese guidelines take a more conservative approach, sug
Recent studies suggest that optimization of testing strategies could improve cost-effectiveness. A probability calculator incorporating polyp count and age has shown promise for identifying patients most likely to benefit from genetic testing[17]. Such tools could help address the challenge of low diagnostic yields in certain patient populations, particularly older patients with fewer adenomas.
The integration of artificial intelligence (AI) and machine learning approaches represents a promising frontier in polyposis genetics. AI algorithms are already showing remarkable success in polyp detection during colonoscopy, with sensitivities approaching 95%-99%[18]. Extension of these technologies to genetic variant interpretation and patient risk stratification could significantly enhance diagnostic accuracy and clinical decision-making. Emerging high-throughput technologies, such as deep mutational scanning (typically used for functional evaluation of single amino acid mutations in a protein of interest), can help solve the VUS challenge via simultaneous and unbiased evaluation of large protein variant libraries, thus generating comprehensive genotype-phenotype functional maps. This opens up the possibility of classifying VUSs that have been observed thus far[19].
Whole-genome sequencing is increasingly replacing WES for comprehensive genetic analysis, offering the advantage of detecting non-coding variants, structural rearrangements, and copy number variations that may be missed by exome-based approaches[18]. The identification of APC promoter mutations and complex structural variants in a recent study by Yang et al[20] demonstrates the importance of comprehensive genomic analysis.
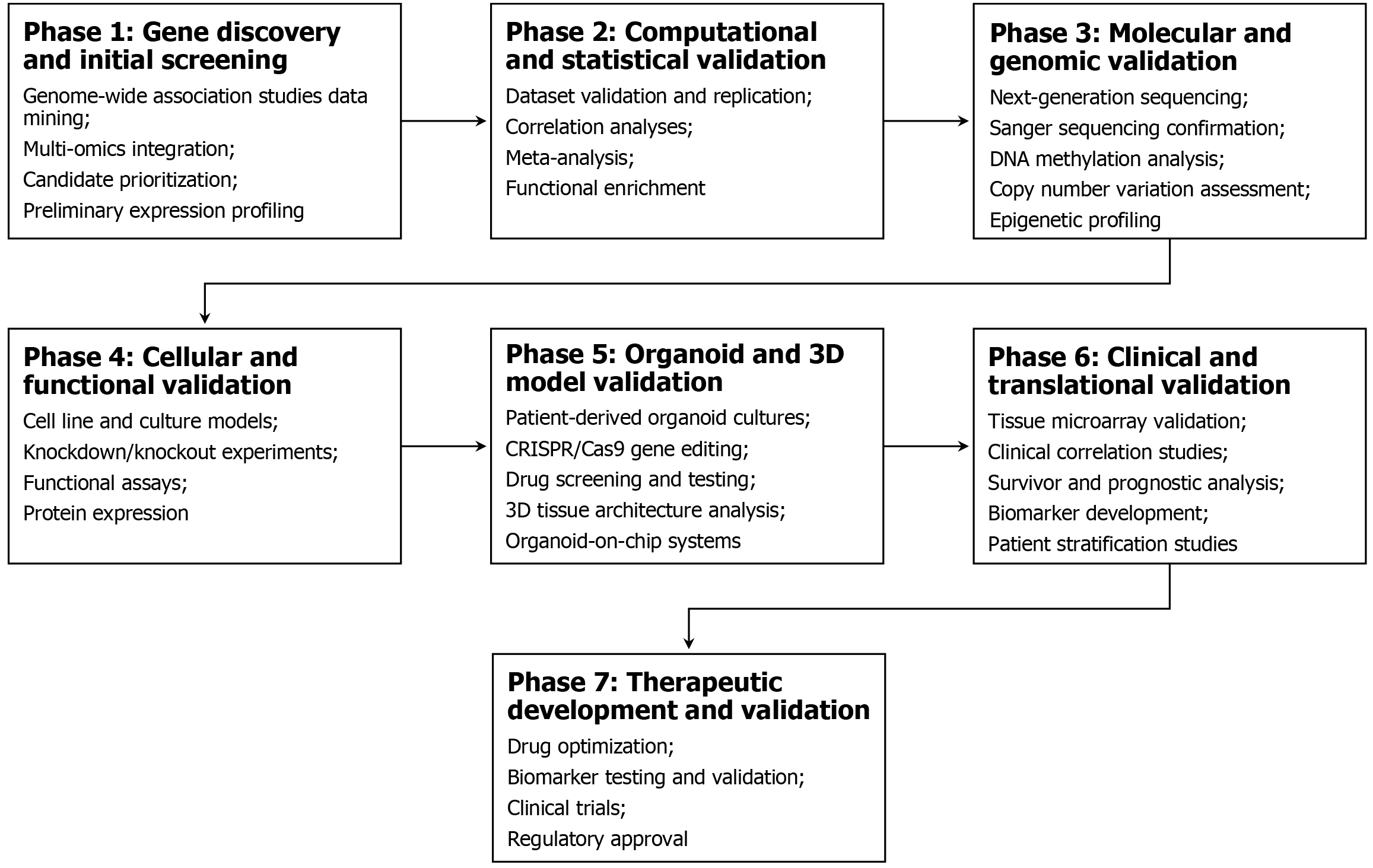
The emerging understanding of the gut microbiome’s role in polyposis represents another exciting avenue. Recent evidence suggests that colibactin-producing Escherichia coli may account for a significant proportion of unexplained adenomatous polyposes, with approximately 30% of such cases harboring APC mutations associated with bacterial genotoxin exposure[4]. Figure 1 describes a multi-omics validation and clinical actionability pipeline for novel candidate genes to bridge the gap between genomic discovery and clinical implementation.
The expansion of the genetic landscape of polyposis has implications for personalized medicine approaches. Genetic testing results increasingly inform surgical timing, surveillance strategies, and family counseling. However, identifying variants in genes with uncertain clinical significance poses challenges for genetic counseling and clinical management.
The study’s identification of variants in genes like FRK, which encodes a tumor suppressor involved in PTEN re
The integration of multi-omic approaches, as demonstrated in recent studies of FAP, may provide a more comprehensive understanding of disease mechanisms and inform personalized treatment strategies. Such approaches could help distinguish passengers from drivers among the numerous variants identified in comprehensive genetic testing[22].
Economic evaluation of expanded genetic testing remains complex. While the cost of sequencing has decreased dramatically, the downstream costs of genetic counseling, increased surveillance, and potential over-treatment of patients with VUS must be considered. Cost-effectiveness analyses of Lynch syndrome screening suggest that targeted approaches with appropriate risk thresholds can achieve favorable cost-effectiveness ratios of USD 8000-26000 per quality-adjusted life year[23].
For polyposis syndromes, the cost-effectiveness equation is complicated by the variable penetrance of newly identified genes and uncertainty regarding appropriate surveillance strategies. Studies suggest that genetic testing yields of > 10% support current guidance for constitutional testing, but this threshold may need refinement as our understanding of genetic architecture evolves[8].
Several critical research priorities emerge from this analysis. First, large-scale population-based studies are needed to establish the true prevalence and penetrance of variants in newly identified polyposis genes. Such studies should include diverse populations to ensure the broad applicability of findings.
Second, functional validation studies are essential for establishing the pathogenic role of candidate variants. The high rate of VUS in current studies underscores the need for robust functional assays and mechanistic studies to support variant classification.
Third, the development of clinical decision support tools incorporating genetic, clinical, and potentially microbiome data could optimize patient management and improve cost-effectiveness. AI-powered risk prediction models could help stratify patients for appropriate testing and surveillance strategies.
Finally, the integration of multi-omic approaches, including transcriptomics, proteomics, and metabolomics, may provide a more comprehensive understanding of disease mechanisms and identify novel therapeutic targets.
The study by Dos Santos et al[1] highlights the growing ability of comprehensive genetic analysis to uncover pathogenic variants in unexplained colorectal polyposis cases, marking significant progress in the field. However, challenges remain in translating these findings into clinical practice due to uncertain variant significance. As technological advancements outpace clinical interpretation, future efforts must prioritize functional validation, clinical correlation, and evidence-based strategies. True success lies not in gene discovery alone, but in improved patient outcomes through precision medicine. Continued multidisciplinary collaboration will be essential to transform genetic insights into effective, personalized prevention and treatment strategies for hereditary colorectal cancer syndromes.
| 1. | Dos Santos W, Pereira AS, Laureano T, de Andrade ES, Reis MT, Garcia FA, Campacci N, Melendez ME, Reis RM, Galvão HC, Palmero EI. Whole-exome sequencing identifies new pathogenic germline variants in patients with colorectal polyposis. World J Gastroenterol. 2025;31:104830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Grover S, Kastrinos F, Steyerberg EW, Cook EF, Dewanwala A, Burbidge LA, Wenstrup RJ, Syngal S. Prevalence and phenotypes of APC and MUTYH mutations in patients with multiple colorectal adenomas. JAMA. 2012;308:485-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 3. | Zaffaroni G, Mannucci A, Koskenvuo L, de Lacy B, Maffioli A, Bisseling T, Half E, Cavestro GM, Valle L, Ryan N, Aretz S, Brown K, Buttitta F, Carneiro F, Claber O, Blanco-Colino R, Collard M, Crosbie E, Cunha M, Doulias T, Fleming C, Heinrich H, Hüneburg R, Metras J, Nagtegaal I, Negoi I, Nielsen M, Pellino G, Ricciardiello L, Sagir A, Sánchez-Guillén L, Seppälä TT, Siersema P, Striebeck B, Sampson JR, Latchford A, Parc Y, Burn J, Möslein G. Updated European guidelines for clinical management of familial adenomatous polyposis (FAP), MUTYH-associated polyposis (MAP), gastric adenocarcinoma, proximal polyposis of the stomach (GAPPS) and other rare adenomatous polyposis syndromes: a joint EHTG-ESCP revision. Br J Surg. 2024;111:znae070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 43] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 4. | Joo JE, Viana-Errasti J, Buchanan DD, Valle L. Genetics, genomics and clinical features of adenomatous polyposis. Fam Cancer. 2025;24:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 5. | Toma C, Díaz-Gay M, Soares de Lima Y, Arnau-Collell C, Franch-Expósito S, Muñoz J, Overs B, Bonjoch L, Carballal S, Ocaña T, Cuatrecasas M, Díaz de Bustamante A, Castells A, Bujanda L, Cubiella J, Balaguer F, Rodríguez-Alcalde D, Fullerton JM, Castellví-Bel S. Identification of a Novel Candidate Gene for Serrated Polyposis Syndrome Germline Predisposition by Performing Linkage Analysis Combined With Whole-Exome Sequencing. Clin Transl Gastroenterol. 2019;10:e00100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 6. | Cornish AJ, Gruber AJ, Kinnersley B, Chubb D, Frangou A, Caravagna G, Noyvert B, Lakatos E, Wood HM, Thorn S, Culliford R, Arnedo-Pac C, Househam J, Cross W, Sud A, Law P, Leathlobhair MN, Hawari A, Woolley C, Sherwood K, Feeley N, Gül G, Fernandez-Tajes J, Zapata L, Alexandrov LB, Murugaesu N, Sosinsky A, Mitchell J, Lopez-Bigas N, Quirke P, Church DN, Tomlinson IPM, Sottoriva A, Graham TA, Wedge DC, Houlston RS. The genomic landscape of 2,023 colorectal cancers. Nature. 2024;633:127-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 86] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 7. | Rotunno M, Barajas R, Clyne M, Hoover E, Simonds NI, Lam TK, Mechanic LE, Goldstein AM, Gillanders EM. A Systematic Literature Review of Whole Exome and Genome Sequencing Population Studies of Genetic Susceptibility to Cancer. Cancer Epidemiol Biomarkers Prev. 2020;29:1519-1534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Mak S, Alexander JL, Clark SK, Hawkins M, Cuthill V, Latchford A, Monahan KJ. The Diagnostic Yield of Genetic Testing in Patients With Multiple Colorectal Adenomas: A Specialist Center Cohort Study. Clin Transl Gastroenterol. 2024;15:e00645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | NCCN. Genetic/Familial High-Risk Assessment: Colorectal, Endometrial, and Gastric. [cited August 27, 2025]. Available from: https://www.nccn.org/guidelines/guidelines-detail?category=2&id=1544. |
| 10. | Terlouw D, Suerink M, Singh SS, Gille HJJP, Hes FJ, Langers AMJ, Morreau H, Vasen HFA, Vos YJ, van Wezel T, Tops CM, Ten Broeke SW, Nielsen M. Declining detection rates for APC and biallelic MUTYH variants in polyposis patients, implications for DNA testing policy. Eur J Hum Genet. 2020;28:222-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. |
Yen T, Stanich PP, Axell L, Patel SG.
|
| 12. | Burke W, Parens E, Chung WK, Berger SM, Appelbaum PS. The Challenge of Genetic Variants of Uncertain Clinical Significance : A Narrative Review. Ann Intern Med. 2022;175:994-1000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 94] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 13. | Johnson H, Hartzfeld D, Peters M, Xiao J, Berksoy E, Kirshner C, Sancar F, Leach B, Cox H, Thompson G, Kong J, Thakkar S, Cuyun CG. SA21 Economic Analysis of Germline Genetic Testing to Assess for Hereditary Colorectal Cancer: A Systematic Review. Value Health. 2024;27:S399. [DOI] [Full Text] |
| 14. | Stjepanovic N, Moreira L, Carneiro F, Balaguer F, Cervantes A, Balmaña J, Martinelli E; ESMO Guidelines Committee. Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol. 2019;30:1558-1571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 163] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 15. | Monahan KJ, Bradshaw N, Dolwani S, Desouza B, Dunlop MG, East JE, Ilyas M, Kaur A, Lalloo F, Latchford A, Rutter MD, Tomlinson I, Thomas HJW, Hill J; Hereditary CRC guidelines eDelphi consensus group. Guidelines for the management of hereditary colorectal cancer from the British Society of Gastroenterology (BSG)/Association of Coloproctology of Great Britain and Ireland (ACPGBI)/United Kingdom Cancer Genetics Group (UKCGG). Gut. 2020;69:411-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 329] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 16. | Zare B, Monahan KJ. Guidelines for Familial Adenomatous Polyposis (FAP): challenges in defining clinical management for a rare disease. Fam Cancer. 2025;24:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 17. | García-Simón N, Valentín F, Royuela A, Hidalgo-Calero B, Blázquez-Martín R, de-Miguel-Reyes M, Sánchez-Zapardiel JM, Adán-Merino L, Rodríguez-Festa A, Gallego-Gil P, Mediavilla-Medel P, Quiñonero-Moreno L, Gutiérrez L, Herreros-de-Tejada A, Sánchez A, Provencio M, Romero A. Optimizing genetic testing strategy for suspected attenuated adenomatous polyposis: effective solutions in public health systems. Clin Transl Oncol. 2025;27:2710-2718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 18. | Wang KW, Dong M. Potential applications of artificial intelligence in colorectal polyps and cancer: Recent advances and prospects. World J Gastroenterol. 2020;26:5090-5100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Maes S, Deploey N, Peelman F, Eyckerman S. Deep mutational scanning of proteins in mammalian cells. Cell Rep Methods. 2023;3:100641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 20. | Yang M, Zhang D, Yuan Z, Chen D, Ju H, Wu B, Pan J, Gu G, Cui Y, Gu Y, Xu D, Yuan Y. Uncovering the genetic variation spectrum of colorectal polyposis from a multicentre cohort in China. NPJ Precis Oncol. 2025;9:75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 21. | Pak CM, Gilmore MJ, Bulkley JE, Chakraborty P, Dagan-Rosenfeld O, Foreman AKM, Gollob MH, Jenkins CL, Katz AE, Lee K, Meeks N, O'Daniel JM, Posey JE, Rego SM, Shah N, Steiner RD, Stergachis AB, Subramanian SL, Trotter T, Wallace K, Williams MS, Goddard KAB, Buchanan AH, Manickam K, Powell B, Ezzell Hunter J; ClinGen Resource. Implementing evidence-based assertions of clinical actionability in the context of secondary findings: Updates from the ClinGen Actionability Working Group. Genet Med. 2024;26:101164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 22. | Esplin ED, Hanson C, Wu S, Horning AM, Barapour N, Nevins SA, Jiang L, Contrepois K, Lee H, Guha TK, Hu Z, Laquindanum R, Mills MA, Chaib H, Chiu R, Jian R, Chan J, Ellenberger M, Becker WR, Bahmani B, Khan A, Michael B, Weimer AK, Esplin DG, Shen J, Lancaster S, Monte E, Karathanos TV, Ladabaum U, Longacre TA, Kundaje A, Curtis C, Greenleaf WJ, Ford JM, Snyder MP. Multiomic analysis of familial adenomatous polyposis reveals molecular pathways associated with early tumorigenesis. Nat Cancer. 2024;5:1737-1753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 23. | Dinh TA, Rosner BI, Atwood JC, Boland CR, Syngal S, Vasen HF, Gruber SB, Burt RW. Health benefits and cost-effectiveness of primary genetic screening for Lynch syndrome in the general population. Cancer Prev Res (Phila). 2011;4:9-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 145] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/