Published online Sep 14, 2025. doi: 10.3748/wjg.v31.i34.108807
Revised: June 23, 2025
Accepted: August 19, 2025
Published online: September 14, 2025
Processing time: 134 Days and 20.8 Hours
Congestive hepatopathy, also known as nutmeg liver, is liver damage secondary to chronic heart failure (HF). Its morphological characteristics in terms of medical imaging are not defined and remain unclear.
To leverage machine learning to capture imaging features of congestive hepatopathy using incidentally acquired computed tomography (CT) scans.
We retrospectively analyzed 179 chronic HF patients who underwent echocardiography and CT within one year. Right HF severity was classified into three grades. Liver CT images at the paraumbilical vein level were used to develop a ResNet-based machine learning model to predict tricuspid regurgitation (TR) severity. Model accuracy was compared with that of six gastroenterology and four radiology experts.
In the included patients, 120 were male (mean age: 73.1 ± 14.4 years). The accuracy of the results predicting TR severity from a single CT image for the machine learning model was significantly higher than the average accuracy of the experts. The model was found to be exceptionally reliable for predicting severe TR.
Deep learning models, particularly those using ResNet architectures, can help identify morphological changes associated with TR severity, aiding in early liver dysfunction detection in patients with HF, thereby improving outcomes.
Core Tip: Using ResNet-based deep learning on paraumbilical vein-level computed tomography images from 179 patients with chronic heart failure, we developed a model to predict tricuspid regurgitation severity. The model outperformed six gastroenterologists and three radiologists, excelling particularly in identifying severe TR. This novel, noninvasive approach captures subtle hepatic congestion features, enabling earlier detection of liver dysfunction secondary to heart failure. Our findings highlight the potential of machine learning to enhance diagnostic accuracy and guide timely intervention in congestive hepatopathy management.
- Citation: Miida S, Kamimura H, Fujiki S, Kobayashi T, Endo S, Maruyama H, Yoshida T, Watanabe Y, Kimura N, Abe H, Sakamaki A, Yokoo T, Tsukada M, Numano F, Kashimura T, Inomata T, Fuzawa Y, Hirata T, Horii Y, Ishikawa H, Nonaka H, Kamimura K, Terai S. Image analysis of cardiac hepatopathy secondary to heart failure: Machine learning vs gastroenterologists and radiologists. World J Gastroenterol 2025; 31(34): 108807
- URL: https://www.wjgnet.com/1007-9327/full/v31/i34/108807.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i34.108807
Nutmeg liver refers to a congested liver in which the central lobular regions appear dark red, whereas the peripheral areas turn yellow, creating a mottled appearance of dark red and yellow regions resembling the cut surface of a nutmeg[1].
Similarly, biochemical test results for liver function often show abnormalities, with a significant correlation between the severity of hepatic dysfunction and heart disease in cases of circulatory failure. Liver function tests have also been suggested in predicting the progression of heart disease[2].
Nutmeg liver in patients who succumbed to HF was first reported in 1833. In the early 1950s, Sherlock et al[3] eva
In 2012, Poelzl et al[4] investigated liver dysfunction in chronic HF, reporting its high prevalence, diverse characteristics, and significant impact on prognosis, thus highlighting an association between impaired liver function and poor outcomes in patients with HF[4].
van Deursen et al[5] examined abnormal liver function in patients with HF and correlated these abnormalities with hemodynamic profiles. Their findings emphasize the role of elevated central venous pressure and cardiac output in liver dysfunction[5].
Cardiac hepatopathy, also known as congestive hepatopathy, is attributed to chronic right HF, resulting in hepatic congestion and hypoxia. The molecular mechanisms involved include hypoxia-inducible factor-1α, oxidative stress, and inflammatory cytokines, such as tumor necrosis factor-α and interleukin-6, contributing to hepatocyte injury and fibrosis. Chronic venous congestion activates the hepatic stellate cells, promoting collagen production and liver fibrosis. These processes highlight the critical roles of systemic congestion, hypoxia-induced damage, and inflammatory signaling in the progression of cardiac hepatopathy[6-8].
Hemodynamic effects on the liver following surgery for congenital heart disease, particularly in relation to Fontan-associated liver disease, have garnered increasing attention. Advances in medical care, including improvements in surgical techniques, have significantly extended the long-term survival of patients with congenital heart disease, leading to a growing number of reports on the development of hepatocellular carcinoma (HCC) in this population[9].
HF can induce liver injury through mechanisms such as ischemia and passive congestion, resulting in conditions like congestive hepatopathy-commonly observed in patients with congenital heart disease and those who have undergone the Fontan procedure. This hepatopathy may progress to fibrosis, cirrhosis, or HCC, highlighting the importance of regular liver surveillance in patients with chronic cardiac disease. Early intervention and optimization of cardiac function can potentially reverse liver damage; however, in cases of end-stage HF unresponsive to medical therapy or mechanical support, combined heart-liver transplantation should be considered[10].
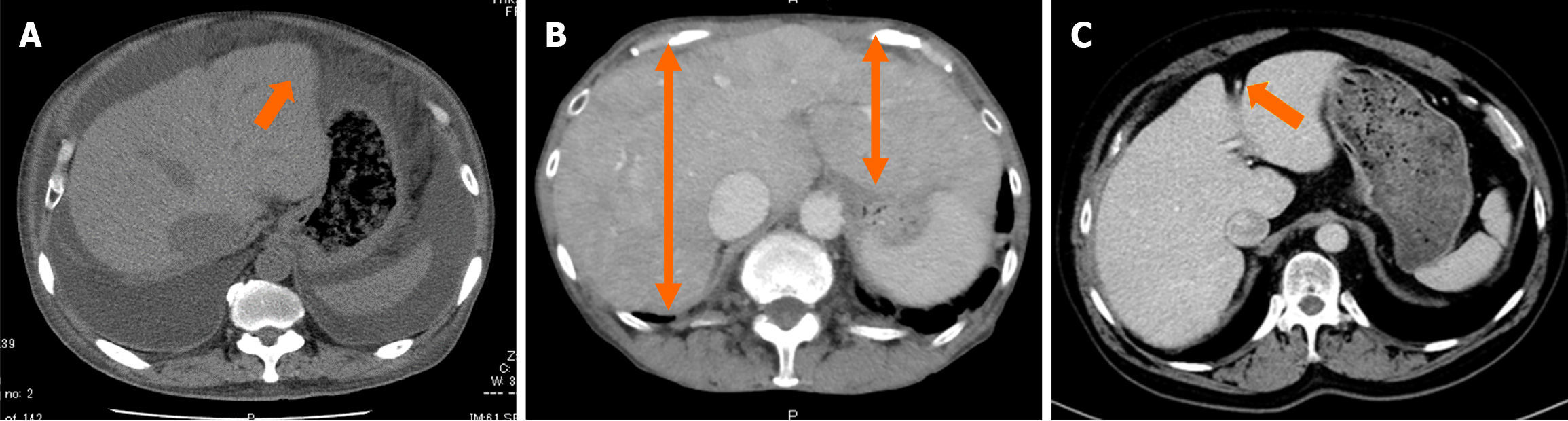
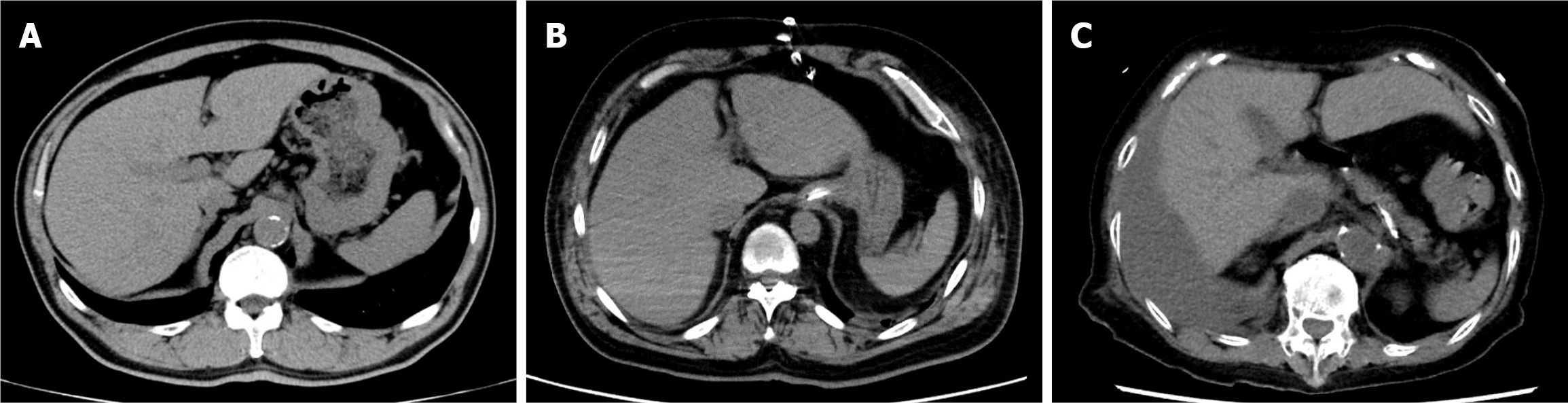
Early detection of cirrhosis and HCC warrants collaboration between cardiologists and hepatologists, with imaging serving as the basis. However, the imaging characteristics of congestive hepatopathy remain poorly understood. Although phrases such as “typical imaging findings: Congestive hepatopathy” exist and certain features are considered characteristic, the geometric definition of morphological changes resulting from congestive hepatomegaly remains challenging. The following features are characteristic of congestive hepatopathy (Figure 1): Enlargement of the hepatic veins and inferior vena cava (IVC); heterogeneous liver enhancement due to congestion; ascites in severe cases; hypoattenuating liver parenchyma on computed tomography (CT) or magnetic resonance imaging (MRI) due to reduced perfusion; dilation of the paraumbilical vein; and slightly blunted edges[11].
Extracting landmarks from CT images is challenging owing to the complexity of anatomical structures and individual variability, warranting advanced algorithms and specialized knowledge. Although the Fourier transform is useful for analyzing periodic patterns, its application to irregular shapes in CT images demands significant adaptation, including converting shape data into a suitable format. Additionally, the large data size of CT images imposes a heavy computational load, making optimization essential for real-time processing and large-scale analysis. Furthermore, inconsistencies attributed to variations in CT scanners and imaging conditions highlight the need for standardization to ensure reliable and consistent results[12,13].
To address these challenges, advancements are warranted in automating landmark extraction using artificial intelligence (AI) and machine learning, developing high-performance image-processing algorithms, and promoting data standardization. These technological advancements are expected to enable more efficient and accurate CT image analyses in the future.
The use of machine learning for imaging-based diagnosis of liver diseases has recently gained significant attention due to the complexity involved in accurately analyzing and interpreting large-scale, high-dimensional imaging data, as well as capturing subtle morphological or functional changes in the liver that may be difficult to detect with traditional approaches[14,15].
In patients diagnosed with HF via echocardiography, congestive hepatopathy identification based on liver morphology in incidentally captured CT images has not been extensively explored but is considered valuable and warrants further investigation.
From a medical imaging perspective, the morphological features of congestive hepatopathy remain unclear. Thus, this study aimed to leverage machine learning to capture imaging features of congestive hepatopathy using incidentally acquired CT scans. After training and validating the models using the test data, the accuracy of the machine learning models was compared with that of specialists. This approach seeks to raise awareness about the importance of liver disease in patients with HF.
All participants provided informed consent. Only anonymized clinical data obtained from patients who consented to the treatment were included in the study. Additionally, the patients were given the option to opt out of the study via an announcement posted on the institution’s homepage. The study design was approved by the Niigata University Ethics Committee (approval number: 2020-0199).
This retrospective, observational, single-center study was conducted at the Niigata University Medical and Dental Hospital. A total of 179 patients with chronic HF who underwent echocardiography between 2011 and 2020 were included. These patients underwent CT within 1 year before or after echocardiography; blood samples were collected within 6 months of imaging. Patients positive for hepatitis virus antibodies or with known liver diseases including alcoholic liver disease were excluded from the study and patients who were receiving outpatient care for liver cirrhosis in the gastroenterology department were excluded.
The severity of right HF was classified into three groups (mild, moderate, and severe) based on a comprehensive echocardiographic assessment. The data were then matched to ensure that there were no significant differences in the left ventricular ejection fraction across the three groups. The cutoffs between the three groups were determined according to the predefined echocardiographic criteria (e.g., right ventricular size, function, or other relevant parameters) as assessed by expert clinicians.
An echocardiographic evaluation was performed according to the American Society of Echocardiography for valvular regurgitation assessment[16].
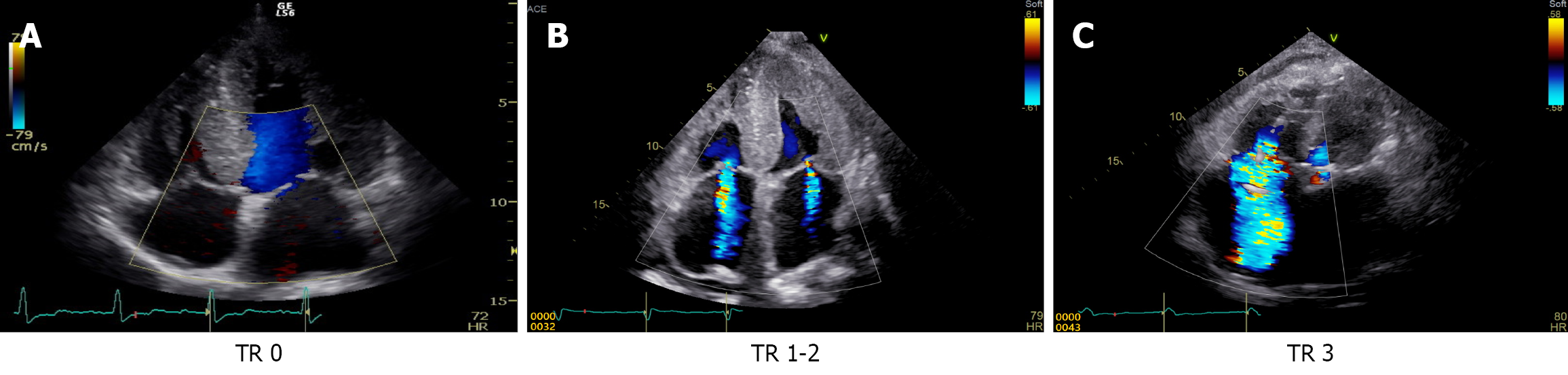
Tricuspid regurgitation (TR) severity is typically graded using guidelines from the American Heart Association and the American Society of Echocardiography. Key parameters included central jet area, vena contracta width, continuous-wave Doppler jet density/contour, and hepatic vein flow. Based on these findings, TR was classified as mild, moderate, and severe. Mild TR involves minimal regurgitant flow, whereas moderate TR exhibits intermediate jets and partial hemo
Echocardiographic evaluation was performed using one of the following ultrasound systems: (1) LOGIQ Series (LOGIQ B, LOGIQ E95; GE Healthcare, Chicago, IL, United States) with onboard software developed by GE Healthcare; and (2) Aplio Series (Aplio; Toshiba Medical Systems, Otawara, Tochigi, Japan) with integrated software developed by Toshiba Medical Systems.
Imaging of the training cohort was conducted on one of the following four CT scanners: (1) SOMATOM Definition Flash (Siemens, Munich, Germany); (2) Aquilion ONE (Canon Medical Systems, Otawara, Tochigi, Japan); (3) Aquilion 64 (Canon Medical Systems); and (4) Ingenuity Elite (Philips, Amsterdam, Netherlands).
All echocardiographic and CT acquisitions followed standard scanning protocols and the acquired images were analyzed according to the manufacturer-recommended software workflows. Such details ensure reproducibility for independent researchers.
These variables were selected owing to their suitability for retrospective CT analysis and their relevance as objective indicators. Laboratory data related to liver reserve function were retrospectively compiled for analysis.
For the analysis, hepatic CT images were chosen based on previous reports suggesting that congestive hepatopathy is better reflected in CT scans. Specifically, liver CT images at the level of the paraumbilical vein were extracted because this level clearly depicts signs of hepatic congestion. Additionally, cross-sectional views that highlighted right lobe hyper
Continuous variables were expressed as medians and interquartile ranges, and differences were assessed using the χ2 test, Fisher’s exact test, Mann-Whitney U test, or Kruskal-Wallis test. All the statistical analyses were performed using IBM SPSS Statistics for Windows (version 22.0; IBM Corp., Armonk, NY, United States). Statistical significance was defined as P < 0.05.
The machine learning model was built and refined using a computer with Windows version 11, an Intel Core i5 2-GHz processor, and RAM of 16 GB. The deep learning-integrated development environment used was NNC version 2.0.0 (Sony Corporation, Tokyo, Japan).
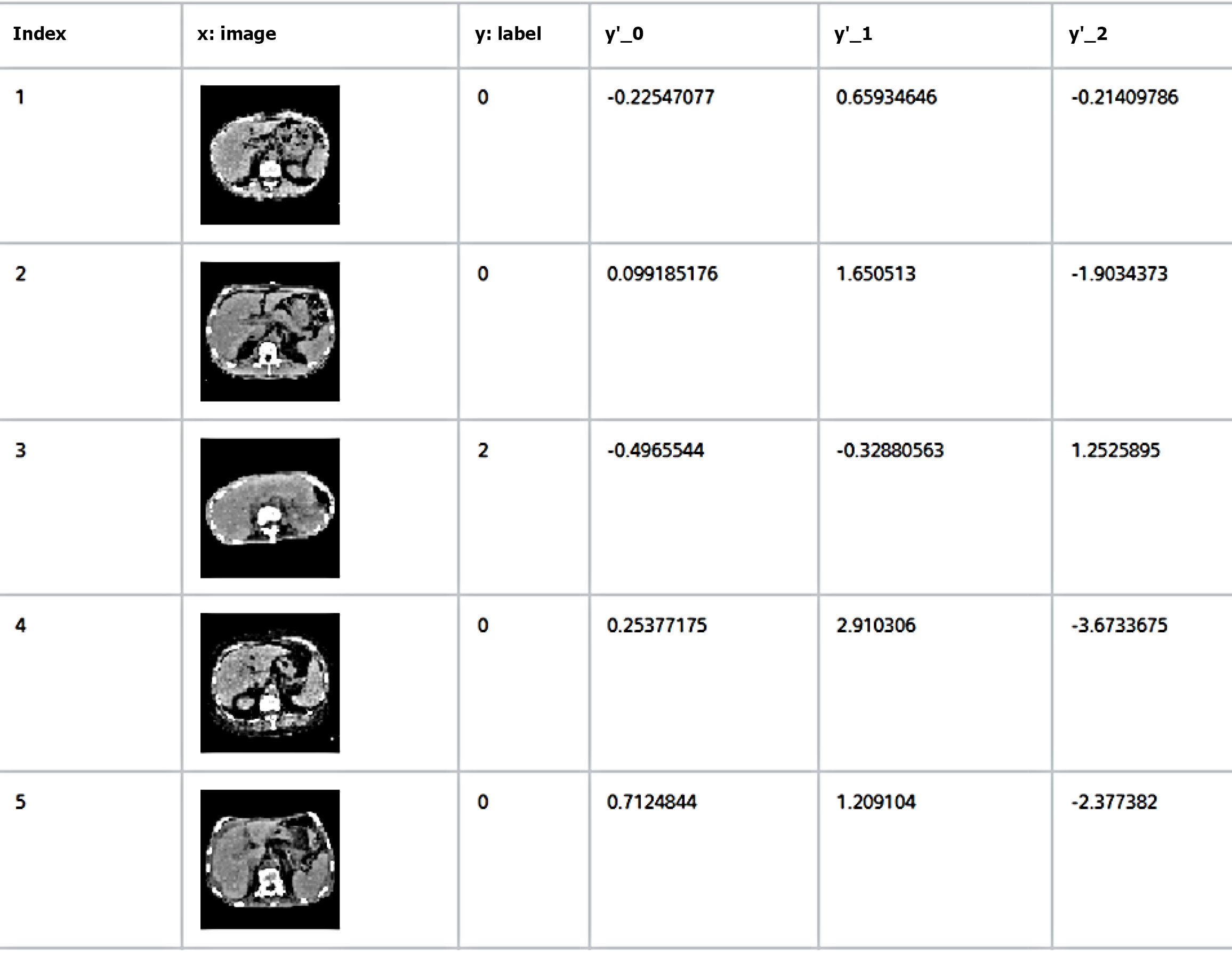
All images were automatically resized to 64 × 64 pixels using NNC, based on the input layer size for this analysis. Liver images from the 179 patients included in the study were categorized into three groups (mild, moderate, and severe) based on the severity of TR assessed via echocardiography.
The dataset was then divided into two subsets: One for training the deep learning model and the other for evaluating its post-training performance. Approximately 80% of the images (143 images) were used as training data, whereas the remaining 20% (36 images) were used as evaluation data. The selection of images for these two groups was performed randomly.
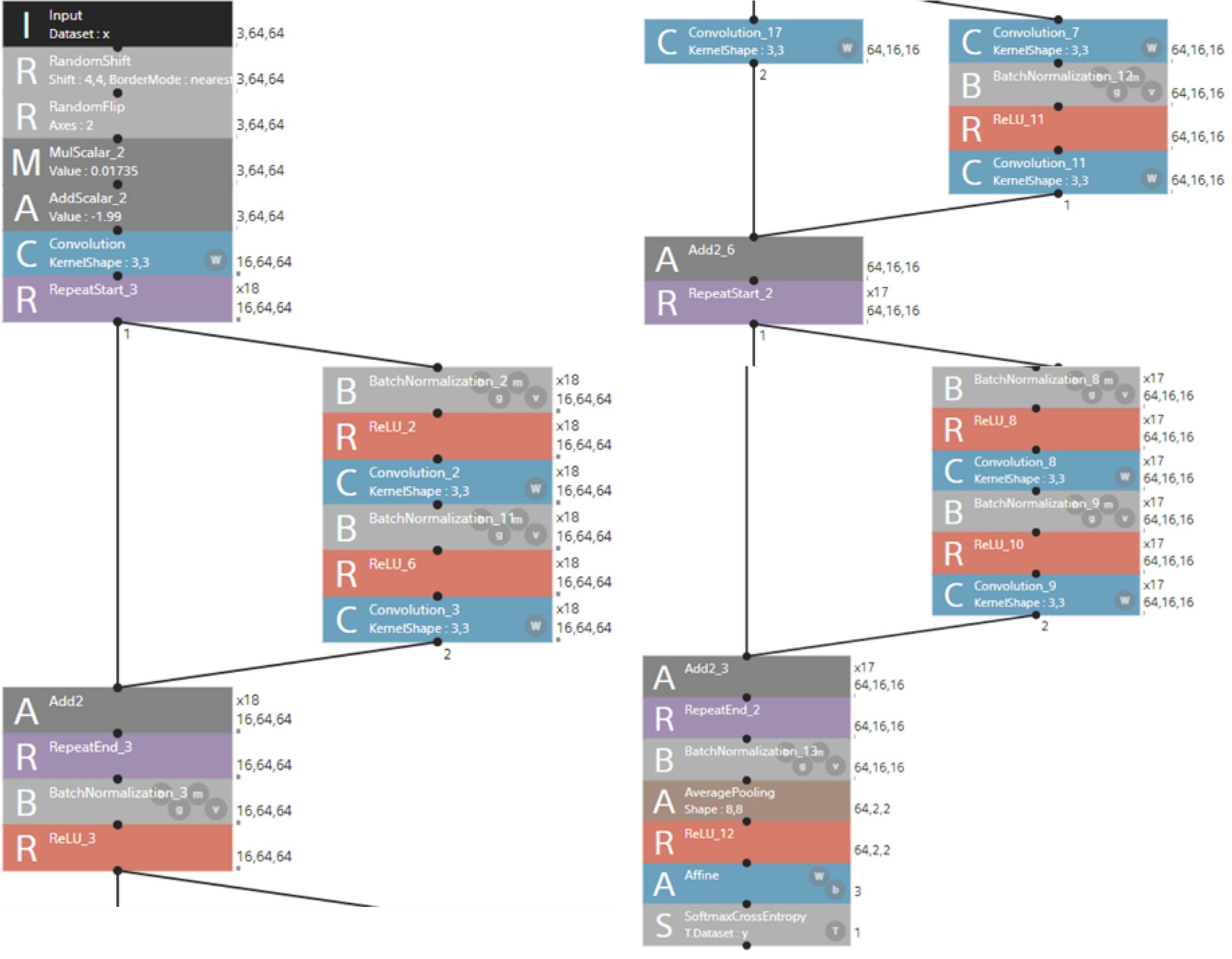
We developed a deep learning model based on a Residual Network-110 (ResNet-110) architecture to classify CT-derived hepatic features associated with TR severity. The input dataset had a shape of 3 × 64 × 643 and underwent preprocessing that included random pixel shifting (± 4 pixels), horizontal flipping (probability = 0.5), and normalization (scaling by 0.01735 followed by a shift of -1.99). All the images were resized to this fixed shape to ensure consistency across samples and scanners.
The model was built using proprietary software from Sony Communications Inc. (Tokyo, Japan), which enables graphical interface-based construction of neural networks without requiring programming skills. The implemented ResNet-110 model comprised 110 convolutional layers with bottleneck residual blocks and identity skip connections to facilitate stable gradient propagation and enable residual learning. The initial processing included a 7 × 7 convolution (stride 2), batch normalization, ReLU activation, and 3 × 3 max pooling. An initial convolutional block (3 × 33 kernel, 18 output channels) was repeated 18 times within the residual structure, followed by downsampling and expansion to 64 × 16 × 16. The final feature maps were subjected to global average pooling to reduce their dimensions to 64 × 2 × 264. Classification was achieved using an affine transformation followed by a softmax function, and the network was trained using a cross-entropy loss function (Figure 3).
The model was trained with a batch size of 32 over 60 epochs using stochastic gradient descent with an initial learning rate of 0.01, momentum of 0.9, and weight decay (L2 regularization) of 1e-4. The learning rate was reduced by a factor of 0.1 every 20 epochs to stabilize convergence.
To address class imbalance in the TR severity classification task, two strategies were employed: (1) Data augmentation through the random transformations described above; and (2) Application of a weighted cross-entropy loss function. Class weights were calculated inversely proportional to the class frequencies to ensure that minority classes were not underrepresented in the optimization process.
When the “learning curve” button was clicked to begin training, the progress of the learning curve was displayed on the graph monitor (training results screen). The training was completed after a specified number of epochs. The evaluation results could be viewed under the “Evaluation” tab.
Evaluation metrics were used to assess the quality of the machine learning model, focusing on the trade-off between precision (sensitivity) and recall (positive predictive value). The F-measure is often employed as a comprehensive evaluation metric, balancing both precision and recall, and is calculated as the harmonic mean.
To evaluate the machine learning model’s accuracy, the results were compared with the assessments of 6 gastroenterologists and 4 radiologists, each with more than 10 years of clinical experience. The experts used the same image pixel resolution (64 × 64 pixels) as the machine learning model for image classification (Figure 4).
The characteristics of the 179 patients included in this study are summarized in Table 1. Univariate analysis revealed significant differences in TR and TR pressure gradients (TRPG). In addition, the diameter of the IVC increased with the severity of TR. In blood tests, gamma-glutamyl transferase (γ-GTP), total bilirubin, creatinine, model for end-stage liver disease (MELD)-XI score, and brain natriuretic peptide (BNP) level showed significant differences with TR severity.
| TR | P value | |||||
| (1) Mild | (2) Moderate | (3) Severe | (1) vs (2) | (1) vs (3) | (2) vs (3) | |
| Blood test results | ||||||
| Sex, male | 34 (64.2) | 45 (68.2) | 41 (68.3) | NS | NS | NS |
| Age (year) | 68 (29-93) | 70 (25-91) | 68 (36-91) | NS | NS | NS |
| AST (IU/L) | 23 (11-83) | 23 (7-772) | 27 (12-67) | NS | NS | NS |
| ALT (IU/L) | 21 (6-107) | 20 (7-459) | 19 (6-71) | NS | NS | NS |
| γ-GTP (IU/L) | 36 (8-562) | 39 (9-425) | 53 (11-377) | NS | NS | 0.039 |
| Alb (g/dL) | 3.8 (2.1-4.9) | 3.8 (1.7-4.9) | 3.7 (1.4-4.4) | NS | NS | NS |
| T-Bil (mg/dL) | 0.6 (0.2-1.9) | 0.8 (0.3-3.7) | 1.1 (0.3-4.1) | NS | 0.025 | NS |
| Cre (mg/dL) | 0.86 (0.41-5.82) | 0.84 (0.38-8.93) | 1.01 (0.4-5.52) | NS | NS | 0.033 |
| eGFR (mL/minute) | 57.8 (2.6-128.8) | 62.8 (4.5-153.0) | 49.2 (16.2-95.0) | NS | NS | NS |
| Plt (× 104/µL) | 20.7 (7.9-49.6) | 19.8 (4.2-40.5) | 22.6 (12.7-32.7) | NS | NS | NS |
| ALBI score | -2.48 (-3.46 to -1.12) | -2.48 (-3.44 to -0.83) | -2.35 (-3.10 to -064) | NS | NS | NS |
| MELD-XI score | 6.39 (-3.12 to 35.9) | 7.49 (-6.62 to 33.9) | 8.96 (-0.15 to 42.9) | NS | 0.024 | NS |
| BNP (pg/dL) | 141 (11.8-2110) | 264 (6.6-3752) | 397 (35.0-3147) | NS | 0.047 | NS |
| Echocardiographic findings | ||||||
| LVDd (mm) | 55.1 (37.4-82.9) | 56.6 (38.7-96.9) | 59.9 (37.4-78.9) | NS | NS | NS |
| EF (%) | 36.8 (10.1-49.7) | 36.4 (10.8-49.8) | 34.7 (17.2-49.9) | NS | NS | NS |
| E/E’ | 15.7 (7.1-54.2) | 17.3 (2.8-50.7) | 15.5 (5.7-46.4) | NS | NS | NS |
| LAD (mm) | 42.8 (25.8-68.9) | 47.0 (24.5-87.1) | 49.1 (28.5-69.7) | NS | NS | NS |
| TRPG (mmHg) | 23.0 (9.7-49.0) | 33.9 (13.0-91.4) | 40.8 (22.2-70.9) | < 0.01 | < 0.01 | < 0.01 |
| IVC (mm) | 14.9 (5.5- 27.0) | 18.0 (2.2-28.2) | 23.0 (13.0-30.0) | < 0.01 | < 0.01 | 0.012 |
Upon admission, the patients were categorized into the following three groups based on the severity of their condition: Mild, moderate, and severe (Figure 5). A comparison of the clinical and echocardiographic parameters among these groups yielded the following findings. No significant differences were observed in the sex distribution, age, some liver function markers (aspartate transaminase, alanine transaminase, and albumin), renal function (estimated glomerular filtration rate), platelet count, or albumin bilirubin score across the groups. However, γ-GTP, total bilirubin, creatinine, MELD-XI score, and BNP level demonstrated significant differences between the severity groups. These blood test parameters were significantly different between the TR severity and TR mild or moderate groups.
Echocardiographic findings revealed no significant differences in the left ventricular end-diastolic diameter, left ventricular ejection fraction, E/E' ratio, or left atrial diameter among the groups. However, the TRPG and IVC diameters demonstrated significant differences between the severity groups. The TRPG increased significantly with disease severity (P < 0.01 for all intergroup comparisons) as did the IVC diameter (P < 0.05 for all intergroup comparisons).
Although most parameters did not vary significantly across the groups, the TRPG and IVC diameter significantly varied with increasing TR severity. These results suggest that TRPG and IVC diameter may serve as useful indicators of disease severity.
The constructed TR severity prediction model was evaluated using 36 test cases to validate its accuracy. The confusion matrix for the three-tier TR severity classification (mild, moderate, and severe) is shown in Table 2. The overall prediction accuracy of the model was calculated as 63.9%, demonstrating relatively high performance. The F-measure analysis revealed that the model’s accuracy improved with increasing TR severity. Specifically, the model tended to classify cases more accurately as the severity progressed from mild to moderate and then to severe. This finding suggests that the model is particularly reliable for predicting severe TR.
| Predicted severity | Recall | ||||
| Mild | Moderate | Severe | |||
| Actual severity | Mild | 6 | 7 | 3 | 0.375 |
| Moderate | 1 | 10 | 2 | 0.769 | |
| Severe | 0 | 0 | 7 | 1 | |
| Precision | 0.857 | 0.588 | 0.583 | ||
| F-measure | 0.522 | 0.667 | 0.737 | ||
The severe class demonstrated the highest recall (1.000), with all cases correctly classified. In contrast, the mild class had a relatively high number of misclassifications, resulting in a lower recall of 0.375. Regarding precision, the mild class achieved the highest value (0.857), whereas the severe class achieved a slightly lower precision (0.583). Comparing the F-measures, the severe class had the highest score (0.737), followed by the moderate (0.667) and mild (0.522) classes.
These results indicate that, although the classification performance for the severe class is strong, there is room for improvement in the classification of the mild class. The classification performance for the moderate class was relatively balanced overall; however, potential exists for further improvement in precision (Table 2).
For comparing the performance of the machine learning model with that of medical experts, hepatology and radiology specialists independently evaluated the same set of 36 cases. Before this evaluation, the specialists reviewed the same 143 hepatic CT images used for training the model to familiarize themselves with the features and patterns associated with TR severity. Subsequently, they predicted the TR severity for the 36 test cases.
The evaluation revealed that the average accuracy of the specialists was 41.4% (individual accuracies of 38.89%, 38.89%, 52.78%, 38.89%, 38.89%, 38.89%, 41.67%, 41.67%, 41.67%, and 41.67%), which was significantly lower than that of the machine learning model (63.9%).
This result highlights the potential of the machine learning model to surpass conventional methods relying on expert knowledge and experience. The ability of the model to process large datasets and recognize patterns with high precision is a significant advantage.
We further explored the potential of a multimodal model combining imaging and biochemical data, given the weak correlations previously reported between TR severity and liver function tests, such as γ-GTP and total bilirubin. As shown in Table 3, when γ-GTP and total bilirubin were incorporated into the model alongside para-umbilical CT feature maps, the classification performance improved. The multimodal model achieved an area under the curve (AUC) of 0.83 (95%CI: 0.78-0.88), with an accuracy of 68.5%, sensitivity of 70.0%, and specificity of 67.8% for distinguishing between the three TR severity classes. These results suggest that integrating selected biomarkers with imaging features can better enhance the predictive performance than using imaging data alone.
| Model | Input data | AUC (95%CI) | Accuracy (%) | Sensitivity (%) | Specificity (%) |
| Image-only model | Para-umbilical CT feature maps | 0.78 (0.72-0.84) | 63.9 | 60.0 | 66.7 |
| Multimodal model | CT features + γ-GTP, total bilirubin | 0.83 (0.78-0.88) | 68.5 | 70.0 | 67.8 |
To assess the generalizability of the model, we conducted 5-fold cross-validation. As shown in Table 4, the average accuracy across all folds was 63.8% ± 0.7%, with a corresponding mean AUC of 0.78 ± 0.01. The performance was consistent across folds, indicating stable classification ability.
| Analysis | Dataset configuration | Accuracy (%) | AUC (95%CI) |
| A 5-fold cross-validation | Fold 1 | 62.8 | 0.77 (0.69-0.85) |
| Fold 2 | 64.2 | 0.79 (0.71-0.87) | |
| Fold 3 | 63.5 | 0.78 (0.70-0.86) | |
| Fold 4 | 64.7 | 0.79 (0.71-0.87) | |
| Fold 5 | 63.9 | 0.78 (0.70-0.86) | |
| mean ± SD | 63.8 ± 0.7 | 0.78 ± 0.01 | |
In addition, a sensitivity analysis was performed to evaluate the effect of varying test set sizes on the model perfor
Given that imaging data were acquired from four different CT scanner models with potential variations in the resolution, contrast protocols, and slice thickness, we performed harmonization using ComBat batch correction to minimize scanner-related bias. Subgroup analyses were conducted to assess model performance by scanner type (Table 5).
| CT scanner (manufacturer & model) | No. of cases (n) | Accuracy (%) | AUC (95%CI) |
| SOMATOM Definition Flash (Siemens, Munich, Germany) | 45 | 64.4 | 0.78 (0.70-0.86) |
| Aquilion ONE (Canon Medical Systems, Otawara, Tochigi, Japan) | 40 | 63.1 | 0.76 (0.68-0.84) |
| Aquilion 64 (Canon Medical Systems, Otawara, Tochigi, Japan) | 35 | 62.9 | 0.77 (0.67-0.87) |
| Ingenuity Elite (Philips, Amsterdam, Netherlands) | 32 | 64.0 | 0.79 (0.69-0.89) |
| Overall mean | 152 | 63.6 | 0.78 (0.74-0.82) |
Following ComBat correction, model accuracy across scanners ranged narrowly from 62.9% to 64.4%, with AUC values between 0.76 and 0.79. The maximum deviation in the accuracy compared to the overall mean (63.6%) was within ± 1.0%, suggesting minimal scanner-specific influence on the model performance after harmonization. These findings indicate that batch correction effectively mitigated inter-scanner variability and preserved the predictive power of the model across different imaging platforms.
ResNets, introduced by He et al[17], was developed to address the degradation problem observed in very deep neural networks, in which the performance may plateau or degrade as the network depth increases owing to vanishing or exploding gradients. ResNets resolve this by incorporating residual learning and shortcut connections, enabling the effective training of very deep networks. Instead of directly mapping input x to output H (x), the network learns a residual function F (x) such that H (x) = F (x) + x. This reformulation allows identity mapping (x) to propagate directly, thereby reducing the optimization complexity and mitigating gradient vanishing issues.
The architecture of ResNets comprises residual blocks that include convolutional layers with batch normalization and ReLU activation functions. The shortcut connections in these blocks bypass one or more layers and directly add the input (xxx) to the output of the residual function F (x). This mechanism ensures effective gradient flow, even in networks with hundreds of layers. ResNets employ a bottleneck design that uses 1 × 1 convolutions for dimensionality reduction and restoration and 3 × 3 convolutions for spatial feature extraction to optimize the computational efficiency in deeper architectures.
ResNets have demonstrated significant advancements in medical imaging, particularly in disease diagnosis and classification. For instance, Abid et al[18] developed a multimodal medical image classification system combining ResNet-50 with a genetic algorithm, achieving an accuracy of 98.61% across diverse medical imaging modalities. Anwar et al[19] reviewed the effectiveness of ResNets in diagnosing various conditions, including lung tumors, skin diseases, breast cancer, and brain disorders. Lin et al[20] proposed a modified ResNet-50 architecture with adaptive learning rates and visualization tools to improve both the diagnostic accuracy and interpretability in grading diabetic retinopathy. These examples highlight the versatility and effectiveness of ResNet diagnostic accuracy advancement and complex medical image analyses.
HF is a rapidly growing public health issue that affects approximately 64 million individuals worldwide. Although the incidence of HF has remained stable globally, with a decline in developed countries, its prevalence continues to rise due to aging populations, improved survival rates following myocardial infarction, and advancements in HF treatment and management[21].
Notably, the incidence of HF with preserved ejection fraction (HFpEF) is increasing and is now recognized as a major contributor to the HF pandemic. HFpEF accounts for nearly half of all HF cases, with an estimated prevalence of 1.1% among adults in developed nations[22]. HFpEF is characterized by impaired left ventricular relaxation, resulting in the chronic elevation of pulmonary venous pressure, which places significant stress on the right ventricle and increases the likelihood of right HF[23].
Fortea et al[24] highlighted that congestive hepatopathy, a liver condition attributed to HF or hemodynamic abnormalities, is primarily from elevated central venous pressure associated with right HF. Chronic hepatic congestion leads to impaired liver blood flow, abnormal liver enzyme levels, fibrosis, and an increased risk of cirrhosis and HCC. Furthermore, the progression of hepatic congestion can affect other organs and cause systemic complications. Early detection and appropriate management, including imaging and blood tests, are critical for improving patient outcomes.
In recent years, liver complications, such as cirrhosis and HCC attributed to hepatic congestion from the Fontan circulation, have been increasingly reported, leading to the recognition of Fontan-associated liver disease[9]. Right ventricular function evaluation is essential, as it is a key prognostic determinant in HFpEF. Morphological liver abnormalities, including umbilical vein dilatation, hepatomegaly, and increased IVC diameter, are commonly observed in patients with portal hypertension[25]. Japan has one of the highest rates of CT globally, with 116 CT scanners per 100000 people, far surpassing other Organization for Economic Co-operation and Development countries[26]. Consequently, incidental liver imaging during cardiac or coronary CT is frequently performed in patients with HF. However, radiologists and cardiologists often focus primarily on the heart, making early detection of liver changes challenging[27].
Machine learning models have shown the potential to match or even surpass the diagnostic accuracy of medical experts in image recognition tasks[28].
The relationship between the heart and liver in patients with HF has been extensively studied, with many studies reporting a close connection. Alvarez and Mukherjee[29] suggested that passive hepatic congestion attributed to elevated central venous pressure could increase the liver enzyme levels and direct and indirect bilirubin levels[30]. End-stage HF is characterized by a cholestatic liver enzyme profile, with elevated γ-GTP and bilirubin levels[30].
In the present study, no strong correlation was observed between TR severity and blood test findings related to liver function or BNP levels, although some significant correlations were found (Table 1). However, the IVC diameter increased with TR severity, suggesting that morphological abnormalities in congested livers detected through medical imaging may precede changes in blood test findings.
Early detection of liver dysfunction is critical for improving outcomes, particularly in patients with cirrhosis, where screening for liver cancer plays a key role. Non-invasive tools such as abdominal ultrasound and serum tumor markers (e.g., alpha-fetoprotein and Protein Induced by Vitamin K Absence-II) are essential for identifying HCC, with advanced imaging modalities such as contrast-enhanced CT or MRI providing a detailed evaluation for high-risk cases. Screening intervals are typically 6 months but may be shortened to 3-4 months for high-risk patients[31].
Early intervention and minimally invasive treatments, such as radiofrequency ablation, have been shown to improve prognosis. Machine learning has advanced liver cancer screening, with models, such as the transformer-based model developed by Sato et al[32], demonstrating superior performance in survival prediction for patients with HCC under
By integrating such innovations with cardiac imaging, the present study contributes to a growing field that aims to unify cross-organ diagnostics via AI-bridging HF and liver dysfunction through imaging-based markers.
The constructed TR severity prediction model demonstrated promising practicality and reliability, particularly for assessing severe cases. This study highlights the potential utility of deep learning models, particularly those using ResNet architectures, for identifying morphological changes associated with TR severity. These findings suggest that such models can serve as effective diagnostic tools, aiding in the early detection of liver dysfunction in patients with HF, thereby improving patient outcomes. These models enhance diagnostic accuracy and reduce the workload of healthcare professionals, thus holding significant value for use in clinical practice (Supplementary Figure 1).
We would like to express our sincere gratitude to the CT and echocardiography technologists of Niigata University Medical and Dental Hospital for their invaluable assistance and to all staff members involved in cardiac surgeries for their unwavering support.
| 1. | Kiernan F. XXIX. The anatomy and physiology of the liver. Phil Trans R Soc. 1833;123:711-770. [DOI] [Full Text] |
| 2. | Evans JM, Zimmerman HJ, Wilmer JG, Thomas LJ, Ethridge CB. Altered liver function of chronic congestive heart failure. Am J Med. 1952;13:704-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 15] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 3. | Sherlock S. The liver in heart failure; relation of anatomical, functional, and circulatory changes. Br Heart J. 1951;13:273-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 199] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 4. | Poelzl G, Ess M, Mussner-Seeber C, Pachinger O, Frick M, Ulmer H. Liver dysfunction in chronic heart failure: prevalence, characteristics and prognostic significance. Eur J Clin Invest. 2012;42:153-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 126] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 5. | van Deursen VM, Damman K, Hillege HL, van Beek AP, van Veldhuisen DJ, Voors AA. Abnormal liver function in relation to hemodynamic profile in heart failure patients. J Card Fail. 2010;16:84-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 212] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 6. | Xanthopoulos A, Starling RC, Kitai T, Triposkiadis F. Heart Failure and Liver Disease: Cardiohepatic Interactions. JACC Heart Fail. 2019;7:87-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 220] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 7. | Kou K, Li S, Qiu W, Fan Z, Li M, Lv G. Hypoxia-inducible factor 1α/IL-6 axis in activated hepatic stellate cells aggravates liver fibrosis. Biochem Biophys Res Commun. 2023;653:21-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 8. | Berezin AA, Obradovic Z, Berezina TA, Boxhammer E, Lichtenauer M, Berezin AE. Cardiac Hepatopathy: New Perspectives on Old Problems through a Prism of Endogenous Metabolic Regulations by Hepatokines. Antioxidants (Basel). 2023;12:516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Asrani SK, Warnes CA, Kamath PS. Hepatocellular carcinoma after the Fontan procedure. N Engl J Med. 2013;368:1756-1757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 10. | Sessa A, Allaire M, Lebray P, Medmoun M, Tiritilli A, Iaria P, Cadranel JF. From congestive hepatopathy to hepatocellular carcinoma, how can we improve patient management? JHEP Rep. 2021;3:100249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 11. | Wells ML, Fenstad ER, Poterucha JT, Hough DM, Young PM, Araoz PA, Ehman RL, Venkatesh SK. Imaging Findings of Congestive Hepatopathy. Radiographics. 2016;36:1024-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 12. | Dujardin JP. A template-dependent semilandmarks treatment and its use in medical entomology. Infect Genet Evol. 2019;70:197-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Zelditch ML, Swiderski DL, Sheets HD. Landmarks and Semilandmarks. In: Zelditch ML, Swiderski DL, Sheets HD. Geometric Morphometrics for Biologists. Amsterdam: Academic Press, 2012. [DOI] [Full Text] |
| 14. | Nam D, Chapiro J, Paradis V, Seraphin TP, Kather JN. Artificial intelligence in liver diseases: Improving diagnostics, prognostics and response prediction. JHEP Rep. 2022;4:100443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 15. | Fazlollahi AM, Bakhaidar M, Alsayegh A, Yilmaz R, Winkler-Schwartz A, Mirchi N, Langleben I, Ledwos N, Sabbagh AJ, Bajunaid K, Harley JM, Del Maestro RF. Effect of Artificial Intelligence Tutoring vs Expert Instruction on Learning Simulated Surgical Skills Among Medical Students: A Randomized Clinical Trial. JAMA Netw Open. 2022;5:e2149008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 16. | Zoghbi WA, Adams D, Bonow RO, Enriquez-Sarano M, Foster E, Grayburn PA, Hahn RT, Han Y, Hung J, Lang RM, Little SH, Shah DJ, Shernan S, Thavendiranathan P, Thomas JD, Weissman NJ. Recommendations for Noninvasive Evaluation of Native Valvular Regurgitation: A Report from the American Society of Echocardiography Developed in Collaboration with the Society for Cardiovascular Magnetic Resonance. J Am Soc Echocardiogr. 2017;30:303-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1533] [Cited by in RCA: 2684] [Article Influence: 298.2] [Reference Citation Analysis (0)] |
| 17. | He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27-30; Las Vegas, NV, United States. IEEE, 2016: 770-778. [RCA] [DOI] [Full Text] [Cited by in Crossref: 72655] [Cited by in RCA: 24717] [Article Influence: 2471.7] [Reference Citation Analysis (0)] |
| 18. | Abid MH, Ashraf R, Mahmood T, Faisal CMN. Multi-modal medical image classification using deep residual network and genetic algorithm. PLoS One. 2023;18:e0287786. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 19. | Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical Image Analysis using Convolutional Neural Networks: A Review. J Med Syst. 2018;42:226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 554] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 20. | Lin CL, Wu KC. Development of revised ResNet-50 for diabetic retinopathy detection. BMC Bioinformatics. 2023;24:157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 21. | Shahim B, Kapelios CJ, Savarese G, Lund LH. Global Public Health Burden of Heart Failure: An Updated Review. Card Fail Rev. 2023;9:e11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 283] [Reference Citation Analysis (0)] |
| 22. | Owan TE, Redfield MM. Epidemiology of diastolic heart failure. Prog Cardiovasc Dis. 2005;47:320-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 319] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 23. | Melenovsky V, Hwang SJ, Lin G, Redfield MM, Borlaug BA. Right heart dysfunction in heart failure with preserved ejection fraction. Eur Heart J. 2014;35:3452-3462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 520] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 24. | Fortea JI, Puente Á, Cuadrado A, Huelin P, Pellón R, González Sánchez FJ, Mayorga M, Cagigal ML, García Carrera I, Cobreros M, Crespo J, Fábrega E. Congestive Hepatopathy. Int J Mol Sci. 2020;21:9420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 25. | Morales A, Hirsch M, Schneider D, González D. Congestive hepatopathy: the role of the radiologist in the diagnosis. Diagn Interv Radiol. 2020;26:541-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Martella M, Lenzi J, Gianino MM. Diagnostic Technology: Trends of Use and Availability in a 10-Year Period (2011-2020) among Sixteen OECD Countries. Healthcare (Basel). 2023;11:2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 27. | Haddadin R, Aboujaoude C, Trad G. Congestive Hepatopathy: A Review of the Literature. Cureus. 2024;16:e58766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 28. | Garcea F, Serra A, Lamberti F, Morra L. Data augmentation for medical imaging: A systematic literature review. Comput Biol Med. 2023;152:106391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 114] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 29. | Alvarez AM, Mukherjee D. Liver abnormalities in cardiac diseases and heart failure. Int J Angiol. 2011;20:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 158] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 30. | Dichtl W, Vogel W, Dunst KM, Grander W, Alber HF, Frick M, Antretter H, Laufer G, Pachinger O, Pölzl G. Cardiac hepatopathy before and after heart transplantation. Transpl Int. 2005;18:697-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Kokudo N, Takemura N, Hasegawa K, Takayama T, Kubo S, Shimada M, Nagano H, Hatano E, Izumi N, Kaneko S, Kudo M, Iijima H, Genda T, Tateishi R, Torimura T, Igaki H, Kobayashi S, Sakurai H, Murakami T, Watadani T, Matsuyama Y. Clinical practice guidelines for hepatocellular carcinoma: The Japan Society of Hepatology 2017 (4th JSH-HCC guidelines) 2019 update. Hepatol Res. 2019;49:1109-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 411] [Article Influence: 58.7] [Reference Citation Analysis (1)] |
| 32. | Sato M, Moriyama M, Fukumoto T, Yamada T, Wake T, Nakagomi R, Nakatsuka T, Minami T, Uchino K, Enooku K, Nakagawa H, Shiina S, Koike K, Fujishiro M, Tateishi R. Development of a transformer model for predicting the prognosis of patients with hepatocellular carcinoma after radiofrequency ablation. Hepatol Int. 2024;18:131-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 33. | Wei H, Zheng T, Zhang X, Wu Y, Chen Y, Zheng C, Jiang D, Wu B, Guo H, Jiang H, Song B. MRI radiomics based on deep learning automated segmentation to predict early recurrence of hepatocellular carcinoma. Insights Imaging. 2024;15:120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 34. | Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S. Deep learning for staging liver fibrosis on CT: a pilot study. Eur Radiol. 2018;28:4578-4585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 35. | Yu Y, Yang Y, Li Q, Yuan J, Zha Y. Predicting metabolic dysfunction associated steatotic liver disease using explainable machine learning methods. Sci Rep. 2025;15:12382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/