Published online Aug 28, 2025. doi: 10.3748/wjg.v31.i32.106424
Revised: May 26, 2025
Accepted: August 1, 2025
Published online: August 28, 2025
Processing time: 118 Days and 19.5 Hours
Helicobacter pylori (H. pylori), a globally prevalent pathogen, is exhibiting increasing rates of antimicrobial resistance. However, clinical implementation of pre-treatment susceptibility testing remains limited due to the organism’s fastidious growth requirements and prolonged culture time.
To propose a novel detection method utilizing antibiotic-supplemented media to inhibit susceptible strains, while resistant isolates were identified through urease-mediated hydrolysis of urea, inducing a phenol red color change for visual confirmation.
Colombia agar was supplemented with urea, phenol red, and nickel chloride, and the final pH was adjusted to 7.35. Antibiotic-selective media were prepared by incorporating amoxicillin (0.5 μg/mL), clarithromycin (2 μg/mL), metronidazole (8 μg/mL), or levofloxacin (2 μg/mL) into separate batches. Gastric antral biopsies were homogenized and inoculated at 1.0 × 105 CFU onto the media, and then incubated under microaerobic conditions at 37 °C for 28-36 hours. Resistance was determined based on a color change from yellow to pink, and the results were validated via broth microdilution according to Clinical and Laboratory Standards Institute guidelines.
After 28-36 hours of incubation, the drug-resistant H. pylori isolates induced a light red color change in the medium. Conversely, susceptible strains (H. pylori 26695 and G27) produced no visible color change. Compared with the conventional 11-day protocol, the novel method significantly reduced detection time. Among 201 clinical isolates, 182 were successfully evaluated using the new method, resulting in a 90.5% detection rate. This was consistent with the 95.5% agreement rate observed when compared with microdilution-based susceptibility testing. The success rate of the novel approach was significantly higher than that of the comparative method (P < 0.01). The accuracy of the new method was comparable to that of the dilution method.
The novel detection method can rapidly detect H. pylori drug resistance within 28-36 hours. With its operational simplicity and high diagnostic performance, it holds strong potential for clinical application in the management of H. pylori antimicrobial resistance.
Core Tip: Helicobacter pylori (H. pylori) drug resistance hinders clinical treatment, with slow, inaccurate detection prolonging therapy and reducing efficacy. This paper presents a novel rapid detection method for H. pylori resistance, identifying it in 28-36 hours - much faster than traditional approaches. It offers high accuracy, reliable resistance assessment, and easy operation, suitable for wide use in clinics and research. This method shows great potential to optimize clinical decisions, improve outcomes, and aid H. pylori infection management.
- Citation: Guan AX, Yang SY, Wu T, Zhou WT, Chen H, Huang ZS, Luo PP, Huang YQ. Novel chromogenic medium-based method for the rapid detection of Helicobacter pylori drug resistance. World J Gastroenterol 2025; 31(32): 106424
- URL: https://www.wjgnet.com/1007-9327/full/v31/i32/106424.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i32.106424
Helicobacter pylori (H. pylori) is a microaerophilic, gram-negative bacterium[1], which has infected more than 50% of the world population[2]. Once an infection with H. pylori occurs, it can lead to gastritis, gastric cancer, and other diseases, posing a serious threat to human health[3]. Current treatment guidelines recommend quadruple therapy, which includes a proton pump inhibitor, bismuth, and two antibiotics[4]. However, with the widespread use of antibiotics, resistance in H. pylori has become increasingly problematic, particularly against metronidazole (MTZ) and clarithromycin (CLR), and resistance rates were reported as high as 71% and 55%, respectively[5,6]. In some areas, the resistance rate for MTZ has reached up to 81.7%[7], resulting in reduced eradication rates. Antimicrobial susceptibility testing is crucial for guiding effective therapies to address the growing challenges associated with drug resistance[8,9]. Currently, the primary methods for detecting H. pylori resistance involve classical culture-based methods. These methods require the collection of gastric mucosal biopsies, followed by isolation and culture to obtain pure bacterial strains. Resistance is therefore assessed using methods, such as the broth or agar dilution method and the Kirby-Bauer (disc diffusion) test[10]. However, such detection methods typically require 11-12 days to complete and are susceptible to procedural errors[11], which may compromise clinical decision-making An alternative strategy involves direct polymerase chain reaction (PCR) amplification of resistance genes from gastric mucosa or fecal samples An alternative strategy involves direct PCR amplification of resistance genes from gastric mucosa or fecal samples[12]. Although this approach is more rapid, the genotypic diversity of resistant H. pylori strains mainly does not correspond fully with phenotypic resistance[13], leading to false-negative results. For instance, CLR resistance in H. pylori is commonly associated with mutations at the A2142G and A2143G sites[14], while resistant strains without these mutations have also been identified[15]. Moreover, mutations in alternative genomic regions can lead to misinterpretation of resistance status. High-throughput genetic testing is conducted by examining stool samples to avoid invasive procedures, while the coincidence rate between genotypes from stool and biopsy samples is approximately 89%[16], indicating limited reliability. Therefore, resistance testing is recommended only for patients with treatment failure, and it remains underutilized. Some scholars have proposed the direct use of biopsy tissues for drug susceptibility testing[17], while the accuracy of this method needs further improvement. Consequently, the development of rapid, accurate, and clinically applicable methods for detecting H. pylori drug resistance essential. This study developed a colorimetric detection method using a specialized culture medium designed to identify H. pylori drug resistance. The approach determined the optimal antibiotic concentrations required to inhibit bacterial growth. In cases where drug-resistant strains proliferate, urease activity increases the medium’s alkalinity, resulting in a color change of phenol red. This change enables rapid identification of resistance. The method appeared fast, simple, accurate, and cost-effective, providing technical support for the clinical application of H. pylori drug resistance testing.
The H. pylori strains, including susceptible strains G27 and 26695, as well as resistant strains 159, 161, 162, 163, 286, 287, 289, and 290, were kindly provided by Professor Hong-Kai Bi from Nanjing Medical University. Furthermore, 201 clinical strains were collected from the Endoscopy Center of the Affiliated Hospital of Youjiang Medical University for Nationalities. All strains were retrieved from a -80 °C ultra-low temperature freezer. The bacterial pellets were resuspended in a Columbia medium (OXOID, Lot: 2179850) and incubated in a tri-gas incubator (85% N2, 5% O2, 10% CO2) at 37 °C for resuscitation. Colonies in the logarithmic growth phase were selected for subculturing.
Amoxicillin (AMX) (Lot: C15338727), CLR (Lot: C114967375), MTZ (Lot: C11.594013), and levofloxacin (LVX) (Lot: C10619762) were each prepared at a concentration of 1 mg/mL for subsequent use. In the first well of a 96-well microtiter plate, 173.6 μL of brain heart infusion (BHI) medium (OXOID, Lot: 7075659) was added, followed by the addition of 6.4 μL of the antimicrobial drug. Serial two-fold dilutions were performed from wells 1 to 11 to achieve final drug concentrations of 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125, 0.0625, and 0.3125 μg/mL. The 12th well served as a drug-free control. A bacterial suspension was prepared by adjusting the OD600 to 0.3 (approximately 1 × 108 CFU/mL), followed by a 10-fold dilution. Ten microliters of the diluted suspension were added to wells 1-10 and 12, yielding a final concentration of 1 × 106 CFUs/mL. Well 11, containing only sterile water and medium, served as the no-bacteria control. The plate was incubated for 72 hours under microaerophilic conditions. Negative controls (containing only sterile water, medium, and drug) and positive controls (containing only medium and bacteria) were included. The minimum inhibitory concentration (MIC) was defined as the lowest antibiotic concentration that completely inhibited visible bacterial growth. If a skipped well was observed, the MIC was determined based on the highest concentration showing complete inhibition.
Evaluation of drug resistance of H. pylori and calculation of the breakpoint range: To assess the breakpoint range, H. pylori suspensions were prepared at concentrations of 1 × 106, 1 × 105, and 1 × 104 CFU/mL. Drug susceptibility to LVX, CLR, MTZ, and AMX was evaluated using the MIC determination method. The fold differences in MICs between bacterial concentrations were calculated to estimate breakpoint variability. According to the Clinical and Laboratory Standards Institute (CLSI) guidelines, the resistance breakpoints for H. pylori at 1 ×106 CFU/mL were defined as 2 μg/mL for LVX, 2 μg/mL for CLR, 8 μg/mL for MTZ, and 0.5 μg/mL for AMX. Then, the range of resistance breakpoints was determined for a bacterial dose of 1 × 105 CFUs.
Verification of the estimated breakpoint value: According to the estimated range of drug resistance breakpoints, Columbia agar was prepared with twofold serial concentrations of each antibiotic to create a solid medium gradient. H. pylori was suspended in BHI broth and adjusted to 1 × 108 CFU/mL, followed by a 10-fold dilution. Ten microliters of the diluted suspension (final concentration 1 × 107 CFU/mL) were inoculated onto each antibiotic-containing medium. Plates were incubated at 37 °C for 3-5 days in a tri-gas incubator (85% N2, 5% O2, 10% CO2). Bacterial growth was monitored and recorded to verify the accuracy of the estimated resistance breakpoints.
Preparation of solutions: The following stock solutions were prepared, sterilized, and stored at -20 °C for future use: 10 mL of 0.2% phenol red solution (Lot: SHBM7900), 50 mL of 20% urea solution (Lot: C15338727), 10 mL of 5% NaOH solution (Lot: 190826978E), and 10 mL of 100 μmol/L nickel chloride (NiCl2) solution (Lot: C12077718).
Determining the urea concentration in the characteristic medium: Columbia medium (2.0 g) was dissolved in 45 mL of purified water, transferred to a conical flask, and sterilized. After cooling to approximately 55 °C, 7 mL of calf serum (Lot: PRXNXQ-500X) and 1% (v/v) of H. pylori selective supplement (Dent additive) were added. Solid media were subsequently prepared with urea concentrations of 0.6 mg/mL, 1.2 mg/mL, and 2.4 mg/mL (scheme 1). H. pylori suspensions (1 × 105 CFU/mL) were inoculated onto the prepared media and incubated at 37 °C in a tri-gas incubator (85% N2, 5% O2, 10% CO2) for 3-5 days. The results were recorded to evaluate the optimal urea concentration.
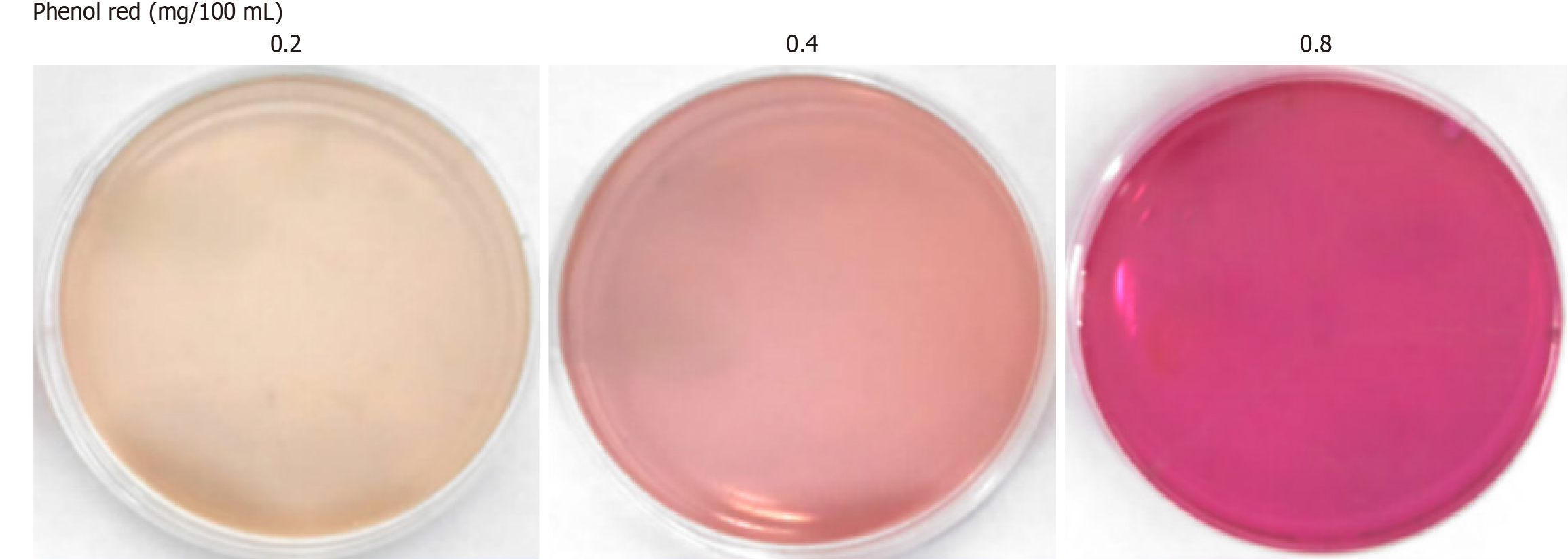
Determining the phenol red concentration in the characteristic medium: In scheme 1, the phenol red solution was added at final concentrations of 0.2 mg/100 mL, 0.4 mg/100 mL, 0.8 mg/100 mL, and 1.6 mg/100 mL, and the solutions were thoroughly mixed. Solid media (scheme 2) were subsequently prepared, and 1 × 105 CFUs/mL of H. pylori were inoculated onto the media from scheme 2. After incubation for 3-5 days, bacterial growth and medium discoloration were evaluated to determine the optimal phenol red concentration.
Adjusting the pH value: As presented in scheme 2, the final pH of the media was adjusted to 7.15, 7.25, 7.35, or 7.45. Solid media (scheme 3) were prepared, inoculated with 1 × 105 CFUs/mL of H. pylori, and incubated for 3-5 days. Observations of bacterial growth and time to color change were utilized to identify the optimal pH for the characteristic medium.
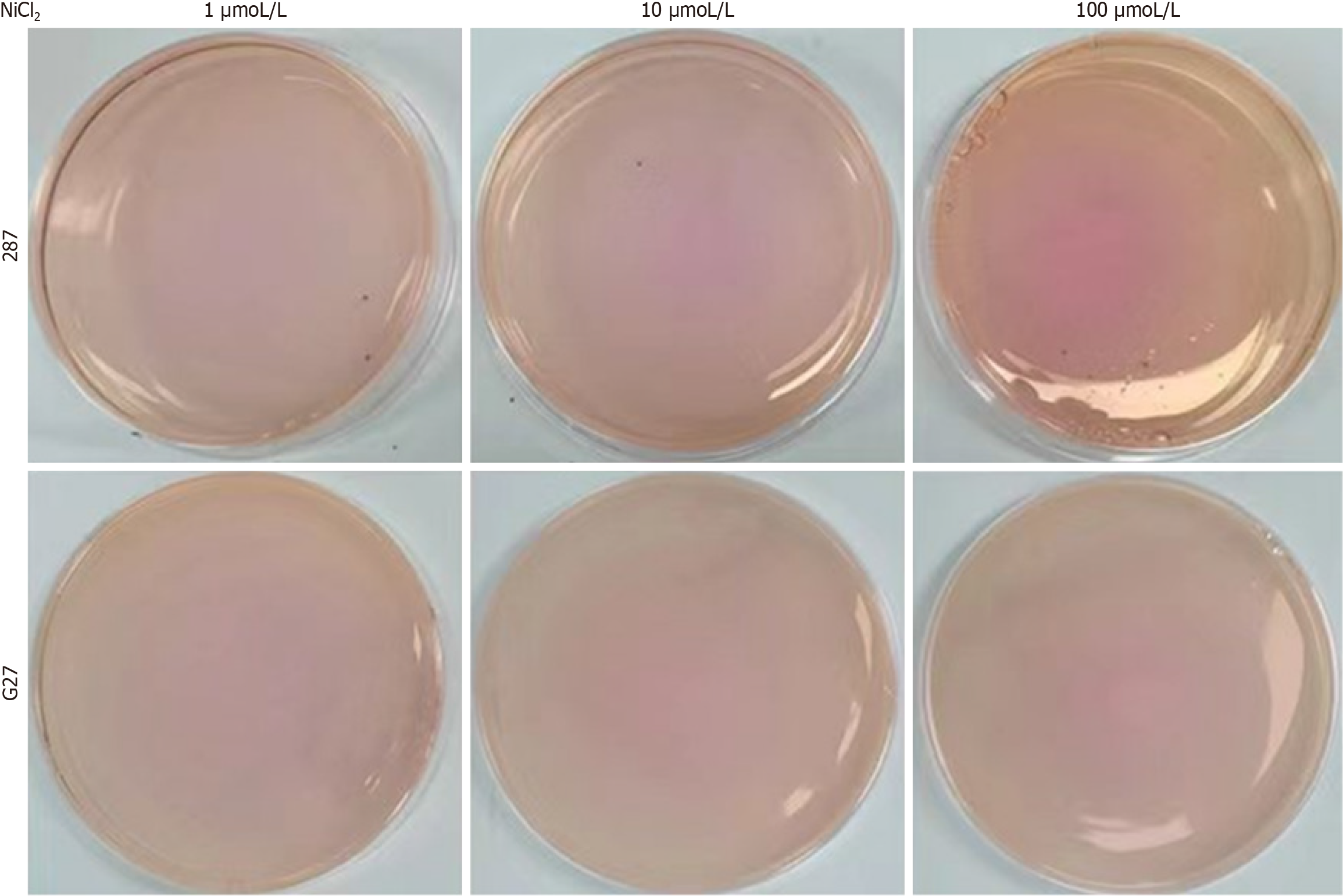
Determination of NiCl2 concentration in the characteristic medium: In scheme 3, NiCl2 solution was added at concentrations of 1 mmol/L, 10 mmol/L, 100 mmol/L, and 1000 mmol/L, which were thoroughly mixed to form a solid medium (scheme 4). Subsequently, 1 × 105 CFUs/mL of H. pylori were inoculated onto the medium prepared in Scheme 4 and incubated for 3-5 days. During this period, a degree of medium discoloration was recorded to evaluate the effect of NiCl2 concentration.
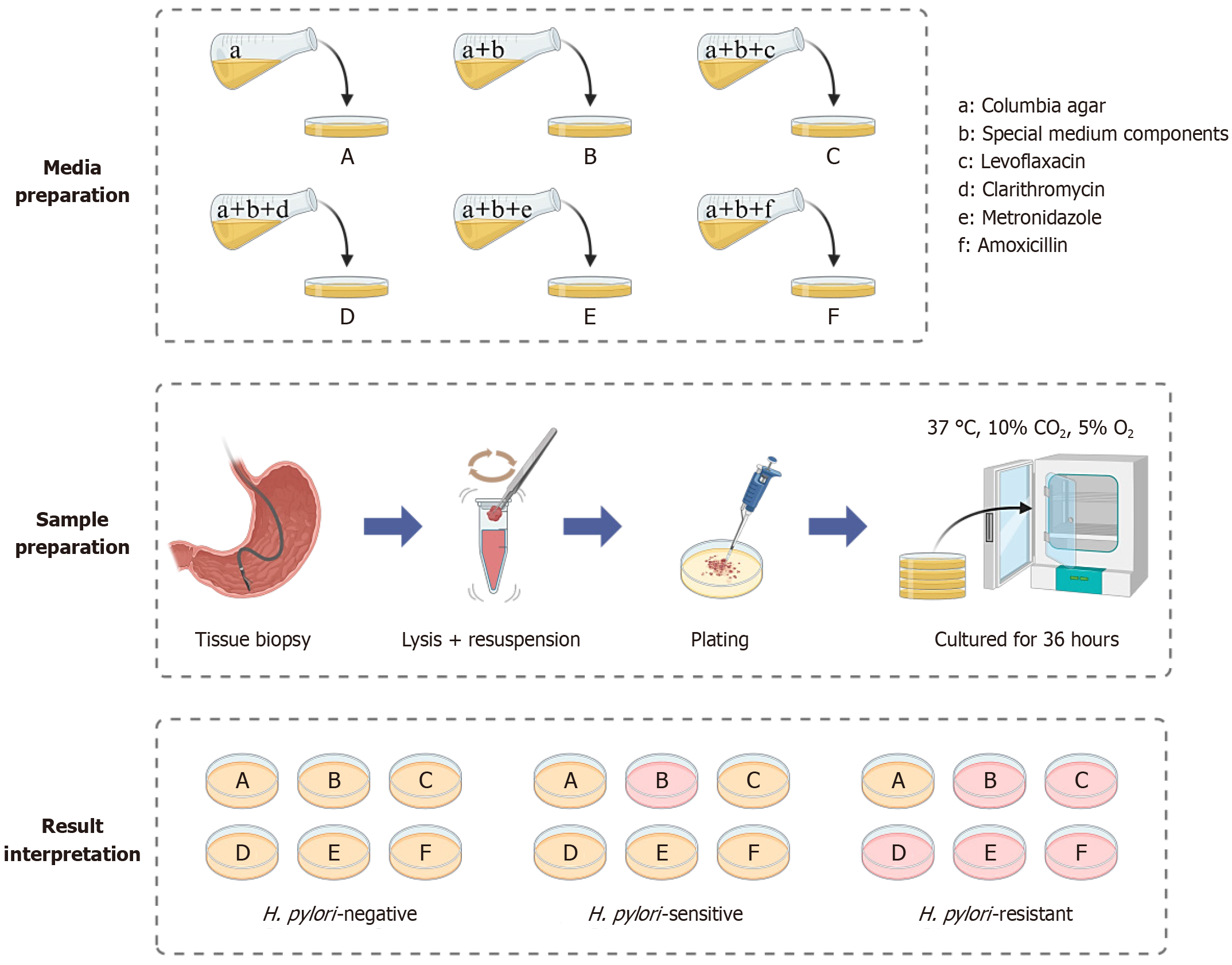
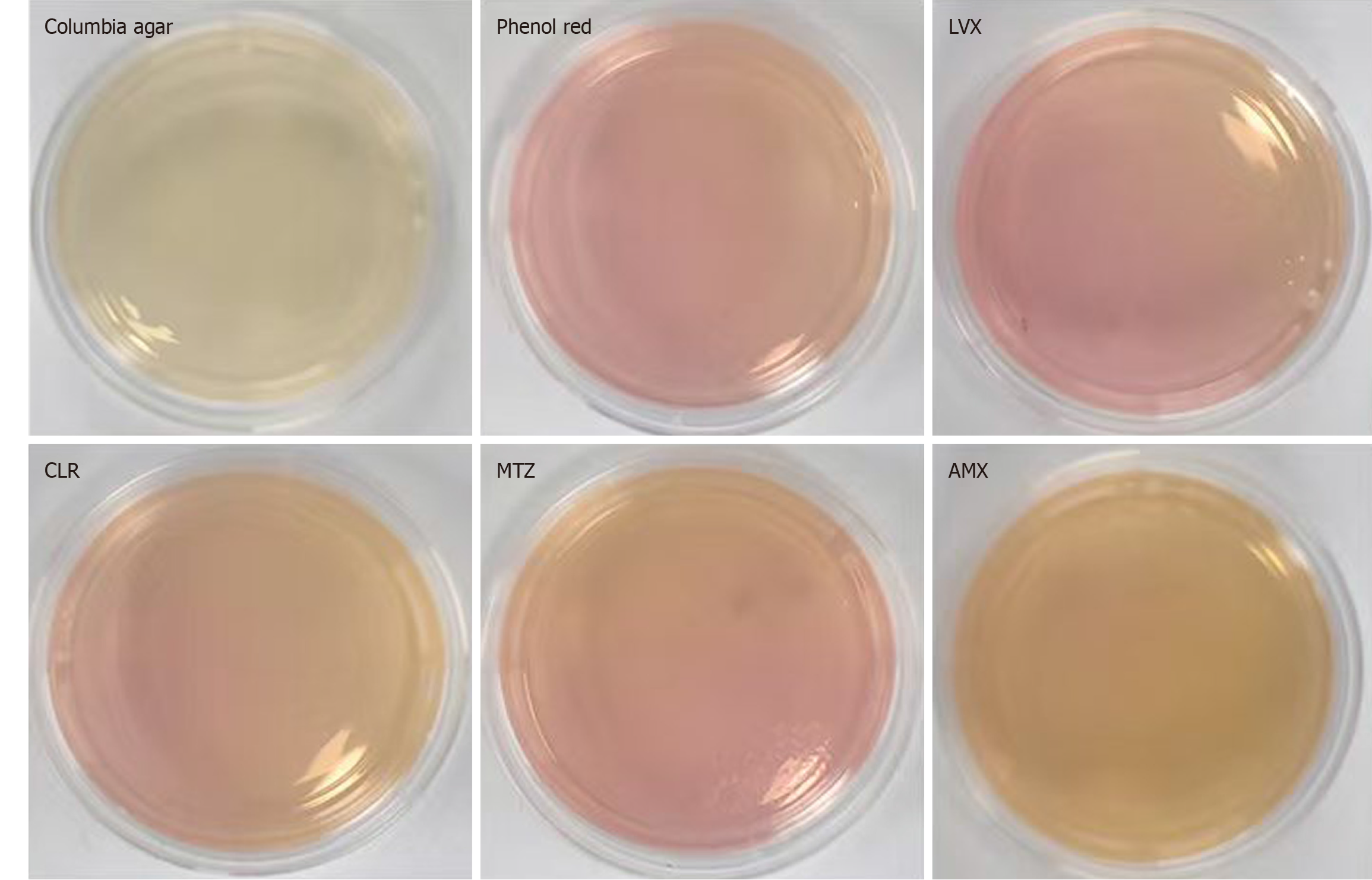
Preparation of a set of characteristic media: A H. pylori selective supplement (Dent) was added to Columbia agar medium to create the base formulation, which was designated as Medium A. Medium B was prepared according to the optimized conditions outlined in scheme 4, incorporating phenol red, urea, NaOH, and NiCl2 at the determined optimal concentrations. Based on medium B, LVX (2 μg/mL), CLR (2 μg/mL), MTZ (8 μg/mL), and AMX (0.5 μg/mL) were added to generate medium C, D, E, and F, respectively. This complete set of characteristic media was assembled for the detection of H. pylori drug resistance (Figure 1). Each medium was inoculated with 10 μL of a bacterial suspension containing 1 × 105 CFUs/mL of H. pylori and incubated at 37 °C for 3-5 days. Bacterial growth and color change of the medium were recorded to assess antimicrobial susceptibility.
Standard H. pylori strain G27 and resistant strains 159, 286, 287, and 290 were harvested and prepared as bacterial suspensions. For each strain, 10 μL of the working suspension (1 × 105 CFUs/mL) was inoculated onto each type of characteristic medium (Figure 1). At designated time points (24 hours, 28 hours, 32 hours, 36 hours, 42 hours, 48 hours, and 72 hours), media plates were removed from the incubator and left at room temperature for 30 minutes to allow for optimal visualization. The presence or absence of bacterial growth and changes in medium coloration were recorded to evaluate resistance profiles.
Preparation of the transfer solution: The transfer solution was prepared by combining BHI, glycerol (30% of the total volume), and H. pylori selective supplement (1% of the total volume). In this experiment, 0.5 milliliter of this transfer solution was placed in a sterile EP tube, to which 6-8 sterile steel beads were, added for later use.
Selection of clinical samples: After obtaining approval from the Ethics Review Committee of the Affiliated Hospital of Youjiang University for Nationalities, patients were selected based on the following criteria: History of receiving antimicrobial drugs, availability of a positive 14c-urea breath test, had stopped taking bismuth or antibiotics for 4 weeks, had stopped taking proton pump inhibitor for 2 weeks, and signing the relevant informed consent form.
Specimen collection: After obtaining the patient’s consent (2024073001), during gastroscopy at the Digestive Endoscopy Center of the Affiliated Hospital of Youjiang Medical University for Nationalities, one tissue sample was collected from both the antrum and gastric body. In cases where lesions with erosion or ulcers were observed, two biopsy samples were obtained. These samples were aseptically transferred to sterile EP tubes and placed on ice, and the subsequent steps were completed as promptly as possible (within 2 hours), ensuring strict aseptic conditions throughout the experiment.
Preparation of the H. pylori suspension: The biopsy tissue in the EP tube was homogenized using a tissue disruptor (300 Hz for 5 minutes), then transferred to a sterile EP tube, and centrifuged at 12000 rpm for 2 minutes. The resulting pellet was resuspended in 200 μL of BHI to prepare a bacterial suspension. The concentration of H. pylori bacteria in the suspension was determined using H. pylori test strips (Supplementary Figure 1), and the concentration was adjusted to approximately 1 × 107 CFUs/mL.
Inoculation and incubation: A total of 10 μL (containing approximately 1 × 105 CFUs) of the H. pylori suspension was added dropwise to a set of characteristic media. These media were subsequently incubated for 28-36 hours, removed, and incubated at room temperature for 30 minutes before the results were interpreted.
For the same clinical, an additional 100 μL of the bacterial suspension was simultaneously inoculated onto antibiotic-free Columbia agar and cultured using conventional methods. Identification tests, including urease, oxidase, and catalase assays, along with PCR analysis, confirmed the isolated bacteria as H. pylori. Following this confirmation, the MIC test was performed to determine whether H. pylori had developed drug resistance.
Various concentrations of H. pylori, including both susceptible and resistant strains, were tested to assess their sensitivity to LVX, CLR, MTZ, and AMX. As the bacterial load decreased from 1 × 106 CFUs/mL to 1 × 105 CFUs/mL, the MICs decreased by 1-4 times (Table 1). According to the CLSI guidelines, when the bacterial load was 1 × 106 CFUs/mL, the resistance breakpoints of H. pylori against these four drugs were set at 2, 2 μg/mL, 8 μg/mL, and 0.5 μg/mL, respectively. Correspondingly, when the bacterial load was reduced to 1 × 105 CFUs/mL, the resistance breakpoints for H. pylori are expected to fall within the following ranges: 0.5-2 μg/mL for LVX and CLR, 1-8 μg/mL for MTZ, and 0.06-0.5 μg/mL for AMX.
| H. pylori | Counts (CFUs) | LVX | CLR | MTZ | AMX |
| G27 | 106 | 0.1250 | 0.0625 | 0.5 | 0.0313 |
| 105 | 0.0625 | 0.0312 | 0.5 | 0.0156 | |
| 104 | 0.0312 | 0.0156 | 0.125 | 0.0078 | |
| 26695 | 106 | 0.1250 | 0.1250 | 0.5 | 0.0625 |
| 105 | 0.0625 | 0.0625 | 0.25 | 0.0325 | |
| 104 | 0.0312 | 0.0312 | 0.125 | 0.0153 | |
| 159 | 106 | 2 | 2 | 4 | 0.1250 |
| 105 | 1 | 2 | 2 | 0.0625 | |
| 104 | 0.5 | 1 | 0.5 | 0.0313 | |
| 161 | 106 | 2 | 2 | 4 | 0.1250 |
| 105 | 2 | 1 | 2 | 0.0625 | |
| 104 | 1 | 0.5 | 1 | 0.0313 | |
| 162 | 106 | 8 | 8 | 2 | 0.1250 |
| 105 | 4 | 4 | 1 | 0.0625 | |
| 104 | 2 | 1 | 0.5 | 0.03125 | |
| 163 | 106 | 16 | 0.5 | 16 | 1 |
| 105 | 4 | 0.25 | 4 | 0.5 | |
| 104 | 2 | 0.125 | 2 | 0.25 | |
| 286 | 106 | 1 | 8 | 32 | 1 |
| 105 | 0.5 | 4 | 16 | 0.5 | |
| 104 | 0.5 | 1 | 4 | 0.5 | |
| 287 | 106 | 4 | 4 | 32 | 0.25 |
| 105 | 2 | 2 | 16 | 0.1250 | |
| 104 | 1 | 8 | 8 | 0.0625 | |
| 289 | 106 | 8 | 32 | 32 | 1 |
| 105 | 4 | 8 | 16 | 0.5 | |
| 104 | 2 | 2 | 8 | 0.125 | |
| 290 | 106 | 32 | 16 | 32 | 4 |
| 105 | 32 | 4 | 16 | 2 | |
| 104 | 16 | 4 | 4 | 1 |
LVX (0.5 μg/mL), CLR (0.5 μg/mL), MTZ (4 μg/mL), and AMX (0.06 μg/mL) were added to the solid medium. Under these conditions, the growth of sensitive H. pylori strains (1 × 105 CFU) was effectively inhibited, whereas drug-resistant strains were not suppressed (Table 2).
| H. pylori | LVX | CLR | MTZ | AMX | ||||||||||||
| 0.25 | 0.5 | 1 | 2 | 0.25 | 0.5 | 1 | 2 | 1 | 2 | 4 | 8 | 0.03 | 0.06 | 0.13 | 0.25 | |
| G27 | + | - | - | - | + | - | - | + | + | + | - | - | + | - | - | - |
| 26695 | + | - | - | - | + | - | - | + | + | + | - | - | + | - | - | - |
| 159 | + | + | - | - | + | + | - | + | + | + | + | - | + | + | - | - |
| 161 | + | + | - | - | + | + | - | + | + | + | + | - | + | + | - | - |
| 162 | + | + | - | - | + | + | - | + | + | + | - | - | + | - | - | - |
| 163 | + | + | - | - | - | - | - | + | + | + | + | - | + | + | - | - |
| 286 | + | + | - | - | + | + | - | + | + | + | + | - | + | + | - | - |
| 287 | + | + | - | - | + | + | - | + | + | + | + | - | + | + | + | - |
| 289 | + | + | + | - | + | + | + | + | + | + | + | - | + | +. | - | - |
| 290 | + | + | + | - | + | + | - | + | + | + | + | + | + | + | - | - |
The addition of 0.60 mg/mL urea did not impede the growth of H. pylori. However, at concentrations between 1.2 mg/mL and 2.4 mg/mL, H. pylori (1 × 105 CFU/mL) exhibited poor growth. In the 0.60-1.2 mg/mL range, further evaluation of 0.8 mg/mL, 0.9 mg/mL, and 1.0 mg/mL urea revealed that 0.8 mg/mL was the optimal concentration (Table 3). When 0.8 mg/100 mL phenol red was added to the medium along with urea, it had no inhibitory or stimulatory effect on H. pylori growth. While it provided optimal colorimetric recognition, its sensitivity was limited. Similarly, 0.2 mg/100 mL phenol red had no influence on H. pylori growth, while both recognition and sensitivity were suboptimal. Finally, 0.4 mg/100 mL phenol red had no adverse effect on H. pylori growth and demonstrated notable recognition and sensitivity (Figure 2, Table 3).
| H. pylori | Urea (mg/mL) | Phenol red (mg/100 mL) | ||||||||
| 0.6 | 0.8 | 0.9 | 1 | 1.2 | 2.4 | 0.2 | 0.4 | 0.8 | 1.6 | |
| G27 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
| 26695 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
| 159 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
| 161 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
| 162 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
| 163 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
| 286 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
| 287 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
| 289 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
| 290 | ++ | ++ | + | + | + | - | 2 | 3 | 1 | 1 |
The sensitivity and visibility of the color change reaction were further enhanced by adjusting the pH of the medium to 7.15-7.35 while maintaining a urea concentration of 0.8 mg/mL and a phenol red concentration of 0.4 mg/100 mL. Various concentrations of NiCl2 were subsequently introduced, resulting in a more notable discoloration reaction. In contrast, NiCl2 did not alter the MIC value (Supplementary Table 1). The optimal outcome was achieved when the NiCl2 concentration was set to 100 mmol/L (Figure 3, Table 4).
| H. pylori | pH | NiCl2 (mmol/L) | ||||
| 7.15 | 7.25 | 7.35 | 1 | 10 | 100 | |
| G27 | 1 | 1 | 2 | + | + | ++ |
| 26695 | 1 | 1 | 2 | + | + | ++ |
| 159 | 1 | 1 | 2 | + | + | ++ |
| 161 | 1 | 1 | 2 | + | + | ++ |
| 162 | 1 | 1 | 2 | + | + | ++ |
| 163 | 1 | 1 | 2 | + | + | ++ |
| 286 | 1 | 1 | 1 | + | + | ++ |
| 287 | 1 | 1 | 2 | + | + | ++ |
| 289 | 1 | 1 | 2 | + | + | ++ |
| 290 | 1 | 1 | 2 | + | + | ++ |
The effects of different concentrations of antibiotics on the growth of H. pylori (sensitive and resistant strains) were consistent between the Columbia medium and the specialty medium. However, the specialty medium incorporated a colorimetric reaction not present in the Columbia medium. Additionally, compared with the antibiotic-free specialty medium, the presence of antibiotics extended the reaction time from approximately 28 hours to around 36 hours (Table 5). Orthogonal experimental design was employed to validate the optimal formulation of the specialty medium (Supplementary Table 2).
| H. pylori | Columbia medium | Characteristic medium | ||||||
| LVX | CLR | MTZ | AMX | LVX | CLR | MTZ | AMX | |
| 0.5 | 0.5 | 4 | 0.0625 | 0.5 | 0.5 | 4 | 0.0625 | |
| G27 | - | - | - | - | × | × | × | × |
| 26695 | - | - | - | - | × | × | × | × |
| 159 | + | + | + | + | √ | √ | √ | √ |
| 161 | + | + | + | + | √ | √ | √ | √ |
| 162 | + | + | + | + | √ | √ | √ | √ |
| 163 | + | + | + | + | √ | √ | √ | √ |
| 286 | + | + | + | + | √ | √ | √ | √ |
| 287 | + | + | + | + | √ | √ | √ | √ |
| 289 | + | + | + | + | √ | √ | √ | √ |
| 290 | + | + | + | + | √ | √ | √ | √ |
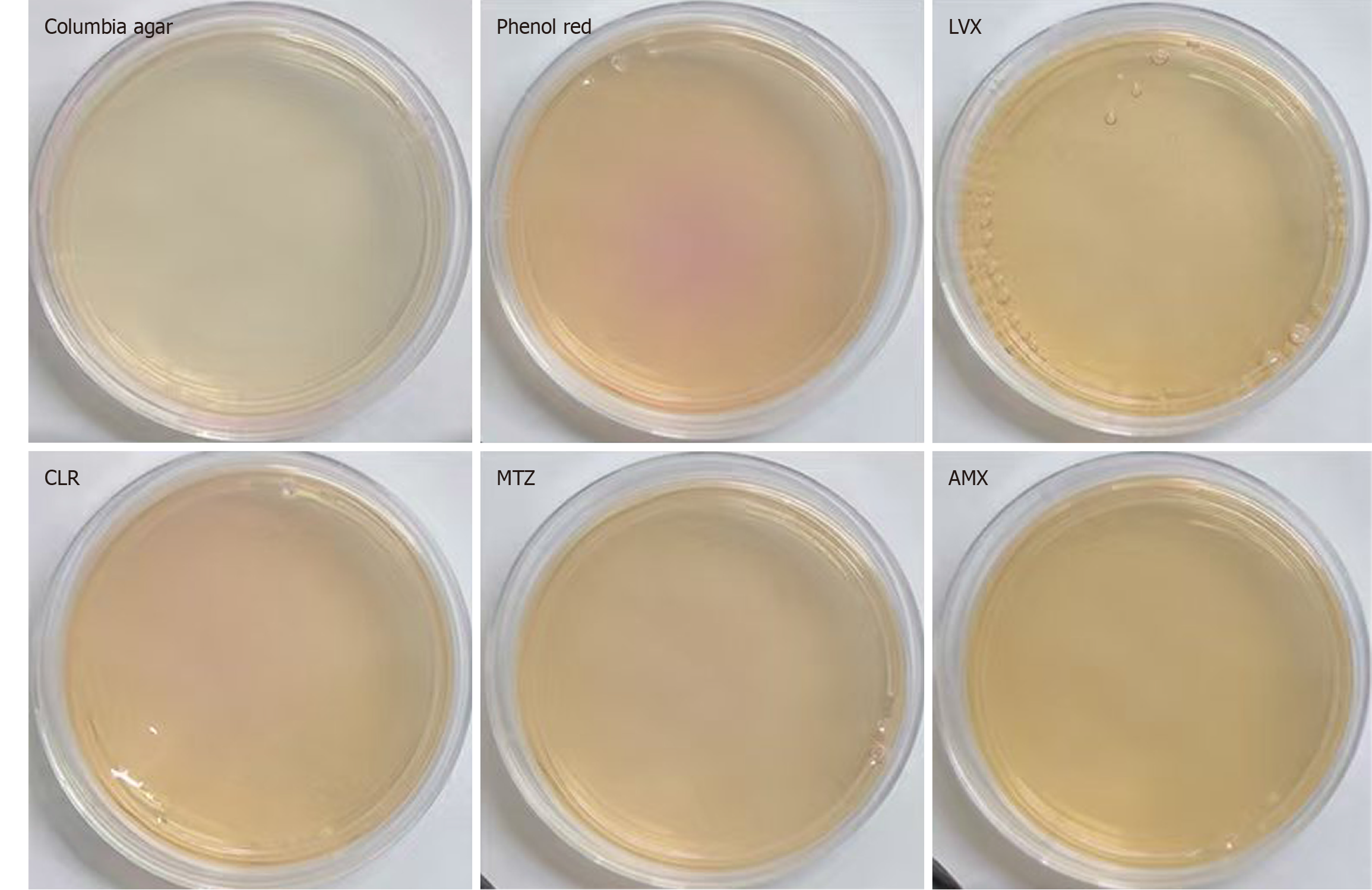
When H. pylori strain G27 at a concentration of 1 × 105 CFU/mL was incubated in the antibiotic-free characteristic medium containing phenol red for 28-36 hours, the medium turned red. However, no color change was found in the presence of antibiotics (LVX, CLR, MTZ, and AMX, Figure 4), indicating that G27 was sensitive to these antibiotics based on the characteristic medium assay. This result was consistent with findings from the microdilution method. Similarly, H. pylori strain 26695 showed concordant results (Supplementary Figure 2). Strain 159 exhibited a color change in media containing CLR, MTZ, and LVX, while showed no color change in the medium supplemented with AMX. These findings indicated that strain 159 appeared resistant to CLR, MTZ, and LVX, while remaining sensitive to AMX (Figure 5). Strains 287 and 290 turned red after 28-36 hours of incubation in both antibiotic-free and antibiotic-containing characteristic media. These findings suggested that strains were resistant to all four tested antibiotics (Supplementary Figures 3 and 4). The results obtained for all the strains tested on the characteristic media were in agreement with the conclusions drawn from the MIC values (Table 1).
In clinical practice, the drug sensitivity of H. pylori was evaluated using both the traditional culture method and the novel method. A total of 201 gastric mucosal samples were collected. Among them, 139 samples were subjected to traditional antimicrobial susceptibility culture, of which 6 samples were contaminated or failed to subculture successfully during the detection process, rendering them untestable. In contrast, the new method was employed to detect drug susceptibility, resulting in the successful culture of H. pylori colonies in 185 samples, and only 3 samples were contaminated by miscellaneous bacteria. Consequently, the success rates of traditional antimicrobial susceptibility testing and the new method were 66.3% and 90.5%, respectively (Table 6).
| Detection method | The number of successful detections | Number of culture failures | Total | Detection success rate (%) |
| Dilution method | 133 | 68 | 201 | 66.3 |
| New approach | 182 | 19 | 201 | 90.5 |
Consistency between the two detection methods: Results from the two detection methods were considered consistent only if all four antibiotic susceptibility tests produced matching outcomes. Any discrepancy in any of the four tests between the methods classified the results as inconsistent. Among 133 samples that met the inclusion criteria, 127 exhibited concordant results, yielding a 95.5% agreement rate. Notably, the novel method demonstrated superior success rates and greater efficiency, providing results within 28-36 hours compared with 11-12 days required by the traditional method (P < 0.001). The difference in the success rates of the two methods was statistically significant (P < 0.01). These data highlight its utility in informing clinical treatment strategies.
Currently, the extensive use of antibiotics has led to severe drug resistance in H. pylori[18]. In 2017, the World Health Organization identified CLR-resistant H. pylori as one of the 12 bacterial pathogens urgently requiring the development of new antibiotics[19]. Hence, several countermeasures have been proposed by scholars to address this challenge. These countermeasures include improving patient adherence, for instance, by combining CLR and AMX into a single capsule to improve the medication experience[20], integrating traditional Chinese and Western medical approaches[21], and exploring emerging therapies, such as nanomaterials, probiotics, and other novel agents that have shown promising efficacy[22-24]. The ability to perform drug resistance testing and subsequently select an appropriate treatment strategy is critical[25,26]. This process is vital to achieve individualized and precise treatments for H. pylori infections. At present, CLSI recommends the use of the international microdilution method for drug resistance testing in H. pylori[27]. Based on the results, the antimicrobial susceptibility of H. pylori can be categorized as susceptible, resistant, or intermediate[28]. However, the microdilution method requires cultivation to obtain pure colonies of H. pylori, which can be challenging and time-consuming, taking up to 9-11 days. This limitation has hindered the widespread adoption of traditional antimicrobial susceptibility testing for H. pylori. Alternative drug resistance detection methods, such as those based on resistance genes, are not as accurate as the microdilution method and may have significant errors. Therefore, the development of faster, more accurate, and easier-to-perform drug resistance detection methods for H. pylori is urgently needed. Such advancements could significantly improve the management and treatment of H. pylori infections, leading to superior patient outcomes and reducing the spread of antibiotic resistance.
Calculations and verification of antibiotic interception were conducted based on resistance breakpoints established by the CLSI and various logarithmic concentrations of antibiotics (1 × 105 CFUs/mL), aiming to enhance detection accuracy and timeliness. By bypassing multiple steps, such as isolation, cultivation, and bacterial enrichment, this method could directly inhibit the growth of susceptible H. pylori strains using targeted antibiotics, thereby improving both efficiency and precision. As H. pylori hydrolyzes urea to produce alkaline substances, and NiCl2 enhances urease activity, a combination of urea, phenol red, and NiCl2 was incorporated into the culture medium to enable a colorimetric response. This modified approach allows for resistance determination during 28-36 hours based on phenol red discoloration, in contrast to the prolonged timeline of traditional antimicrobial susceptibility testing. To validate this method, a comparative analysis of 133 clinical samples was performed, which were also assessed via the standard microdilution method. The results demonstrated a high concordance rate of 95.5%, supporting the reliability and feasibility of the method as a rapid alternative for antimicrobial susceptibility testing in H. pylori.
A key limitation of this study is its concentration on only four commonly used antibiotics (AMX, CLR, MTZ, and LVX). Future investigations should expand the antibiotic panel to include emerging therapeutic agents (e.g., tetracycline and rifabutin). Although the current sample size (n = 201) was regarded as representative, larger multicenter studies with broader geographic diversity are necessary to validate the generalizability of the findings. Another notable limitation is the invasive nature of the method, as it requires endoscopic sampling of gastric mucosa. While non-invasive tests, such as stool-based detection of H. pylori resistance genes, have gained clinical acceptance, these approaches are mainly limited by interference from commensal gut microbiota and a restricted range of detectable resistance markers. For patients with refractory H. pylori infection, invasive methods remain essential for obtaining accurate antimicrobial resistance profiles, with most patients prioritizing diagnostic precision over the inconvenience of invasiveness. Thus, by enabling targeted antimicrobial therapy, this approach holds significant translational potential for improving H. pylori treatment outcomes. Therefore, this study proposes a novel chromogenic medium-based method for the rapid detection of H. pylori drug resistance, demonstrating significant innovation and clinical application potential.
The novel detection method can rapidly identify H. pylori drug resistance within 28-36 hours. It is characterized by high accuracy, operational simplicity, and broad clinical applicability. This approach may provide a practical diagnostic tool for managing H. pylori infections and hold remarkable promise for advancing clinical strategies to eliminate antimicrobial resistance.
We would like to express our gratitude to Professor Bi Hongkai from Nanjing Medical University for providing the Helicobacter pylori strains. The authors wish to thank the participants of the study and the staff at the Affiliated Hospital of Youjiang University for Nationalities for their dedication to patient recruitment and data collection. We also thank Dr. Gguang-Zi Qi for statistical consultation.
| 1. | Rojas-Rengifo DF, Mendoza B, Jaramillo C, Rodríguez-Urrego PA, Vera-Chamorro JF, Alvarez J, Delgado MDP, Jimenez-Soto LF. Helicobacter pylori culture as a key tool for diagnosis in Colombia. J Infect Dev Ctries. 2019;13:720-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 2. | FitzGerald R, Smith SM. An Overview of Helicobacter pylori Infection. Methods Mol Biol. 2021;2283:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 3. | Usui Y, Taniyama Y, Endo M, Koyanagi YN, Kasugai Y, Oze I, Ito H, Imoto I, Tanaka T, Tajika M, Niwa Y, Iwasaki Y, Aoi T, Hakozaki N, Takata S, Suzuki K, Terao C, Hatakeyama M, Hirata M, Sugano K, Yoshida T, Kamatani Y, Nakagawa H, Matsuda K, Murakami Y, Spurdle AB, Matsuo K, Momozawa Y. Helicobacter pylori, Homologous-Recombination Genes, and Gastric Cancer. N Engl J Med. 2023;388:1181-1190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 161] [Article Influence: 53.7] [Reference Citation Analysis (1)] |
| 4. | Zagari RM, Frazzoni L, Marasco G, Fuccio L, Bazzoli F. Treatment of Helicobacter pylori infection: a clinical practice update. Minerva Med. 2021;112:281-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 5. | Medakina I, Tsapkova L, Polyakova V, Nikolaev S, Yanova T, Dekhnich N, Khatkov I, Bordin D, Bodunova N. Helicobacter pylori Antibiotic Resistance: Molecular Basis and Diagnostic Methods. Int J Mol Sci. 2023;24:9433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 6. | Ansari S, Yamaoka Y. Helicobacter pylori Infection, Its Laboratory Diagnosis, and Antimicrobial Resistance: a Perspective of Clinical Relevance. Clin Microbiol Rev. 2022;35:e0025821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 76] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 7. | Shu X, Ye D, Hu C, Peng K, Zhao H, Li H, Jiang M. Alarming antibiotics resistance of Helicobacter pylori from children in Southeast China over 6 years. Sci Rep. 2022;12:17754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 8. | Brennan D, O'Morain C, McNamara D, Smith SM. Molecular Detection of Antibiotic-Resistant Helicobacter pylori. Methods Mol Biol. 2021;2283:29-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 9. | Takayama T, Suzuki H, Okada K, Akiyama S, Narasaka T, Maruo K, Sakamoto T, Seo E, Tsuchiya K. The Optimal Cut-off of the Latex Immunoassay (LZ Test) for Helicobacter pylori Infection Based on the Stool Antigen Test and Helicobacter pylori-associated Gastritis. Intern Med. 2022;61:2103-2109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Shakir SM, Otiso J, Keller G, Heule HV, Osborn LJ, Cole N, Schuetz AN, Richter SS, Couturier MR. Multicenter Evaluation of a Gradient Diffusion Method for Antimicrobial Susceptibility Testing of Helicobacter pylori. Microbiol Spectr. 2022;10:e0211121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 11. | Zhao Y, Li Y, Luan Z, Ma C, Yang L, Zhang W, Shi C. Establishment of a TaqMan-MGB probe multiplex real-time PCR system for one-step levofloxacin and clarithromycin resistant Helicobacter pylori detection. J Microbiol Methods. 2022;192:106393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 12. | Tian XL, Song ZQ, Suo BJ, Zhou LY, Li CL, Zhang YX. [Comparison of Epsilometer test and agar dilution method in detecting the sensitivity of Helicobacter pylori to metronidazole]. Beijing Da Xue Xue Bao Yi Xue Ban. 2023;55:934-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 13. | Peng X, Song Z, He L, Lin S, Gong Y, Sun L, Zhao F, Gu Y, You Y, Zhou L, Zhang J. Gastric Juice-Based Real-Time PCR for Tailored Helicobacter Pylori Treatment: A Practical Approach. Int J Med Sci. 2017;14:595-601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Saranathan R, Levi MH, Wattam AR, Malek A, Asare E, Behin DS, Pan DH, Jacobs WR Jr, Szymczak WA. Helicobacter pylori Infections in the Bronx, New York: Surveying Antibiotic Susceptibility and Strain Lineage by Whole-Genome Sequencing. J Clin Microbiol. 2020;58:e01591-e01519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 15. | Miftahussurur M, Shrestha PK, Subsomwong P, Sharma RP, Yamaoka Y. Emerging Helicobacter pylori levofloxacin resistance and novel genetic mutation in Nepal. BMC Microbiol. 2016;16:256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Clines N, Beckman E. Development of a high throughput human stool specimen processing method for a molecular Helicobacter pylori clarithromycin resistance assay. PLoS One. 2019;14:e0224356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Lin Y, Shao Y, Yan J, Ye G. Antibiotic resistance in Helicobacter pylori: From potential biomolecular mechanisms to clinical practice. J Clin Lab Anal. 2023;37:e24885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 18. | Bujanda L, Nyssen OP, Ramos J, Bordin DS, Tepes B, Perez-Aisa A, Pavoni M, Castro-Fernandez M, Lerang F, Leja M, Rodrigo L, Rokkas T, Kupcinskas J, Jonaitis L, Shvets O, Gasbarrini A, Simsek H, Phull PS, Buzás GM, Machado JC, Boltin D, Boyanova L, Tonkić A, Marlicz W, Venerito M, Vologzanina L, Fadieienko GD, Fiorini G, Resina E, Muñoz R, Cano-Català A, Puig I, García-Morales N, Hernández L, Moreira L, Megraud F, Morain CO, Montes M, Gisbert JP; Hp-EuReg investigators; Hp-EuReg investigators. Effectiveness of Helicobacter pylori Treatments According to Antibiotic Resistance. Am J Gastroenterol. 2024;119:646-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 19. | Nista EC, Pellegrino A, Giuli L, Candelli M, Schepis T, De Lucia SS, Ojetti V, Franceschi F, Gasbarrini A. Clinical Implications of Helicobacter pylori Antibiotic Resistance in Italy: A Review of the Literature. Antibiotics (Basel). 2022;11:1452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Huguet JM, Ferrer-Barceló L, Suárez P, Barcelo-Cerda S, Sempere J, Saracino IM, Fiorini G, Vaira D, Pérez-Aísa Á, Jonaitis L, Tepes B, Castro-Fernandez M, Pabón-Carrasco M, Keco-Huerga A, Voynovan I, Lucendo AJ, Lanas Á, Martínez-Domínguez SJ, Alfaro Almajano E, Rodrigo L, Vologzanina L, Bordin DS, Gasbarrini A, Babayeva G, Lerang F, Leja M, Kupčinskas J, Rokkas T, Marcos-Pinto R, Meštrović A, Gridnyev O, Phull PS, Smith SM, Boltin D, Buzás GM, Kral J, Şimşek H, Matysiak-Budnik T, Milivojevic V, Marlicz W, Venerito M, Boyanova L, Doulberis M, Capelle LG, Cano-Català A, Moreira L, Nyssen OP, Mégraud F, O'Morain C, Gisbert JP; Hp‐EuReg Investigators. Role of compliance in Helicobacter pylori eradication treatment: Results of the European Registry on H. pylori management. United European Gastroenterol J. 2024;12:691-704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 21. | Zhong M, Sun Q, Ren B, Yu C, Zhou S, Gao Q, Wang X, Yuan C, Lu J, Peng Q, Zeng M, Song H. A comparison of the efficacy and safety of Chinese patent medicine combined with Western medicine for Helicobacter pylori-related gastric ulcer: A systematic review and network meta-analysis. Medicine (Baltimore). 2025;104:e41137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 22. | Roszczenko-Jasińska P, Wojtyś MI, Jagusztyn-Krynicka EK. Helicobacter pylori treatment in the post-antibiotics era-searching for new drug targets. Appl Microbiol Biotechnol. 2020;104:9891-9905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 23. | Viazis N, Argyriou K, Kotzampassi K, Christodoulou DK, Apostolopoulos P, Georgopoulos SD, Liatsos C, Giouleme O, Koustenis K, Veretanos C, Stogiannou D, Moutzoukis M, Poutakidis C, Mylonas II, Tseti I, Mantzaris GJ. A Four-Probiotics Regimen Combined with A Standard Helicobacter pylori-Eradication Treatment Reduces Side Effects and Increases Eradication Rates. Nutrients. 2022;14:632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 24. | Bai X, Zhu M, He Y, Wang T, Tian D, Shu J. The impacts of probiotics in eradication therapy of Helicobacter pylori. Arch Microbiol. 2022;204:692. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 25. | Fayed B, Jagal J, Cagliani R, Kedia RA, Elsherbeny A, Bayraktutan H, Khoder G, Haider M. Co-administration of amoxicillin-loaded chitosan nanoparticles and inulin: A novel strategy for mitigating antibiotic resistance and preserving microbiota balance in Helicobacter pylori treatment. Int J Biol Macromol. 2023;253:126706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 26. | Fallone CA, Moss SF, Malfertheiner P. Reconciliation of Recent Helicobacter pylori Treatment Guidelines in a Time of Increasing Resistance to Antibiotics. Gastroenterology. 2019;157:44-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 211] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 27. | Hakemi Vala M, Eyvazi S, Goudarzi H, Sarie HR, Gholami M. Evaluation of Clarithromycin Resistance Among Iranian Helicobacter pylori Isolates by E-Test and Real-Time Polymerase Chain Reaction Methods. Jundishapur J Microbiol. 2016;9:e29839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Kouitcheu Mabeku LB, Eyoum Bille B, Tepap Zemnou C, Tali Nguefack LD, Leundji H. Broad spectrum resistance in Helicobacter pylori isolated from gastric biopsies of patients with dyspepsia in Cameroon and efflux-mediated multiresistance detection in MDR isolates. BMC Infect Dis. 2019;19:880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/