Published online May 28, 2025. doi: 10.3748/wjg.v31.i20.106747
Revised: April 12, 2025
Accepted: May 12, 2025
Published online: May 28, 2025
Processing time: 83 Days and 3.2 Hours
Hepatobiliary and pancreatic (HBP) cancers are among the most aggressive malig
Core Tip: Hepatobiliary and pancreatic (HBP) cancers are highly aggressive, with recurrence and metastasis driven by tumor heterogeneity and drug resistance, posing significant treatment challenges. Current personalized and accurate prediction models are lacking. Patient-derived organoids (PDOs) tumor, three-dimensional in vitro models from patient tumor tissues, show over 70% cultivation success and over 90% predictive accuracy. However, PDOs face limitations in simulating the tumor microenvironment despite advances in co-culture and microfluidic techniques. Additionally, PDOs' potential in predicting multi-drug therapy efficacy requires further assessment. This review outlines the applications and challenges of organoid models in HBP cancer research.
- Citation: Hu JW, Pan YZ, Zhang XX, Li JT, Jin Y. Applications and challenges of patient-derived organoids in hepatobiliary and pancreatic cancers. World J Gastroenterol 2025; 31(20): 106747
- URL: https://www.wjgnet.com/1007-9327/full/v31/i20/106747.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i20.106747
Hepatobiliary and pancreatic (HBP) cancers are highly aggressive, with some of the highest incidence and mortality rates worldwide[1]. According to the latest cancer statistics, the 5-year relative survival rates for pancreatic cancer and liver cancer (including intrahepatic bile duct cancer) between 2013 and 2019 were 13% and 22%, respectively, making them the lowest and second-lowest survival rates among all cancer types[2]. Typically, HBP cancers progress rapidly and are associated with a high risk of recurrence and metastasis, owing to their high heterogeneity and resistance to therapy, creating significant challenges for treatment[3-5]. Although emerging therapies, such as immune checkpoint inhibitors and targeted therapies, have led to some improvements in survival outcomes, only a small number of patients currently benefit from them[6]. Especially in cholangiocarcinoma and pancreatic cancer, conventional chemotherapy remains the mainstream treatment option, highlighting the need to further optimize treatment strategies[7,8]. Current approaches to predicting tumor response to treatment are still quite limited. One of the more popular methods involves using NGS to identify tumor-related gene mutations and molecular features, which can help guide treatment decisions for certain patients. However, given that the underlying mechanisms and therapeutic targets for HBP cancers are still being ex
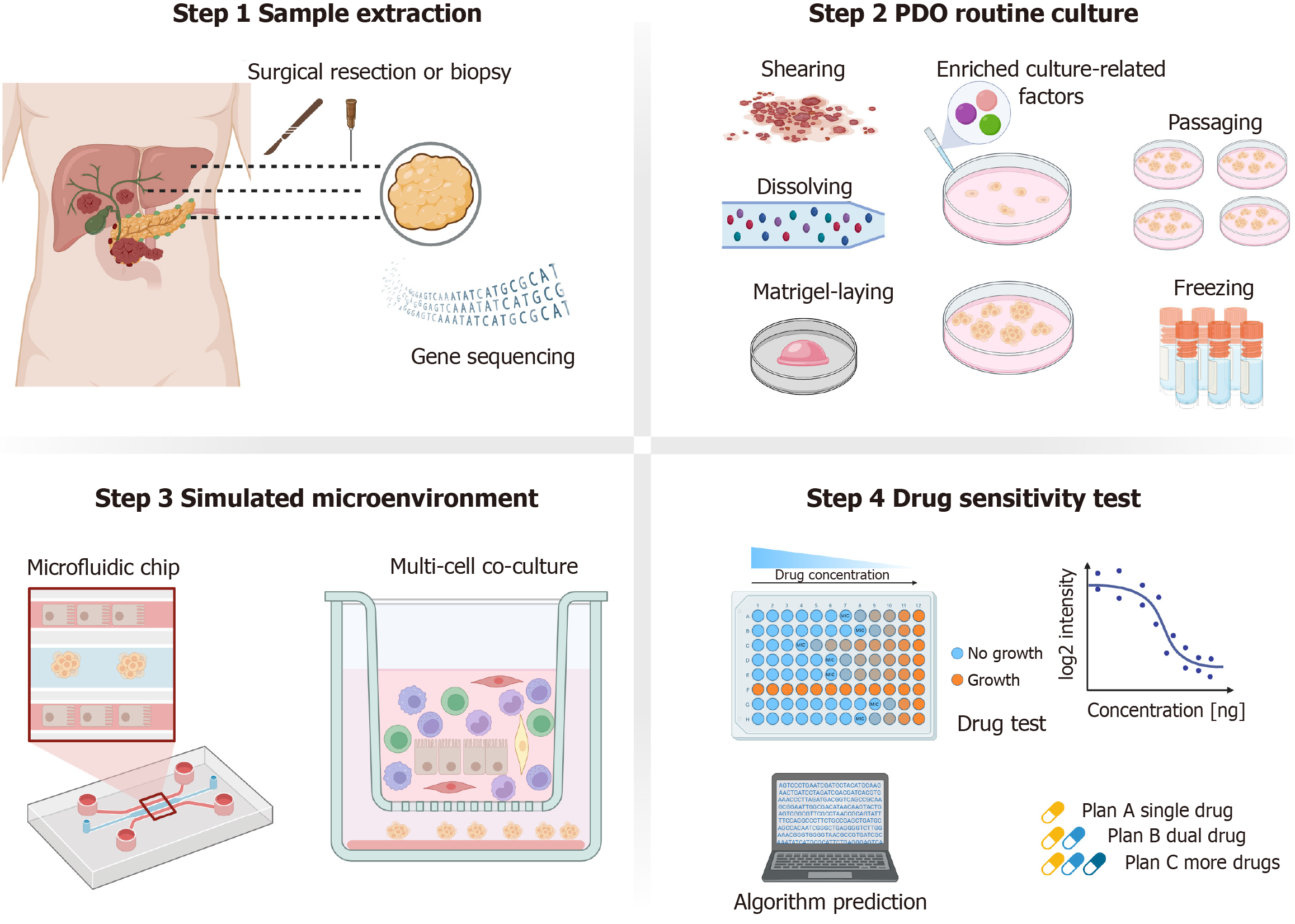
Patient-derived organoids (PDOs) tumor are three-dimensional (3D) cell culture models derived from patient tissue, preserving the genomic and pathological characteristics of the original tumor while closely preserving its biological features and tissue structure in vivo. These models are promising means of informing clinical treatment decisions[10,11]. Organoids were first described in 2009[12], and PDOs were successfully established in 2011[13]; now, PDOs are widely applied in cancer research. PDOs typically require only a small number of cells initially and can proliferate rapidly under specific conditions, greatly reducing both culture time and costs[14]. Furthermore, PDOs maintain genetic stability and demonstrate strong modelling accuracy and reliability, even during long-term culture[15]. These organoids have proven invaluable in investigating disease mechanisms and conducting high-throughput drug screening as approaches towards advancing precision medicine[16]. While the average success rate for generating cancer organoids exceeds 70%[17], success rates vary significantly across different types of PDOs. For instance, the success rate for generating colon cancer organoids can reach up to 90%[18], whereas it remains below 20% for prostate cancer[19]. Although the different tumor culture rates are related to their invasiveness and degree of differentiation, considering that most current culture techniques are derived from colon cancer, there is still a need to improve corresponding culture methods for other tumors to meet the demands of future clinical applications[13].
Recent studies into HBP cancer have utilized PDOs to identify tumor-associated molecular markers[20], assess meta
This review aims to summarize the current applications of HBP tumor organoids in clinical treatment, analyze existing challenges, and discuss future research directions. Specifically, based on the team's previous research, it analyzes the factors contributing to the current variations in culture success rates, uncovers the limitations of microenvironment simulation and combination drug therapy prediction, and provides preliminary insights into potential solutions, such as co-culture models and microfluidic chips.
The success rates for developing liver cancer organoid cultures are reported to be from 26% to 75.6% (Table 1)[11,14,22,25,30-44], with half of the studies reporting rates below 50%. In contrast to other gastrointestinal cancers, liver cancer organoids exhibit lower success rates, which might be due to the absence of epithelial stem cell characteristics in hepatocellular carcinoma (HCC): This is known to impact the proliferation of HCC cells in organoid culture systems[22]. Furthermore, the success rate of organoid culture is linked with the degree of differentiation and the proliferation rate of the tumor itself. Some studies have indicated that only moderately-to-poorly differentiated liver cancers with a Ki-67 index greater than 5 are likely to successfully form organoids[30,31]. In addition, tumor heterogeneity and the specific sampling region also play a role in influencing culture outcomes[11]. There is also evidence reporting a 26% success rate for liver cancer biopsy cultures, which is lower than that for surgical resection samples, possibly due to the insufficient amount of fresh tissue available, which limits the number of initial cells for culture[45]. Altogether, while liver cancer organoid models have proven to be accurate in predicting tumor behavior[17,22,31], further validation of the accuracy of the prediction is crucial. Additionally, there is a need to further increase the number of cohorts to better guide clinical drug treatment based on organoid drug sensitivity results. Additionally, HCC patients show low sensitivity to chemo
| Tumor type | Sample source | Success rate (sample size) | Drug testing | Time | Quality grade | Ref. |
| HCC, cholangiocarcinoma, combined hepatocellular cholangiocarcinoma | Res | 25.0% (3/12), 75.0% (3/4), 100.0% (2/2) | 29 anti-cancer compounds | 2017 | Low | Broutier et al[31] |
| HCC, ICC | Bx | 33.0% (8/24), 60.0% (3/5) | Sorafenib | 2018 | Low | Nuciforo et al[30] |
| HCC, ICC | Res | 40.9% (27/66) | Common clinical regimens | 2024 | Medium | Rao et al[22] |
| HCC, ICC | Res | 75.6% (399/528) | Common clinical regimens | 2024 | High | Yang et al[11] |
| HCC, ICC | Res | > 60% (64/N/A) | 301 compounds | 2024 | High | Walz et al[47] |
| HCC | Res or Bx | 54.5% (12/22) | Common clinical regimens | 2023 | Medium | Zou et al[32] |
| ICC, GBC, PDAC | Res | 50% (3/6), 20% (1/5), 50% (1/2) | 339 anti-cancer compounds | 2019 | Low | Saito et al[35] |
| GBC | Res | 12.2% (5/41) | 29 anti-cancer compounds | 2022 | Low | Yuan et al[36] |
| ICC | Res or Bx | 69.5% (16/23) | Gemcitabine and cisplatin | 2023 | Medium | Lee et al[33] |
| ECC, GBC, ICC | Res, Res, Res or Bx | 81.3% (13/16), 40.0% (4/10), 82.6% (38/46) | Common clinical regimens | 2023 | Medium | Ren et al[34] |
| GBC, ECC | Res | 85.7% (6/7) | Common clinical regimens | 2021 | Low | Wang et al[25] |
| PDAC | Res | 80.0% (8/10) | N/A | 2015 | Low | Boj et al[37] |
| PDAC | Res or Bx | 73% (101/138) | Fluorouracil | 2018 | High | Tiriac et al[38] |
| PC, ECC | Res or Bx | 62.6% (52/83) | Common clinical regimens | 2019 | High | Driehuis et al[43] |
| IPMN | Res | 81.3% (13/16) | N/A | 2021 | Medium | Beato et al[39] |
| PDAC | Res or Bx | 37%-60% (36/N/A) | Common clinical regimens | 2024 | Medium | Kim et al[40] |
| PC | Bx | 48.1% (13/27) | Common clinical regimens | 2023 | Medium | Choi et al[42] |
| PC | Bx | 93.3% (14/15) | Common clinical regimens | 2024 | Medium | Yang et al[14] |
| Cholangiocarcinoma, GBC, perihilar cholangiocarcinoma, PDAC, IPMN | Res | 60.0% (12/20), 100.0% (6/6), 100.0% (2/2), 40.0% (8/20), 33.3% (2/6) | Integrin-linked kinase inhibitor | 2021 | Medium | Shiihara et al[41] |
Based on recent studies, the success rate of culturing biliary tract cancer organoids has increased to 69.5%-74.4%[33,34], which is a significant improvement compared to the rates of 12.2%-36.4% reported in earlier studies[35,36]. The challenge in culturing biliary tract cancer organoids is primarily attributed to the small size of surgical resection samples. In addition, these resection samples often contain a high proportion of non-cancerous stromal cells, such as fibroblasts, which reduce the relative number of tumor cells, leading to contamination of the organoid culture. The success of culturing organoids can be enhanced further by microscopically isolating and extracting tumor organoids from the contaminating non-tumor cells[35]. In terms of culture sources, cholangiocarcinoma organoids are primarily cultured from solid tissue samples. Unlike bladder cancer PDOs, which are cultured from urine, no studies have reported culturing cholangiocarcinoma organoids from liquid biopsy sources, such as bile[47]. Regarding organoid function, cholangiocarcinoma organoids have shown a 92.3% accuracy in predicting drug sensitivity to clinical treatments[34]. Moreover, organoid morphology can be used to classify cholangiocarcinoma subtypes, helping to guide treatment decisions based on the gene expression profiles of the organoids[33].
The success rate for establishing organoid culture with pancreatic cancer tissue ranges from 37% to 93% (Table 1)[11,14,22,25,30-44], with approximately 50% of studies reporting success rates of around 80%[37-39]. It has been found that tumor size, whether from surgical resection or biopsy, is a limiting factor for successful pancreatic cancer organoid culture[40]. Additionally, specimens obtained after neoadjuvant chemotherapy are particularly challenging to grow in organoid culture, often resulting in little-to-no growth or very slow growth[41]. Furthermore, pancreatic cancer organoids have been successfully cultured from ascitic fluid and other liquid biopsy sources, with a success rate of 48.1%. Drug sensitivity testing has shown that clinical responses observed in patient-derived samples can be replicated[42]. However, the limited availability of clinical data on treatment regimens and patient responses has hindered the predictive accuracy of pancreatic organoid models[43]. Despite this, existing data still support the potential of organoids in guiding pan
The success rates of HBP cancer organoid cultures reported in the literature vary significantly[19]. This study summarizes key findings in the culture of HBP tumor organoids and drug sensitivity testing (Table 1)[11,14,22,25,30-44]. Based on the number of successfully cultured tumor organoids, we have provided a simple rating for the current research on HBP tumor organoids. Studies with fewer than 10 successful cultures are classified as having low prevalence, those with 10–50 successful cultures are considered to have moderate prevalence, and studies with more than 50 successful cultures are considered to have high prevalence. The culture success rates of HBP tumor organoids are influenced by factors such as specimen source (biopsy, surgical resection, post-chemotherapy), specimen properties (size, differentiation, invasiveness), sample size, and culture conditions[33]. Furthermore, the definition of cultural success varies. Typically, successful organoid culture is defined by the manifestation of appropriate organoid morphology, pathological features, and genomic consistency with the parental tissue. However, some studies define success as stable and continuous passaging for over a year[35]. Different studies also use varying methods to calculate success rates. Some calculate success based on the number of cultured tissues, while others use the number of patients as the denominator. When multiple samples are taken from the same patient, the calculation of culture success rates may differ[30]. However, continuous advancements in culture techniques have markedly improved the overall success rates of organoids compared to earlier studies[44]. Furthermore, although the efficacy of using organoid-based drug sensitivity molecular features and direct drug sensitivity results to guide therapeutic strategies has been partially validated, higher quality evidence is still required to further confirm the clinical reliability of these approaches[11,34,48]. As organoid culture methodologies continue to expand, this issue is expected to be addressed progressively.
In the HBP cancer microenvironment, the interactions between cells, including immune cells and stromal cells[49], play a crucial role in tumor initiation and progression[50,51]. Additionally, the spatial distribution differences of these cells within tissues also contribute to tumor heterogeneity. Despite originating from the same primary tumor, tissues from different tumor regions exhibit varying proportions of cellular components, leading to the formation of heterogeneous organoids[10,52]. When tumor PDOs are initially cultured, they retain components of the surrounding microenvi
A relevant strategy to simulate the tumor microenvironment is co-culturing PDOs with tumor-associated cells (Table 2)[27,32,51,58-73]. The cell types currently utilized in co-cultures of HBP tumor organoids include macrophages, lympho
| Tumor cells | Co-culture cells | Ratio (co-culture cells: Tumor cells) | Culture time (day) | Pattern | Ref. |
| HCC | T cells | 10:1 | 1 | Direct 3D co-culture | Liu et al[62] |
| HCC, intrahepatic cholangiocarcinoma | CAFs | 2:1 | 10-14 | Transwell culture | Liu et al[51] |
| HCC | PBMC, MSC | 30:1:10 | 7 | Chip | Zou et al[32] |
| HCC | T cells | 10:1 | 1 | Direct 3D co-culture | Zou et al[63] |
| Cholangiocarcinoma | T cells | 25–50:1 | 7 | Direct 3D co-culture | Zhou et al[64] |
| Extrahepatic cholangiocarcinoma | TAMs | 1:5 | 14 | Transwell culture | Guo et al[27] |
| PDAC | MSC, T cells | N/A | 5 | Direct 3D co-culture | Tai et al[65] |
| PDAC | CAFs | 2:1 | 5 | Transwell culture | Tao et al[60] |
| PDAC | TAMs | 1:1 | 6 | ALI culture | Tabe et al[58] |
| PDAC | MSC, endothelial cells | 60:21:30 | 7 | ALI culture | Takeuchi et al[61] |
| PDAC | CAFs | 1:1 | 10 | Transwell culture | Sheng et al[66] |
| PDAC | T cells | 10:1 | 3 | Hydrogel chip | Lahusen et al[67] |
| PC | TAMs | 3:1 | N/A | Transwell culture | Jiang et al[59] |
| PDAC | NK cells | 2-5:1 | 3 | Transwell culture | Beelen et al[68] |
| PC | CAFs | 1:1 | 3 | Transwell culture | Schuth et al[69] |
| PDAC | CAFs | 20:1 | 10 | Transwell culture | Shinkawa et al[70] |
| PDAC | T cells | 1:1 | 14 | Direct 3D co-culture | Meng et al[71] |
| PDAC | PBMC, NK cells | N/A | 7–14 | Direct 3D co-culture | Marcon et al[72] |
| PDAC | CAFs, T cells | N/A | 3 | Direct 3D co-culture | Tsai et al[73] |
Currently, most studies are limited to co-cultures involving a single cell type[60], with research on multi-cellular co-cultures being scarce[61,74,75]. Co-culture systems are primarily divided into two formats: (1) Direct 3D co-culture; and (2) Indirect co-culture, which includes Transwell chambers, air–liquid interface, and microfluidic chips. Organoid-on-chip models are 3D, micro-engineered systems that incorporate multi-cellular layers, tissue interfaces, and continuous perfusion chambers for human microvascular networks. These systems optimize nutrient and oxygen delivery while facilitating waste removal[76]. In addition to optimizing co-culture conditions for tumor organoids and associated cells, organoid-on-chip models can also enable dynamic drug delivery. A microfluidic chip designed by Zou et al[32], which co-cultures HCC organoids with macrophages, has been shown to increase the success rate of HCC organoid culture while reducing the amount of culture medium required and lowering the time and costs associated with high-throughput drug screening. Furthermore, it demonstrated more accurate predictions of drug responses. Vascularized organoids are commonly generated in vitro by co-culturing tumor organoids with endothelial cells using microfluidic platforms[16], and these models are applied to study tumor metastasis and targeted therapies[54]. The study also demonstrated that co-culturing HCC organoids with endothelial cells resulted in the upregulation of angiogenic signals, such as monocyte chemoattractant protein-1, thereby influencing tumor progression[74].
Patients with HBP cancer experience high rates of recurrence and metastasis, coupled with poor sensitivity to chemothe
Given the aggressive nature of HBP tumors, monotherapy is typically ineffective, and combination therapies are the standard treatment approach[7,24,80]. Identifying optimal drug combinations that benefit patients is a key therapeutic strategy. However, organoids currently cannot predict sensitivity to combination therapies directly[81]. Encouragingly, by integrating algorithms with PDO monotherapy drug sensitivity results and treatment regimens in practice, data models have been developed, demonstrating that better therapeutic outcomes are associated with higher matching between combination drug regimens and sensitive drug phenotype[82]. For example, in the treatment of PDAC, patients who received well-matched neoadjuvant and adjuvant chemotherapy exhibited significant improvements in both re
The clinical application of HBP cancer PDOs depends on the maturation of organoid culture technologies. Optimizing culture medium components and refining culture techniques are essential strategies aimed at improving organoid culture efficiency[83,84]. To further enhance the clinical utility of PDOs and reduce reliance on tissue samples will be a key focus for the translational application of HBP tumor organoids, particularly enabling PDOs derived from biopsy samples for large-scale drug sensitivity testing. In support of this, there are studies that report the successful culture of tumor organoids from ascitic fluid and pleural effusions[42,85]. In the future, HBP tumor organoids may be derived from liquid biopsy samples, such as peripheral blood and bile, thus expanding the clinical diagnostic capabilities and applications of organoids[86]. Although studies also indicate that the cell count in ascitic or pleural fluid is relatively low, with tumor cells not being detected in approximately 46% of the fluid samples[42].
With the rapid advancement of medical biotechnology, the development of new biomaterial platforms is expected to synergize with microfluidic chips and organoid culture technologies. The integration of organoids with biotechnological advances will improve culture efficiency, reduce costs, and provide technical support for high-throughput drug screening[87,88]. Establishing standardized and automated systems for sampling, culturing, and testing will not only mitigate the impact of HBP tumor heterogeneity but also minimize operational errors, laying the foundation for large-scale applications of tumor organoids[32,35,89]. Moreover, novel imaging techniques, such as real-time imaging and droplet assays, will provide more accurate observations of tumor cell–immune cell interactions, enhancing the precision of tumor microenvironment simulations within organoids[90]. In addition, the introduction of next-generation sequencing will allow deeper exploration of the genomic landscape of tumors. By combining drug sensitivity data derived from these technologies with clinical data, these technologies will aid in identifying molecular signatures and therapeutic targets associated with drug sensitivity, ultimately guiding personalized treatment strategies[91-93]. Even more promising is the potential for artificial intelligence to further integrate the aforementioned materials, imaging, and sequencing techniques. The non-invasive, automated organoid evaluation process developed with artificial intelligence could enhance large-scale organoid analysis capabilities and significantly improve the accuracy of clinical drug predictions[94,95]. The break
HBP cancer organoids have emerged as crucial tools for high-throughput drug screening and precision medicine. The success rate of organoid culture currently varies significantly, and the cultivation techniques need further improvement. Additionally, standardized protocols for cultivation still need to be established. While the drug prediction capabilities of organoids have been preliminarily validated, further high-quality evidence is required to strengthen their application, especially in predicting multi-drug combinations. The tumor microenvironment and vascularization are features that are challenging to model and, therefore, remain major limitations in improving the accuracy of organoid-based predictions. Co-culture models and microfluidic chip technologies show promise as potential solutions, although they are still in the early stages of development. In the future, with continuous integration of genomics, engineering, and information technologies, organoids are expected to become a powerful tool in clinical diagnostics and therapy.
| 1. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 12654] [Article Influence: 6327.0] [Reference Citation Analysis (6)] |
| 2. | Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2279] [Cited by in RCA: 6058] [Article Influence: 3029.0] [Reference Citation Analysis (4)] |
| 3. | Dilly J, Hoffman MT, Abbassi L, Li Z, Paradiso F, Parent BD, Hennessey CJ, Jordan AC, Morgado M, Dasgupta S, Uribe GA, Yang A, Kapner KS, Hambitzer FP, Qiang L, Feng H, Geisberg J, Wang J, Evans KE, Lyu H, Schalck A, Feng N, Lopez AM, Bristow CA, Kim MP, Rajapakshe KI, Bahrambeigi V, Roth JA, Garg K, Guerrero PA, Stanger BZ, Cristea S, Lowe SW, Baslan T, Van Allen EM, Mancias JD, Chan E, Anderson A, Katlinskaya YV, Shalek AK, Hong DS, Pant S, Hallin J, Anderes K, Olson P, Heffernan TP, Chugh S, Christensen JG, Maitra A, Wolpin BM, Raghavan S, Nowak JA, Winter PS, Dougan SK, Aguirre AJ. Mechanisms of Resistance to Oncogenic KRAS Inhibition in Pancreatic Cancer. Cancer Discov. 2024;14:2135-2161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 147] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 4. | Xu Y, Wang X, Li Y, Mao Y, Su Y, Mao Y, Yang Y, Gao W, Fu C, Chen W, Ye X, Liang F, Bai P, Sun Y, Li S, Xu R, Tian R. Multimodal single cell-resolved spatial proteomics reveal pancreatic tumor heterogeneity. Nat Commun. 2024;15:10100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 22] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 5. | Guo DZ, Zhang X, Zhang SQ, Zhang SY, Zhang XY, Yan JY, Dong SY, Zhu K, Yang XR, Fan J, Zhou J, Huang A. Single-cell tumor heterogeneity landscape of hepatocellular carcinoma: unraveling the pro-metastatic subtype and its interaction loop with fibroblasts. Mol Cancer. 2024;23:157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 35] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 6. | Oh DY, He AR, Bouattour M, Okusaka T, Qin S, Chen LT, Kitano M, Lee CK, Kim JW, Chen MH, Suksombooncharoen T, Ikeda M, Lee MA, Chen JS, Potemski P, Burris HA 3rd, Ostwal V, Tanasanvimon S, Morizane C, Zaucha RE, McNamara MG, Avallone A, Cundom JE, Breder V, Tan B, Shimizu S, Tougeron D, Evesque L, Petrova M, Zhen DB, Gillmore R, Gupta VG, Dayyani F, Park JO, Buchschacher GL Jr, Rey F, Kim H, Wang J, Morgan C, Rokutanda N, Żotkiewicz M, Vogel A, Valle JW. Durvalumab or placebo plus gemcitabine and cisplatin in participants with advanced biliary tract cancer (TOPAZ-1): updated overall survival from a randomised phase 3 study. Lancet Gastroenterol Hepatol. 2024;9:694-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 158] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 7. | Conroy T, Pfeiffer P, Vilgrain V, Lamarca A, Seufferlein T, O'Reilly EM, Hackert T, Golan T, Prager G, Haustermans K, Vogel A, Ducreux M; ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org. Pancreatic cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34:987-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 343] [Article Influence: 114.3] [Reference Citation Analysis (0)] |
| 8. | Vogel A, Bridgewater J, Edeline J, Kelley RK, Klümpen HJ, Malka D, Primrose JN, Rimassa L, Stenzinger A, Valle JW, Ducreux M; ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org. Biliary tract cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34:127-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 412] [Article Influence: 137.3] [Reference Citation Analysis (0)] |
| 9. | Abou-Alfa GK, Macarulla T, Javle MM, Kelley RK, Lubner SJ, Adeva J, Cleary JM, Catenacci DV, Borad MJ, Bridgewater J, Harris WP, Murphy AG, Oh DY, Whisenant J, Lowery MA, Goyal L, Shroff RT, El-Khoueiry AB, Fan B, Wu B, Chamberlain CX, Jiang L, Gliser C, Pandya SS, Valle JW, Zhu AX. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21:796-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 797] [Article Influence: 132.8] [Reference Citation Analysis (0)] |
| 10. | Driehuis E, Kretzschmar K, Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc. 2020;15:3380-3409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 441] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 11. | Yang H, Cheng J, Zhuang H, Xu H, Wang Y, Zhang T, Yang Y, Qian H, Lu Y, Han F, Cao L, Yang N, Liu R, Yang X, Zhang J, Wu J, Zhang N. Pharmacogenomic profiling of intra-tumor heterogeneity using a large organoid biobank of liver cancer. Cancer Cell. 2024;42:535-551.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 76] [Reference Citation Analysis (0)] |
| 12. | Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4394] [Cited by in RCA: 5468] [Article Influence: 321.6] [Reference Citation Analysis (0)] |
| 13. | Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762-1772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2253] [Cited by in RCA: 2866] [Article Influence: 191.1] [Reference Citation Analysis (0)] |
| 14. | Yang JL, Zhang JF, Gu JY, Gao M, Zheng MY, Guo SX, Zhang T. Strategic insights into the cultivation of pancreatic cancer organoids from endoscopic ultrasonography-guided biopsy tissue. World J Gastroenterol. 2024;30:4532-4543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 15. | Luo X, Gong Y, Gong Z, Fan K, Suo T, Liu H, Ni X, Ni X, Abudureyimu M, Liu H. Liver and bile duct organoids and tumoroids. Biomed Pharmacother. 2024;178:117104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Xu H, Jiao D, Liu A, Wu K. Tumor organoids: applications in cancer modeling and potentials in precision medicine. J Hematol Oncol. 2022;15:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 185] [Reference Citation Analysis (0)] |
| 17. | Airola C, Pallozzi M, Cesari E, Cerrito L, Stella L, Sette C, Giuliante F, Gasbarrini A, Ponziani FR. Hepatocellular-Carcinoma-Derived Organoids: Innovation in Cancer Research. Cells. 2024;13:1726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 18. | van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1313] [Cited by in RCA: 1830] [Article Influence: 183.0] [Reference Citation Analysis (0)] |
| 19. | Thorel L, Perréard M, Florent R, Divoux J, Coffy S, Vincent A, Gaggioli C, Guasch G, Gidrol X, Weiswald LB, Poulain L. Patient-derived tumor organoids: a new avenue for preclinical research and precision medicine in oncology. Exp Mol Med. 2024;56:1531-1551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 84] [Reference Citation Analysis (0)] |
| 20. | Zhou L, He L, Liu CH, Qiu H, Zheng L, Sample KM, Wu Q, Li J, Xie K, Ampuero J, Li Z, Lv D, Liu M, Romero-Gómez M, Hu Y, Tang H. Liver cancer stem cell dissemination and metastasis: uncovering the role of NRCAM in hepatocellular carcinoma. J Exp Clin Cancer Res. 2023;42:311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 21. | Zhang Q, Wei T, Jin W, Yan L, Shi L, Zhu S, Bai Y, Zeng Y, Yin Z, Yang J, Zhang W, Wu M, Zhang Y, Peng G, Roessler S, Liu L. Deficiency in SLC25A15, a hypoxia-responsive gene, promotes hepatocellular carcinoma by reprogramming glutamine metabolism. J Hepatol. 2024;80:293-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 22. | Rao J, Song C, Hao Y, Chen Z, Feng S, Xu S, Wu X, Xuan Z, Fan Y, Li W, Li J, Ren Y, Li J, Cheng F, Gu Z. Leveraging Patient-Derived Organoids for Personalized Liver Cancer Treatment. Int J Biol Sci. 2024;20:5363-5374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 23. | Polak R, Zhang ET, Kuo CJ. Cancer organoids 2.0: modelling the complexity of the tumour immune microenvironment. Nat Rev Cancer. 2024;24:523-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 137] [Reference Citation Analysis (0)] |
| 24. | Singal AG, Llovet JM, Yarchoan M, Mehta N, Heimbach JK, Dawson LA, Jou JH, Kulik LM, Agopian VG, Marrero JA, Mendiratta-Lala M, Brown DB, Rilling WS, Goyal L, Wei AC, Taddei TH. AASLD Practice Guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatology. 2023;78:1922-1965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 900] [Cited by in RCA: 1156] [Article Influence: 385.3] [Reference Citation Analysis (23)] |
| 25. | Wang Z, Guo Y, Jin Y, Zhang X, Geng H, Xie G, Ye D, Yu Y, Liu D, Zhou D, Li B, Luo Y, Peng S, Li J. Establishment and drug screening of patient-derived extrahepatic biliary tract carcinoma organoids. Cancer Cell Int. 2021;21:519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Wang Z, Jin Y, Guo Y, Tan Z, Zhang X, Ye D, Yu Y, Peng S, Zheng L, Li J. Conversion Therapy of Intrahepatic Cholangiocarcinoma Is Associated with Improved Prognosis and Verified by a Case of Patient-Derived Organoid. Cancers (Basel). 2021;13:1179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Guo Y, Li Q, Ye Q, Jin Y, Yu Y, Zhang X, Xi L, Wang Y, Wu D, Pan Y, Wei S, Li Q, Wang H, Li J. Construction and drug screening of Co-culture system using extrahepatic cholangiocarcinoma organoids and tumor-associated macrophages. Heliyon. 2024;10:e36377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 28. | Guo Y, Miao S, Jin Y, Li Q, Wang Y, Zhang X, Li J. Tumor-associated macrophages contribute to cholangiocarcinoma progression and chemoresistance through activation of ID1. Ann Hepatol. 2024;30:101773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 29. | Wang Z, Yu Y, Wu P, Ye Q, Guo Y, Zhang X, Xi L, Li Q, Jin Y, Zhou D, Luo Y, Peng S, Li J. Lactate promotes the growth of patient-derived organoids from hepatopancreatobiliary cancers via ENO1/HIF1α pathway and does not affect their drug sensitivities. Cell Death Discov. 2022;8:214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 30. | Nuciforo S, Fofana I, Matter MS, Blumer T, Calabrese D, Boldanova T, Piscuoglio S, Wieland S, Ringnalda F, Schwank G, Terracciano LM, Ng CKY, Heim MH. Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep. 2018;24:1363-1376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 351] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 31. | Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, Georgakopoulos N, Koo BK, Dietmann S, Davies SE, Praseedom RK, Lieshout R, IJzermans JNM, Wigmore SJ, Saeb-Parsy K, Garnett MJ, van der Laan LJ, Huch M. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424-1435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 905] [Cited by in RCA: 1059] [Article Influence: 117.7] [Reference Citation Analysis (0)] |
| 32. | Zou Z, Lin Z, Wu C, Tan J, Zhang J, Peng Y, Zhang K, Li J, Wu M, Zhang Y. Micro-Engineered Organoid-on-a-Chip Based on Mesenchymal Stromal Cells to Predict Immunotherapy Responses of HCC Patients. Adv Sci (Weinh). 2023;10:e2302640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 51] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 33. | Lee HS, Han DH, Cho K, Park SB, Kim C, Leem G, Jung DE, Kwon SS, Kim CH, Jo JH, Lee HW, Song SY, Park JY. Integrative analysis of multiple genomic data from intrahepatic cholangiocarcinoma organoids enables tumor subtyping. Nat Commun. 2023;14:237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 34. | Ren X, Huang M, Weng W, Xie Y, Wu Y, Zhu S, Zhang Y, Li D, Lai J, Shen S, Lin J, Kuang M, Li X, Yu J, Xu L. Personalized drug screening in patient-derived organoids of biliary tract cancer and its clinical application. Cell Rep Med. 2023;4:101277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 35. | Saito Y, Muramatsu T, Kanai Y, Ojima H, Sukeda A, Hiraoka N, Arai E, Sugiyama Y, Matsuzaki J, Uchida R, Yoshikawa N, Furukawa R, Saito H. Establishment of Patient-Derived Organoids and Drug Screening for Biliary Tract Carcinoma. Cell Rep. 2019;27:1265-1276.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 175] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 36. | Yuan B, Zhao X, Wang X, Liu E, Liu C, Zong Y, Jiang Y, Hou M, Chen Y, Chen L, Zhang Y, Wang H, Fu J. Patient-derived organoids for personalized gallbladder cancer modelling and drug screening. Clin Transl Med. 2022;12:e678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 37. | Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Öhlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FH, Molenaar IQ, Borel Rinkes IH, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RG, Clevers H, Tuveson DA. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 1666] [Article Influence: 138.8] [Reference Citation Analysis (0)] |
| 38. | Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, Froeling FEM, Burkhart RA, Denroche RE, Jang GH, Miyabayashi K, Young CM, Patel H, Ma M, LaComb JF, Palmaira RLD, Javed AA, Huynh JC, Johnson M, Arora K, Robine N, Shah M, Sanghvi R, Goetz AB, Lowder CY, Martello L, Driehuis E, LeComte N, Askan G, Iacobuzio-Donahue CA, Clevers H, Wood LD, Hruban RH, Thompson E, Aguirre AJ, Wolpin BM, Sasson A, Kim J, Wu M, Bucobo JC, Allen P, Sejpal DV, Nealon W, Sullivan JD, Winter JM, Gimotty PA, Grem JL, DiMaio DJ, Buscaglia JM, Grandgenett PM, Brody JR, Hollingsworth MA, O'Kane GM, Notta F, Kim E, Crawford JM, Devoe C, Ocean A, Wolfgang CL, Yu KH, Li E, Vakoc CR, Hubert B, Fischer SE, Wilson JM, Moffitt R, Knox J, Krasnitz A, Gallinger S, Tuveson DA. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018;8:1112-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 817] [Article Influence: 102.1] [Reference Citation Analysis (0)] |
| 39. | Beato F, Reverón D, Dezsi KB, Ortiz A, Johnson JO, Chen DT, Ali K, Yoder SJ, Jeong D, Malafa M, Hodul P, Jiang K, Centeno BA, Abdalah MA, Balasi JA, Tassielli AF, Sarcar B, Teer JK, DeNicola GM, Permuth JB, Fleming JB. Establishing a living biobank of patient-derived organoids of intraductal papillary mucinous neoplasms of the pancreas. Lab Invest. 2021;101:204-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 40. | Kim SC, Seo HY, Lee JO, Maeng JE, Shin YK, Lee SH, Jang JY, Ku JL. Establishment, characterization, and biobanking of 36 pancreatic cancer organoids: prediction of metastasis in resectable pancreatic cancer. Cell Oncol (Dordr). 2024;47:1627-1647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 41. | Shiihara M, Ishikawa T, Saiki Y, Omori Y, Hirose K, Fukushige S, Ikari N, Higuchi R, Yamamoto M, Morikawa T, Nakagawa K, Hayashi H, Mizuma M, Ohtsuka H, Motoi F, Unno M, Okamura Y, Kinoshita K, Furukawa T. Development of a system combining comprehensive genotyping and organoid cultures for identifying and testing genotype-oriented personalised medicine for pancreatobiliary cancers. Eur J Cancer. 2021;148:239-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 42. | Choi W, Kim YH, Woo SM, Yu Y, Lee MR, Lee WJ, Chun JW, Sim SH, Chae H, Shim H, Lee KS, Kong SY. Establishment of Patient-Derived Organoids Using Ascitic or Pleural Fluid from Cancer Patients. Cancer Res Treat. 2023;55:1077-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 43. | Driehuis E, van Hoeck A, Moore K, Kolders S, Francies HE, Gulersonmez MC, Stigter ECA, Burgering B, Geurts V, Gracanin A, Bounova G, Morsink FH, Vries R, Boj S, van Es J, Offerhaus GJA, Kranenburg O, Garnett MJ, Wessels L, Cuppen E, Brosens LAA, Clevers H. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A. 2019;116:26580-26590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 340] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 44. | Zhu Y, Tang S, Yuan Q, Fu J, He J, Liu Z, Zhao X, Li Y, Zhao Y, Zhang Y, Zhang X, Zhang Y, Zhu Y, Wang W, Zheng B, Wu R, Wu T, Yang S, Qiu X, Shen S, Hu J, Chen L, Wang Y, Wang H, Gao D, Chen L. Integrated characterization of hepatobiliary tumor organoids provides a potential landscape of pharmacogenomic interactions. Cell Rep Med. 2024;5:101375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 45. | Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, McNary TJ, Churakova Y, Cheung C, Triscott J, Pisapia D, Rao R, Mosquera JM, Robinson B, Faltas BM, Emerling BE, Gadi VK, Bernard B, Elemento O, Beltran H, Demichelis F, Kemp CJ, Grandori C, Cantley LC, Rubin MA. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017;7:462-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 675] [Cited by in RCA: 746] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 46. | Galle PR, Finn RS, Qin S, Ikeda M, Zhu AX, Kim TY, Kudo M, Breder V, Merle P, Kaseb A, Li D, Mulla S, Verret W, Xu DZ, Hernandez S, Ding B, Liu J, Huang C, Lim HY, Cheng AL, Ducreux M. Patient-reported outcomes with atezolizumab plus bevacizumab versus sorafenib in patients with unresectable hepatocellular carcinoma (IMbrave150): an open-label, randomised, phase 3 trial. Lancet Oncol. 2021;22:991-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 261] [Article Influence: 52.2] [Reference Citation Analysis (1)] |
| 47. | Walz S, Pollehne P, Geng R, Schneider J, Maas M, Aicher WK, Stenzl A, Amend B, Harland N. A Protocol for Organoids from the Urine of Bladder Cancer Patients. Cells. 2023;12:2188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 48. | Beutel AK, Schütte L, Scheible J, Roger E, Müller M, Perkhofer L, Kestler AMTU, Kraus JM, Kestler HA, Barth TFE, Lemke J, Kornmann M, Ettrich TJ, Gout J, Seufferlein T, Kleger A. A Prospective Feasibility Trial to Challenge Patient-Derived Pancreatic Cancer Organoids in Predicting Treatment Response. Cancers (Basel). 2021;13:2539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 49. | van Tienderen GS, Groot Koerkamp B, IJzermans JNM, van der Laan LJW, Verstegen MMA. Recreating Tumour Complexity in a Dish: Organoid Models to Study Liver Cancer Cells and their Extracellular Environment. Cancers (Basel). 2019;11:1706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 50. | Fu Y, Guo X, Sun L, Cui T, Wu C, Wang J, Liu Y, Liu L. Exploring the role of the immune microenvironment in hepatocellular carcinoma: Implications for immunotherapy and drug resistance. Elife. 2024;13:e95009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 51. | Liu J, Li P, Wang L, Li M, Ge Z, Noordam L, Lieshout R, Verstegen MMA, Ma B, Su J, Yang Q, Zhang R, Zhou G, Carrascosa LC, Sprengers D, IJzermans JNM, Smits R, Kwekkeboom J, van der Laan LJW, Peppelenbosch MP, Pan Q, Cao W. Cancer-Associated Fibroblasts Provide a Stromal Niche for Liver Cancer Organoids That Confers Trophic Effects and Therapy Resistance. Cell Mol Gastroenterol Hepatol. 2021;11:407-431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 155] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 52. | Sevic I, Spinelli FM, Cantero MJ, Reszegi A, Kovalszky I, García MG, Alaniz L. The Role of the Tumor Microenvironment in the Development and Progression of Hepatocellular Carcinoma. In: Hepatocellular Carcinoma [Internet]. Brisbane (AU): Codon Publications; 2019-Oct-24. [PubMed] [DOI] [Full Text] |
| 53. | Amato F, Rae C, Prete MG, Braconi C. Cholangiocarcinoma Disease Modelling Through Patients Derived Organoids. Cells. 2020;9:832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 54. | Du Y, Wang YR, Bao QY, Xu XX, Xu C, Wang S, Liu Q, Liu F, Zeng YL, Wang YJ, Liu W, Liu Y, Yu SX, Chen YC, Wang C, Zhang W, Gao H, Luo H, Liu B, Jing G, Guo M, Chen FX, Liu YJ. Personalized Vascularized Tumor Organoid-on-a-Chip for Tumor Metastasis and Therapeutic Targeting Assessment. Adv Mater. 2025;37:e2412815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 27] [Article Influence: 27.0] [Reference Citation Analysis (1)] |
| 55. | Luo Q, Wang J, Zhao W, Peng Z, Liu X, Li B, Zhang H, Shan B, Zhang C, Duan C. Vasculogenic mimicry in carcinogenesis and clinical applications. J Hematol Oncol. 2020;13:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 228] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 56. | Oria VO, Erler JT. Tumor Angiocrine Signaling: Novel Targeting Opportunity in Cancer. Cells. 2023;12:2510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 57. | Fischer A, Alsina-Sanchis E. Disturbed endothelial cell signaling in tumor progression and therapy resistance. Curr Opin Cell Biol. 2024;86:102287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 58. | Tabe S, Takeuchi K, Aoshima K, Okumura A, Yamamoto Y, Yanagisawa K, Eto R, Matsuo M, Ueno Y, Konishi T, Furukawa Y, Yamaguchi K, Morinaga S, Miyagi Y, Ohtsuka M, Tanimizu N, Taniguchi H. A pancreatic cancer organoid incorporating macrophages reveals the correlation between the diversity of tumor-associated macrophages and cancer cell survival. Biomaterials. 2025;314:122838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 59. | Jiang S, Deng T, Cheng H, Liu W, Shi D, Yuan J, He Z, Wang W, Chen B, Ma L, Zhang X, Gong P. Macrophage-organoid co-culture model for identifying treatment strategies against macrophage-related gemcitabine resistance. J Exp Clin Cancer Res. 2023;42:199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 52] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 60. | Tao J, Gu Y, Zhang Z, Weng G, Liu Y, Ren J, Shi Y, Qiu J, Wang Y, Su D, Wang R, Fu Y, Liu T, Ye L, Luo W, Chen H, Yang G, Cao Z, Huang H, Xiao J, Ren B, You L, Zhang T, Zhao Y. CALB2 drives pancreatic cancer metastasis through inflammatory reprogramming of the tumor microenvironment. J Exp Clin Cancer Res. 2024;43:277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 61. | Takeuchi K, Tabe S, Takahashi K, Aoshima K, Matsuo M, Ueno Y, Furukawa Y, Yamaguchi K, Ohtsuka M, Morinaga S, Miyagi Y, Yamaguchi T, Tanimizu N, Taniguchi H. Incorporation of human iPSC-derived stromal cells creates a pancreatic cancer organoid with heterogeneous cancer-associated fibroblasts. Cell Rep. 2023;42:113420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 62. | Liu T, Tan J, Wu M, Fan W, Wei J, Zhu B, Guo J, Wang S, Zhou P, Zhang H, Shi L, Li J. High-affinity neoantigens correlate with better prognosis and trigger potent antihepatocellular carcinoma (HCC) activity by activating CD39(+)CD8(+) T cells. Gut. 2021;70:1965-1977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 63. | Zou F, Tan J, Liu T, Liu B, Tang Y, Zhang H, Li J. The CD39(+) HBV surface protein-targeted CAR-T and personalized tumor-reactive CD8(+) T cells exhibit potent anti-HCC activity. Mol Ther. 2021;29:1794-1807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 64. | Zhou G, Lieshout R, van Tienderen GS, de Ruiter V, van Royen ME, Boor PPC, Magré L, Desai J, Köten K, Kan YY, Ge Z, Campos Carrascosa L, Geuijen C, Sprengers D, van der Laan LJW, Verstegen MMA, Kwekkeboom J. Modelling immune cytotoxicity for cholangiocarcinoma with tumour-derived organoids and effector T cells. Br J Cancer. 2022;127:649-660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 65. | Tai TS, Hsu DW, Yang YS, Tsai CY, Shi JW, Wu CH, Hsu SC. IL-10RA governor the expression of IDO in the instruction of lymphocyte immunity. Br J Cancer. 2025;132:126-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 66. | Sheng N, Shindo K, Ohuchida K, Shinkawa T, Zhang B, Feng H, Yamamoto T, Moriyama T, Ikenaga N, Nakata K, Oda Y, Nakamura M. TAK1 Promotes an Immunosuppressive Tumor Microenvironment through Cancer-Associated Fibroblast Phenotypic Conversion in Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2024;30:5138-5153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 67. | Lahusen A, Cai J, Schirmbeck R, Wellstein A, Kleger A, Seufferlein T, Eiseler T, Lin YN. A pancreatic cancer organoid-in-matrix platform shows distinct sensitivities to T cell killing. Sci Rep. 2024;14:9377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 68. | Beelen NA, Aberle MR, Bruno V, Olde Damink SWM, Bos GMJ, Rensen SS, Wieten L. Antibody-dependent cellular cytotoxicity-inducing antibodies enhance the natural killer cell anti-cancer response against patient-derived pancreatic cancer organoids. Front Immunol. 2023;14:1133796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 69. | Schuth S, Le Blanc S, Krieger TG, Jabs J, Schenk M, Giese NA, Büchler MW, Eils R, Conrad C, Strobel O. Patient-specific modeling of stroma-mediated chemoresistance of pancreatic cancer using a three-dimensional organoid-fibroblast co-culture system. J Exp Clin Cancer Res. 2022;41:312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 107] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 70. | Shinkawa T, Ohuchida K, Mochida Y, Sakihama K, Iwamoto C, Abe T, Ideno N, Mizuuchi Y, Shindo K, Ikenaga N, Moriyama T, Nakata K, Oda Y, Nakamura M. Subtypes in pancreatic ductal adenocarcinoma based on niche factor dependency show distinct drug treatment responses. J Exp Clin Cancer Res. 2022;41:89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 71. | Meng Q, Xie S, Gray GK, Dezfulian MH, Li W, Huang L, Akshinthala D, Ferrer E, Conahan C, Perea Del Pino S, Grossman J, Elledge SJ, Hidalgo M, Muthuswamy SK. Empirical identification and validation of tumor-targeting T cell receptors from circulation using autologous pancreatic tumor organoids. J Immunother Cancer. 2021;9:e003213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 72. | Marcon F, Zuo J, Pearce H, Nicol S, Margielewska-Davies S, Farhat M, Mahon B, Middleton G, Brown R, Roberts KJ, Moss P. NK cells in pancreatic cancer demonstrate impaired cytotoxicity and a regulatory IL-10 phenotype. Oncoimmunology. 2020;9:1845424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 73. | Tsai S, McOlash L, Palen K, Johnson B, Duris C, Yang Q, Dwinell MB, Hunt B, Evans DB, Gershan J, James MA. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer. 2018;18:335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 316] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 74. | Lim JTC, Kwang LG, Ho NCW, Toh CCM, Too NSH, Hooi L, Benoukraf T, Chow PK, Dan YY, Chow EK, Toh TB, Fong ELS. Hepatocellular carcinoma organoid co-cultures mimic angiocrine crosstalk to generate inflammatory tumor microenvironment. Biomaterials. 2022;284:121527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 75. | Tanaka C, Furihata K, Naganuma S, Ogasawara M, Yoshioka R, Taniguchi H, Furihata M, Taniuchi K. Establishment of a mouse model of pancreatic cancer using human pancreatic cancer cell line S2-013-derived organoid. Hum Cell. 2022;35:735-744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 76. | Del Piccolo N, Shirure VS, Bi Y, Goedegebuure SP, Gholami S, Hughes CCW, Fields RC, George SC. Tumor-on-chip modeling of organ-specific cancer and metastasis. Adv Drug Deliv Rev. 2021;175:113798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 77. | Xu Y, Le J, Qin J, Zhang Y, Yang J, Chen Z, Li C, Qian X, Zhang A. Decoding the microbiota metabolome in hepatobiliary and pancreatic cancers: Pathways to precision diagnostics and targeted therapeutics. Pharmacol Res. 2024;208:107364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 78. | Chen H, Liu H, Zhang X, Wang S, Liu C, An K, Liu R, Tian X. Diversified applications of hepatocellular carcinoma medications: molecular-targeted, immunotherapeutic, and combined approaches. Front Pharmacol. 2024;15:1422033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 79. | Song T, Kong B, Liu R, Luo Y, Wang Y, Zhao Y. Bioengineering Approaches for the Pancreatic Tumor Organoids Research and Application. Adv Healthc Mater. 2024;13:e2300984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 80. | Benson AB, D'Angelica MI, Abrams T, Abbott DE, Ahmed A, Anaya DA, Anders R, Are C, Bachini M, Binder D, Borad M, Bowlus C, Brown D, Burgoyne A, Castellanos J, Chahal P, Cloyd J, Covey AM, Glazer ES, Hawkins WG, Iyer R, Jacob R, Jennings L, Kelley RK, Kim R, Levine M, Palta M, Park JO, Raman S, Reddy S, Ronnekleiv-Kelly S, Sahai V, Singh G, Stein S, Turk A, Vauthey JN, Venook AP, Yopp A, McMillian N, Schonfeld R, Hochstetler C. NCCN Guidelines® Insights: Biliary Tract Cancers, Version 2.2023. J Natl Compr Canc Netw. 2023;21:694-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 135] [Reference Citation Analysis (0)] |
| 81. | Benboubker V, Ramzy GM, Jacobs S, Nowak-Sliwinska P. Challenges in validation of combination treatment strategies for CRC using patient-derived organoids. J Exp Clin Cancer Res. 2024;43:259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 82. | Nicolson NG, Tandurella JA, Wu LW, Patel J, Morris E, Seppälä TT, Guinn S, Zlomke H, Shubert CR, Lafaro KJ, Burns WR, Cameron JL, He J, Fertig EJ, Jaffee EM, Zimmerman JW, Burkhart RA. Patient-derived Organoid Pharmacotyping As A Predictive Tool for Therapeutic Selection in Pancreatic Ductal Adenocarcinoma. Ann Surg. 2024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 83. | Lv J, Du X, Wang M, Su J, Wei Y, Xu C. Construction of tumor organoids and their application to cancer research and therapy. Theranostics. 2024;14:1101-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 47] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 84. | Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490-1494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 473] [Cited by in RCA: 439] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 85. | Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, Wongvipat J, Kossai M, Ramazanoglu S, Barboza LP, Di W, Cao Z, Zhang QF, Sirota I, Ran L, MacDonald TY, Beltran H, Mosquera JM, Touijer KA, Scardino PT, Laudone VP, Curtis KR, Rathkopf DE, Morris MJ, Danila DC, Slovin SF, Solomon SB, Eastham JA, Chi P, Carver B, Rubin MA, Scher HI, Clevers H, Sawyers CL, Chen Y. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 882] [Cited by in RCA: 1201] [Article Influence: 100.1] [Reference Citation Analysis (0)] |
| 86. | Xiang D, He A, Zhou R, Wang Y, Xiao X, Gong T, Kang W, Lin X, Wang X; PDO-based DST Consortium, Liu L, Chen YG, Gao S, Liu Y. Building consensus on the application of organoid-based drug sensitivity testing in cancer precision medicine and drug development. Theranostics. 2024;14:3300-3316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 46] [Reference Citation Analysis (0)] |
| 87. | Lai Benjamin FL, Lu Rick X, Hu Y, Davenport HL, Dou W, Wang EY, Radulovich N, Tsao MS, Sun Y, Radisic M. Recapitulating pancreatic tumor microenvironment through synergistic use of patient organoids and organ-on-a-chip vasculature. Adv Funct Mater. 2020;30:2000545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 88. | Song T, Zhang H, Luo Z, Shang L, Zhao Y. Primary Human Pancreatic Cancer Cells Cultivation in Microfluidic Hydrogel Microcapsules for Drug Evaluation. Adv Sci (Weinh). 2023;10:e2206004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 89. | LeSavage BL, Suhar RA, Broguiere N, Lutolf MP, Heilshorn SC. Next-generation cancer organoids. Nat Mater. 2022;21:143-159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 343] [Cited by in RCA: 297] [Article Influence: 74.3] [Reference Citation Analysis (0)] |
| 90. | Kim SE, Yun S, Doh J, Kim HN. Imaging-Based Efficacy Evaluation of Cancer Immunotherapy in Engineered Tumor Platforms and Tumor Organoids. Adv Healthc Mater. 2024;13:e2400475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 91. | Zhao Y, Li ZX, Zhu YJ, Fu J, Zhao XF, Zhang YN, Wang S, Wu JM, Wang KT, Wu R, Sui CJ, Shen SY, Wu X, Wang HY, Gao D, Chen L. Single-Cell Transcriptome Analysis Uncovers Intratumoral Heterogeneity and Underlying Mechanisms for Drug Resistance in Hepatobiliary Tumor Organoids. Adv Sci (Weinh). 2021;8:e2003897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 92. | Shi X, Li Y, Yuan Q, Tang S, Guo S, Zhang Y, He J, Zhang X, Han M, Liu Z, Zhu Y, Gao S, Wang H, Xu X, Zheng K, Jing W, Chen L, Wang Y, Jin G, Gao D. Integrated profiling of human pancreatic cancer organoids reveals chromatin accessibility features associated with drug sensitivity. Nat Commun. 2022;13:2169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 93. | Wang W, Yuan T, Ma L, Zhu Y, Bao J, Zhao X, Zhao Y, Zong Y, Zhang Y, Yang S, Qiu X, Shen S, Wu R, Wu T, Wang H, Gao D, Wang P, Chen L. Hepatobiliary Tumor Organoids Reveal HLA Class I Neoantigen Landscape and Antitumoral Activity of Neoantigen Peptide Enhanced with Immune Checkpoint Inhibitors. Adv Sci (Weinh). 2022;9:e2105810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 94. | Deininger L, Jung-Klawitter S, Mikut R, Richter P, Fischer M, Karimian-Jazi K, Breckwoldt MO, Bendszus M, Heiland S, Kleesiek J, Opladen T, Kuseyri Hübschmann O, Hübschmann D, Schwarz D. An AI-based segmentation and analysis pipeline for high-field MR monitoring of cerebral organoids. Sci Rep. 2023;13:21231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 95. | Yang R, Du Y, Kwan W, Yan R, Shi Q, Zang L, Zhu Z, Zhang J, Li C, Yu Y. A quick and reliable image-based AI algorithm for evaluating cellular senescence of gastric organoids. Cancer Biol Med. 2023;20:519-536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/