Published online Apr 28, 2011. doi: 10.3748/wjg.v17.i16.2096
Revised: September 18, 2010
Accepted: September 25, 2010
Published online: April 28, 2011
AIM: To evaluate the usefulness of differentially expressed proteins from colorectal cancer (CRC) tissues for differentiating cancer and normal tissues.
METHODS: A Proteomic approach was used to identify the differentially expressed proteins between CRC and normal tissues. The proteins were extracted using Tris buffer and thiourea lysis buffer (TLB) for extraction of aqueous soluble and membrane-associated proteins, respectively. Chemometrics, namely principal component analysis (PCA) and linear discriminant analysis (LDA), were used to assess the usefulness of these proteins for identifying the cancerous state of tissues.
RESULTS: Differentially expressed proteins identified were 37 aqueous soluble proteins in Tris extracts and 24 membrane-associated proteins in TLB extracts. Based on the protein spots intensity on 2D-gel images, PCA by applying an eigenvalue > 1 was successfully used to reduce the number of principal components (PCs) into 12 and seven PCs for Tris and TLB extracts, respectively, and subsequently six PCs, respectively from both the extracts were used for LDA. The LDA classification for Tris extract showed 82.7% of original samples were correctly classified, whereas 82.7% were correctly classified for the cross-validated samples. The LDA for TLB extract showed that 78.8% of original samples and 71.2% of the cross-validated samples were correctly classified.
CONCLUSION: The classification of CRC tissues by PCA and LDA provided a promising distinction between normal and cancer types. These methods can possibly be used for identification of potential biomarkers among the differentially expressed proteins identified.
- Citation: Yeoh LC, Dharmaraj S, Gooi BH, Singh M, Gam LH. Chemometrics of differentially expressed proteins from colorectal cancer patients. World J Gastroenterol 2011; 17(16): 2096-2103
- URL: https://www.wjgnet.com/1007-9327/full/v17/i16/2096.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i16.2096
Proteomic research has made great achievements in biomarker discovery, especially when incorporated with high-throughput analytical tools and technology, for example 2D-PAGE and LC-MS/MS[1]. Two-dimensional gel electrophoresis is a fundamental tool for protein analysis to detect alterations in protein expression between control and disease states of cells, which can lead to the discovery of various biomarkers that contribute to pathogenesis or carcinogenesis[2]. Biomarkers can be used to discriminate variables for subsequent classification of normal and diseased groups[3]. The complexity of variables generated by mass spectra, microarray and immunohistochemistry often requires advanced statistical techniques or chemometrics to evaluate their clinical value.
Multivariate analyses including the dimension reduction method known as principal component analysis (PCA), and classification methods such as linear discriminant analysis (LDA) are often employed in proteomic studies. PCA reduces the number of variables for further data analysis and interpretation while identifying the variables that retain most of the data variance[4]. A principal component (PC) is defined as a new variable to explain the maximum amount of variance in the original data and corresponds to a linear combination of the original variables. PCs are presented orthogonally to each other, which provides a more effective representation of the data than the original variables[2]. LDA is a multivariate technique to classify observations into groups or categories. LDA forms new variables from the original data and identifies the variables that provide the best discrimination between the groups[5].
Djidja et al[6] have used a novel approach that combines matrix-assisted laser desorption ionization-ion mobility separation-mass spectrometry (MALDI-IMS-MS) and PCA-discriminant analysis (PCA-DA) to generate tumor classification models based on pancreatic cancer protein patterns. Furthermore, Kamath et al[7] have used PCA-based k-nearest neighbor analysis to classify normal and cancerous autofluorescence spectra of colonic mucosal tissues. Zwielly et al[8] have investigated the use of Fourier transform infrared microscopy for colon cancer diagnosis. Their model uses PCA to define spectral changes among normal and cancerous human biopsied colon tissues. Ragazzi et al[9] have reported the use of multivariate techniques on plasma proteins to diagnose colorectal cancer (CRC). The plasma protein profile generated by MALDI-MS is analyzed by PCA and LDA to discriminate ionic species from normal subjects and CRC patients.
In this study, we carried out the comparison of 2-D images of cancerous and normal colorectal tissues. The differentially expressed proteins from Tris and thiourea lysis buffer (TLB) extractions were respectively tested on a PCA-LDA model to find out the possibility of using protein expression to classify the disease and non-disease tissues of CRC.
Matching pairs of normal colonic mucosa and cancerous colonic tissue (located 10 cm from each other) from 26 CRC patients were collected after surgery at the Penang General Hospital, Penang, Malaysia. The study was approved by the Human Ethical Committee of Universiti Sains Malaysia. Informed written consent was received from all patients before the study was conducted. Prior to surgery, the patients did not receive preoperative neoadjuvant chemotherapy and radiotherapy. The tissues were confirmed as cancerous and normal, respectively, by the hospital’s pathologist. The cancerous tissues were classified using the TNM system. Surgically removed samples were stored at -80°C until use.
The method of protein analysis was as described in Yeoh et al[10]. Frozen tissue (250 mg) was rinsed in distilled water to remove cell debris and excess blood. The tissues were homogenized in ice-cold Tris buffer (0.5 g tissue/mL buffer) [40 mmol/L Tris and 1 × Protease Inhibitor Cocktail (Sigma, St Louis, MO, USA)] and centrifuged at 12 000 rpm for 15 min at 18°C. The supernatant was recovered and labeled as Tris extract. The pellet was subjected to further extraction using TLB (1 g tissue/1 mL buffer) [8 mol/L urea, 2 mol/L thiourea, 4% (w/v) CHAPS, 0.4% (w/v) carrier ampholytes and 50 mmol/L dithiothreitol] and centrifuged at 12 000 rpm for 15 min at 18°C. The supernatant was recovered and labeled as TLB extract. The extracts were subjected to 2D gel separation on 11 cm ReadyStrip™ IPG strip (linear pH 4-7, Bio-Rad, USA) followed by separation on 10% (w/v) PAGE at a constant voltage of 200 V. The gels were stained with Coomassie Blue. The images obtained were analyzed by PDQuest version 7.3 (Bio-Rad). Comparison of the protein expression levels was carried out between cancerous and normal tissues. Differentially expressed proteins were defined as proteins with a spot intensity that was 1.5-fold higher or lower in cancerous tissues when compared to that in the corresponding normal tissues. A differentially expressed protein was defined as upregulated when it was found at greater intensity in cancerous tissue than in the corresponding normal tissue. The downregulated proteins were detected at greater intensity in normal tissues than in the corresponding CRC cancerous tissues.
The differentially expressed proteins were excised from the gel and subjected to in-gel digestion using trypsin and the tryptic peptides were analyzed by LC/MS/MS using an electrospray ionization ion trap mass analyzer (Agilent Technologies, Santa Clara, CA, USA). The MS/MS data were subjected to the MASCOT protein database search engine for protein identification. The identities of a few proteins (dependent on the availability of antibodies) were further confirmed using western blotting.
The differential expression of the proteins was tested by the paired Student’s t test that is included in PDQuest, to determine their statistical significance (P < 0.05). For PCA and LDA, the protein spot intensities were exported out from PDQuest and imported into SPSS version 15.0 (Chicago, IL, USA) to perform multivariate analyses. Protein spot intensities were used as variables.
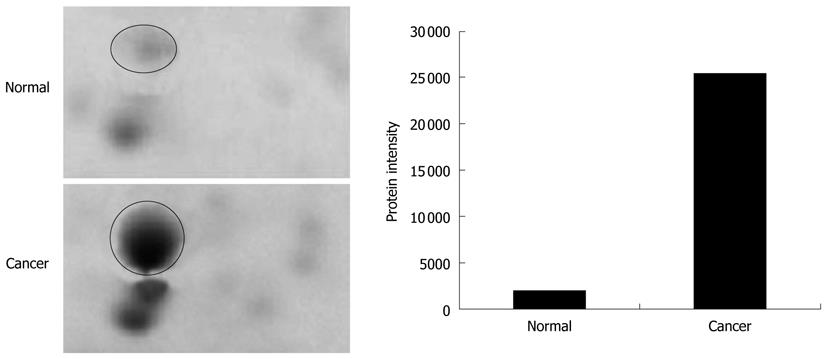
The tissues specimens from each patient were collected in pairs of cancerous and normal tissues. Table 1 shows the details of the tissues used in the analysis. The tissues were subjected to a sequential extraction method to extract aqueous soluble proteins and membrane-associated proteins in two different fractions using Tris and TLB, respectively. Tables 2 and 3 show the 37 and 24 differentially expressed proteins identified in Tris and TLB extracts, respectively. The average fold change indicates the degree of differentiation in expression levels of the protein in cancerous tissues compared to normal tissues in all the patients tested, where a positive sign indicates a greater expression level in cancerous tissues, whereas a negative sign indicates a greater expression level in normal tissues. The MOWSE score refers to the score values given by the MASCOT search. Tables 4 and 5 show the mean intensity of spots and SD, and percentage coefficient of variation (%CV) of spot intensity of differentially expressed proteins in all patients for Tris and TLB extracts, respectively. An example of the differentially expressed protein, as represented by different intensities of protein spots between normal and cancerous tissues for glutathione S-transferase P (GST-P), is shown in Figure 1; the bar chart was plotted according to the intensity of the respective protein spots. GST-P was detected as upregulated in cancerous tissues.
| Patient No. | Age (yr) | Race | Sex | pTNM | Stage | Degree of differentiation | Tumor location |
| 1 | 62 | Malay | Male | pT3N1Mx | IIIB | MD | Sigmoid colon |
| 2 | 79 | Malay | Male | pT2NoM0 | I | MD | Descending colon |
| 3 | 74 | Malay | Male | pT3N0M0 | IIA | MD | Ascending colon |
| 4 | - | Malay | Male | pT3N2Mx | IIIC | MD | Rectum |
| 5 | 37 | Malay | Male | pT3N0M0 | IIA | MD | Transverse colon |
| 6 | 58 | Malay | Female | pT3N0Mx | IIA | MD | Recto-sigmoid |
| 7 | 59 | Malay | Female | pT4N2Mx | IIIC | MD | Ileocecal |
| 8 | 69 | Malay | Male | pT3N0Mx | IIA | MD | Sigmoid colon |
| 9 | 63 | Malay | Female | pT3N0Mx | IIA | MD | Recto-sigmoid |
| 10 | 84 | Chinese | Female | pT4N0M0 | IIB | MD | Rectum |
| 11 | 58 | Chinese | Male | pT3N0Mx | IIA | MD | Recto-sigmoid |
| Spot No. | Protein name | Swissprot No.1 | MOWSE score2 | MW (Da) | pI | Sequence coverage (%) | GRAVY | Average fold change3 |
| 1 | Proteasome subunit β type 6 | P28072 | 134 | 25573 | 4.80 | 16 | 0.034 | -2.967 |
| 2 | 14-3-3 protein ζ | P63104 | 336 | 35567 | 6.97 | 40 | -0.744 | 11.659 |
| 3 | Tropomyosin α-3C-like protein | A6NL28 | 127 | 27407 | 4.71 | 31 | -0.992 | 44.183 |
| 4 | Rho GDP-dissociation inhibitor 1 | P52565 | 167 | 23120 | 5.03 | 29 | -0.700 | -7.607 |
| 5 | 14-3-3 protein ζ | P63104 | 282 | 27919 | 4.73 | 16 | -0.621 | 4.127 |
| 6 | Tubulin β-2C chain | P68371 | 524 | 50304 | 4.83 | 40 | -0.362 | -52.184 |
| 7 | Cathepsin B | P07858 | 74 | 22981 | 5.20 | 18 | -0.433 | 33.149 |
| 8 | Rho GDP-dissociation inhibitor 2 | P52566 | 48 | 22901 | 5.10 | 18 | -0.799 | -10.625 |
| 9 | SEC13 homolog | P55735 | 78 | 36040 | 5.22 | 9 | -0.372 | 6.873 |
| 10 | Hsc70-interacting protein | P50502 | 164 | 28464 | 8.92 | 21 | -0.653 | 20.959 |
| 11 | Apolipoprotein A-I | P02647 | 143 | 30777 | 5.56 | 26 | -0.717 | -4.478 |
| 12 | Proteasome subunit α type 3 | P25788 | 201 | 15958 | 6.82 | 41 | 0.008 | 4.249 |
| 13 | Actin, cytoplasmic 2 | P63261 | 105 | 26169 | 5.65 | 14 | -0.156 | 28.601 |
| 14 | 60 kDa heat shock protein | P10809 | 151 | 61348 | 5.70 | 14 | -0.074 | 131.219 |
| 15 | Peroxiredoxin-2 | P32119 | 283 | 21935 | 5.67 | 42 | -0.210 | 1.250 |
| 16 | Guanine nucleotide binding protein subunit β 2 | P62879 | 112 | 37954 | 5.60 | 11 | -0.183 | -14.442 |
| 17 | F-actin-capping protein subunit β | P47756 | 259 | 34187 | 6.02 | 37 | -0.574 | 33.554 |
| 18 | GST-P | P09211 | 730 | 23442 | 5.44 | 60 | -0.131 | 4.834 |
| 19 | Haptoglobin-related protein | P00739 | 49 | 39529 | 6.42 | 3 | -0.308 | 56.209 |
| 20 | Cathepsin Z | Q9UBR2 | 100 | 27787 | 5.48 | 15 | -0.545 | -60.766 |
| 21 | F-actin-capping protein subunit β | P47756 | 245 | 21280 | 7.93 | 34 | -0.540 | 13.278 |
| 22 | Actin-related protein 3 | P61158 | 148 | 47704 | 5.61 | 27 | -0.271 | 15.881 |
| 23 | Abhydrolase domain-containing protein 14B | Q96IU4 | 200 | 25429 | 6.82 | 26 | -0.023 | 0.765 |
| 24 | Nucleoside diphosphate kinase A | P15531 | 87 | 19873 | 5.42 | 36 | -0.075 | 73.120 |
| 25 | L-lactate dehydrogenase B chain | P07195 | 228 | 36928 | 5.71 | 14 | 0.056 | 3.513 |
| 26 | Fibrinogen β chain | P02675 | 151 | 56624 | 8.54 | 22 | -0.758 | 41.329 |
| 27 | Leukocyte elastase inhibitor | P30740 | 170 | 42857 | 5.90 | 11 | -0.249 | 10.458 |
| 28 | PDI A3 | P30101 | 674 | 57202 | 5.98 | 35 | -0.506 | 7.579 |
| 29 | Gelsolin | P06396 | 238 | 86103 | 5.90 | 20 | -0.415 | -11.917 |
| 30 | Heat shock 27 kDa protein | P04792 | 256 | 22840 | 5.98 | 47 | -0.567 | -1.508 |
| 31 | DJ-1 protein | Q99497 | 122 | 20079 | 6.33 | 54 | 0.004 | 4.981 |
| 32 | Fibrinogen β chain | P02675 | 75 | 56624 | 8.54 | 22 | -0.758 | -72.722 |
| 33 | Selenium-binding protein 1 | Q13228 | 502 | 52971 | 5.93 | 21 | -0.254 | -26.544 |
| 34 | Selenium-binding protein 1 | Q13228 | 592 | 52938 | 5.93 | 30 | -0.254 | 27.403 |
| 35 | Selenium-binding protein 1 | Q13228 | 979 | 52938 | 5.93 | 37 | -0.254 | -1.887 |
| 36 | Leukotriene A-4 hydrolase | P09960 | 215 | 69792 | 5.80 | 22 | -0.259 | 29.759 |
| 37 | Proteasome subunit α type 6 | P60900 | 71 | 20988 | 8.57 | 39 | -0.247 | 0.768 |
| Spot No. | Protein name | SwissProt No.1 | MOWSE score2 | MW (Da) | pI | Sequence coverage (%) | GRAVY | Average fold change3 |
| 1 | Tropomyosin α-4 chain | P67936 | 139 | 28506 | 4.67 | 33 | -1.033 | -51.151 |
| 2 | Putative tropomyosin α-3-chain-like protein | A6NL28 | 53 | 27407 | 4.71 | 25 | -0.992 | 4.922 |
| 3 | GC1q-R, mitochondrial | Q07021 | 123 | 31768 | 4.74 | 20 | -0.461 | -3.333 |
| 4 | Calreticulin | P27797 | 73 | 47092 | 4.30 | 11 | -1.191 | 1.394 |
| 5 | Prohibitin | P35232 | 421 | 29890 | 5.57 | 41 | 0.024 | 0.032 |
| 6 | Heat shock 70 kDa protein | P11021 | 775 | 72488 | 5.07 | 42 | -0.487 | -32.940 |
| 7 | Tubulin β-2C chain | P68371 | 299 | 48142 | 4.70 | 25 | -0.347 | -9.060 |
| 8 | PDI | P07237 | 266 | 57510 | 4.82 | 42 | -0.450 | -1.515 |
| 9 | ATP synthase subunit β, mitochondrial | P06576 | 1096 | 56559 | 5.26 | 43 | 0.018 | -15.661 |
| 10 | ATP synthase D chain | O75947 | 117 | 18406 | 5.22 | 32 | -0.569 | -5.129 |
| 11 | Chloride intracellular channel protein 1 | O00299 | 299 | 27123 | 5.09 | 30 | -0.293 | 20.288 |
| 12 | Tubulin α-1 chain | Q71U36 | 61 | 50800 | 4.94 | 6 | -0.229 | -30.291 |
| 13 | Apolipoprotein A-I | P02647 | 129 | 28078 | 5.27 | 37 | -0.840 | 78.135 |
| 14 | Actin, cytoplasmic 2 | P63261 | 52 | 42009 | 5.31 | 4 | -0.205 | -26.716 |
| 15 | Actin, aortic smooth muscle | P62736 | 261 | 42154 | 5.23 | 21 | -0.233 | 46.181 |
| 16 | Stomatin-like protein 2 | Q9UJZ1 | 151 | 38644 | 6.88 | 28 | -0.161 | -29.709 |
| 17 | 60 kDa heat shock protein, mitochondrial | P10809 | 451 | 61386 | 5.70 | 28 | -0.074 | 14.023 |
| 18 | Triosephosphate isomerase | P60174 | 167 | 26828 | 6.51 | 24 | -0.126 | 16.757 |
| 19 | Annexin A5 | P08758 | 195 | 35994 | 4.94 | 39 | -0.330 | -2.019 |
| 20 | Cytochrome b-c1 complex subunit 1, mitochondrial | P31930 | 96 | 53342 | 5.94 | 18 | -0.141 | 13.151 |
| 21 | Annexin A3 | P12429 | 140 | 36396 | 5.63 | 22 | -0.430 | 31.244 |
| 22 | Annexin A4 | P09525 | 165 | 35983 | 5.85 | 33 | -0.447 | 11.890 |
| 23 | α-enolase | P06733 | 143 | 47385 | 6.99 | 12 | -0.226 | 85.960 |
| 24 | Lamin-A/C | P02545 | 198 | 65192 | 6.40 | 25 | -0.947 | -3.378 |
| Protein spot No. | Intensity of spots(mean ± SD) | % CV of spot intensity |
| 1 | 2565.84 ± 2247.86 | 87.60 |
| 2 | 3865.47 ± 3766.11 | 97.42 |
| 3 | 2424.01 ± 1847.71 | 76.23 |
| 4 | 4957.17 ± 2923.49 | 58.97 |
| 5 | 3901.55 ± 3900.52 | 99.97 |
| 6 | 2105.64 ± 2444.14 | 116.08 |
| 7 | 2572.91 ± 1765.28 | 68.61 |
| 8 | 2959.95 ± 2177.86 | 73.58 |
| 9 | 2478.29 ± 1697.98 | 68.51 |
| 10 | 1253.48 ± 1472.88 | 117.50 |
| 11 | 3373.93 ± 2451.35 | 72.66 |
| 12 | 3247.26 ± 2519.26 | 77.58 |
| 13 | 9413.58 ± 10 685.11 | 113.51 |
| 14 | 2735.49 ± 2665.85 | 97.45 |
| 15 | 8354.35 ± 4824.59 | 57.75 |
| 16 | 7370.39 ± 7935.67 | 100.34 |
| 17 | 14 200.72 ± 16 194.91 | 114.04 |
| 18 | 6254.81 ± 5105.54 | 81.63 |
| 19 | 14 364.73 ± 10 849.77 | 75.53 |
| 20 | 10 753.33 ± 14 509.06 | 134.93 |
| 21 | 5171.49 ± 3304.12 | 63.89 |
| 22 | 3230.12 ± 1905.24 | 58.98 |
| 23 | 2114.69 ± 1164.19 | 55.05 |
| 24 | 2331.41 ± 2122.56 | 91.04 |
| 25 | 9254.07 ± 4830.01 | 52.19 |
| 26 | 9118.41 ± 9336.23 | 102.39 |
| 27 | 3750.45 ± 3869.35 | 103.17 |
| 28 | 8098.16 ± 5450.79 | 67.31 |
| 29 | 3984.55 ± 2658.12 | 66.71 |
| 30 | 4236.70 ± 4229.74 | 99.84 |
| 31 | 3932.80 ± 2507.88 | 63.77 |
| 32 | 1681.49 ± 2019.10 | 120.08 |
| 33 | 6600.04 ± 4860.85 | 73.65 |
| 34 | 3121.51 ± 2694.58 | 86.32 |
| 35 | 8587.77 ± 5871.40 | 68.37 |
| 36 | 939.46 ± 1682.25 | 179.07 |
| 37 | 3780.67 ± 1967.05 | 52.03 |
| Protein spot No. | Intensity of spots(mean ± SD) | % CV of spot intensity |
| 1 | 10 918.80 ± 8005.09 | 73.31 |
| 2 | 8516.42 ± 7898.33 | 92.74 |
| 3 | 3986.45 ± 3471.51 | 87.08 |
| 4 | 36 146.18 ± 24 859.84 | 68.78 |
| 5 | 13 329.50 ± 7123.20 | 53.44 |
| 6 | 4091.51 ± 4636.51 | 113.32 |
| 7 | 6512.40 ± 6048.73 | 92.88 |
| 8 | 13 401.28 ± 8031.43 | 59.93 |
| 9 | 24 196.99 ± 14 907.64 | 61.61 |
| 10 | 4861.29 ± 4327.71 | 89.02 |
| 11 | 4128.52 ± 3764.18 | 91.18 |
| 12 | 3522.46 ± 2821.84 | 80.11 |
| 13 | 9624.81 ± 8295.52 | 86.19 |
| 14 | 5407.19 ± 5270.17 | 97.47 |
| 15 | 4683.89 ± 6994.94 | 149.34 |
| 16 | 2633.26 ± 2593.91 | 98.51 |
| 17 | 10 104.77 ± 10 369.91 | 102.62 |
| 18 | 16 086.82 ± 19 928.39 | 123.88 |
| 19 | 6791.99 ± 5063.21 | 74.55 |
| 20 | 7596.19 ± 4759.49 | 62.66 |
| 21 | 2685.37 ± 3298.54 | 122.84 |
| 22 | 5022.01 ± 3735.74 | 74.39 |
| 23 | 5957.62 ± 7526.42 | 124.65 |
| 24 | 2323.67 ± 2269.62 | 97.67 |
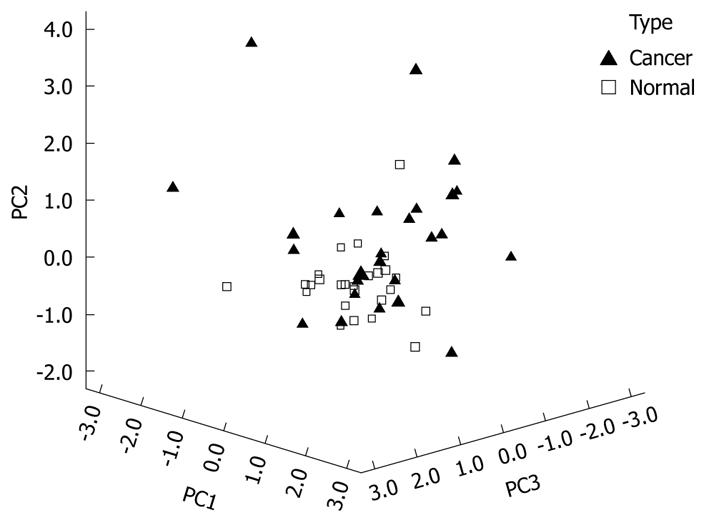
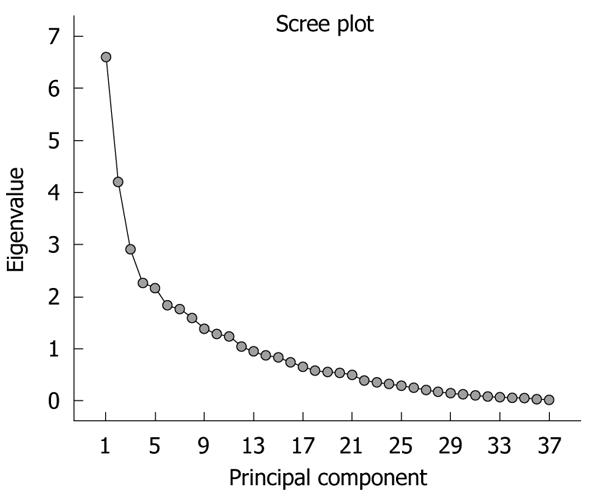
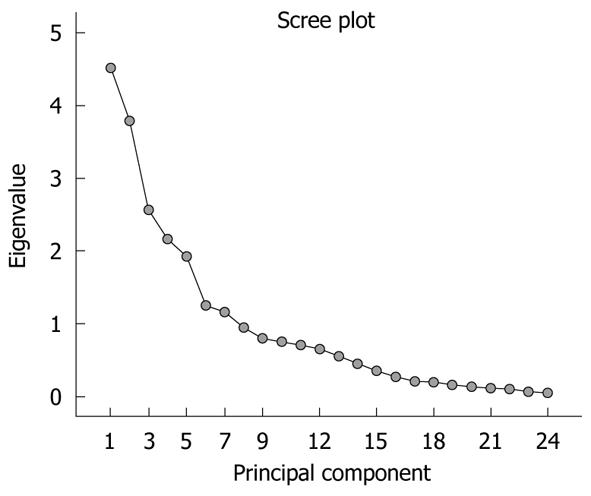
The significance of the expression levels of the differentially expressed proteins in both Tris and TLB extracts was analyzed by Student’s t test. After univariate analysis was performed, the normalized intensities of 37 differentially expressed protein spots in Tris extracts were subjected to PCA. The PCA reduced the original data to 12 PCs based on an eigenvalue of > 1, and these 12 PCs contributed 76.43% of the total data variance of the Tris extract data. Figure 2 shows the 3D PC plot with the x- y- and z-axes representing the first, second and third PC number. The variables that had the highest loadings were those that contributed most to the differentiation of the disease state. Figure 3 shows the scree plot of Tris extracts. Six PCs were chosen and these components contributed 53.97% of the total variance of the Tris extract data. Table 6 shows the LDA results for Tris extract proteins, where 22 out of 26 original normal tissues, and 21 out of 26 original cancer tissues were correctly classified. In cross-validated samples, 22 out of 26 normal tissues and 21 out of 26 cancer tissues were correctly classified. Both original and cross-validation samples had an average 82.7% correct classification.
| Type | Predicted group membership | % correct classification | |
| Cancer | Normal | ||
| Original count | |||
| Cancer (26) | 21 | 5 | 82.7 |
| Normal (26) | 4 | 22 | |
| Cross-validated count | |||
| Cancer (26) | 21 | 5 | 82.7 |
| Normal (26) | 4 | 22 | |
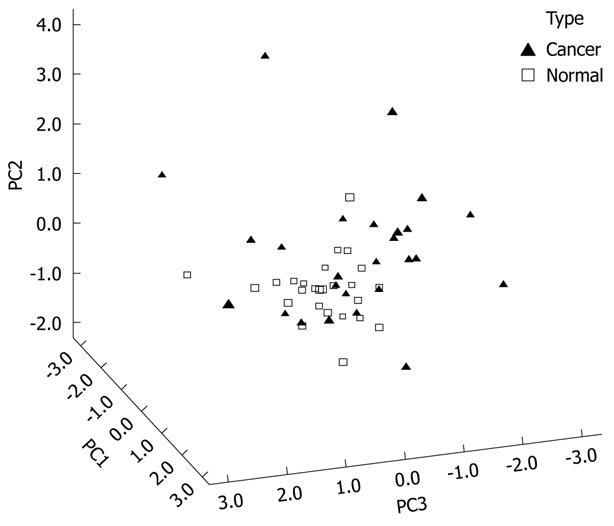
Figure 4 shows the 3D view of the PCs plot for the TLB extract. PCA reduced the original data of the TLB extract to seven PCs based on an eigenvalue one of > 1, and the seven PCs accounted for 72.46% of the total data variance. The 3D view indicates that tissues can be grouped according to CRC disease state. Figure 5 shows the scree plot of the TLB extracts. Six PCs were chosen based on the slope of scree plot, which contributed 67.61% of the total data variance of TLB extracts. Table 7 shows the LDA results of TLB extracts, where 22 out of 26 original normal tissues, and 19 out of 26 original cancerous tissues were correctly classified. In cross-validated samples, 21 out of 26 normal tissues and 16 out of 26 cancerous tissues were correctly classified. The average percentages of correct classification for original and cross-validation samples were 78.8% and 71.2%, respectively.
| Type | Predicted group membership | % correct classification | |
| Cancer | Normal | ||
| Original count | |||
| Cancer (26) | 19 | 7 | 78.8 |
| Normal (26) | 4 | 22 | |
| Cross-validated count | |||
| Cancer (26) | 16 | 10 | 71.2 |
| Normal (26) | 5 | 21 | |
The expression levels of the differentially expressed protein between colorectal cancerous and normal tissues were analyzed using PCA based on a multivariate analysis approach, to assess their usefulness in classifying colorectal tissues as cancerous or normal. The differentially expressed proteins identified showed good consistency in their expression levels in cancerous and normal tissues. The proteins were extracted in two fractions according to their polarities. In the PCA-LDA model, the selected proteins from the first few PCs were able to discriminate colorectal tissues with and without CRC.
A scree plot was derived by plotting the eigenvalues against the PC number. The shape of the plot was used to evaluate the number of PCs to be retained. In general, the point at which the scree plot straightens out indicates the number of PCs to be extracted[11]. Cross-validation is a method to estimate the accuracy of a predicted classification model if performed using new future data sets (samples); this is because a classification model is considered incomplete until the prediction error is estimated[12]. One method of cross validation is leave-one-out cross-validation, where one sample from the data set of N samples is removed, the discriminant rule is recalibrated, and a classification model is built based on the remaining N - 1 data. The one sample that is left out is classified in this model and the process repeated N times[12].
PCA and LDA results from Tris extract indicated that six out of 37 proteins were reliable to determine the tissues with CRC. The proteins comprised five upregulated proteins, namely GST-P, tropomyosin α-3C-like protein, F-actin capping protein subunit β, selenium binding protein 1 and DJ-1 protein, and one downregulated protein, namely, proteasome subunit β type 6. DJ-1 protein and GST-P contributed the most to the first PC based on the weight of their loadings. This was followed by the tropomyosin α-3C-like protein and proteasome subunit β type 6 that contributed to the second PC, while F-actin capping protein subunit β and selenium binding protein 1 contributed to the third PC. The initial PCA reduced the original data and therefore enabled LDA to be carried out because LDA is sensitive to the number of variables. In LDA, the six PCs chosen were shown to be capable of predicting whether the tissues were with or without CRC. Two-way validation by using original and cross-validation analyses was applied to validate the state of the tissues, where the cancerous and normal tissues were classified correctly at 82.7% for both original and cross-validation samples.
Two proteins that contributed most to PC1 in Tris extract were DJ-1 and GST. DJ-1 is a putative oncoprotein that is able to transform cells with H-Ras[13]. Overexpression of DJ-1 activates protein kinase B, which subsequently increases cell survival. Furthermore, increased DJ-1 expression also activates Nrf2 (nuclear factor erythroid 2-related factor), which in turn increases expression of antioxidant enzymes that confer a survival advantage to tumor cells[14]. Upregulation of DJ-1 protein in esophageal squamous cell carcinoma is correlated with lymph node metastasis[15]. Although there is no reported role of DJ-1 in CRC, its upregulation in CRC is undeniable, and we have shown that its expression can be used to discriminate between CRC cancerous and normal tissues.
GST catalyzes the conjugation of reduced glutathione to electrophiles[16]. GST functions to remove peroxides from endogenous compounds such as lipids and DNA[17]. Overexpression of GST-P1 in CRC may be involved in cell proliferation, differentiation and apoptosis[18]. GST-P1 is overexpressed in liver cancer cells[19].
In TLB extract, six of the 24 differentially expressed proteins identified were found to be useful in discriminating CRC cancerous from normal tissues. These proteins were protein disulfide isomerase (PDI), complement component 1 Q subcomponent-binding protein (GC1q-R), chloride intracellular channel protein 1, triosephosphate isomerase, annexin A5 and actin cytoplasmic 2. All the proteins were downregulated in TLB extracts, except chloride intracellular channel protein 1 and triosephosphate isomerase. PDI and GC1q-R contributed the most to the first PC based on the weight of their loadings. This was followed by the chloride intracellular channel protein 1 and triosephosphate isomerase that contributed the most to the second PC, while annexin A5 and actin cytoplasmic 2 contributed most to the third PC. In LDA, the six PCs that explained 67.61% of the total variance were able to distinguish CRC cancerous from normal tissues. The leave-one-out cross-validation obtained 71.2% correct classification of normal and cancerous tissues. The value for original grouped samples was higher with 78.8% correct classification.
Two proteins that contributed most to PC1 in TLB extracts were PDI and GC1q-R. PDI catalyzes the formation and breakage of disulfide bonds between two cysteine residues[20]. PDI regulates cell transformation and intracellular and extracellular redox activities via its reductase activity[21]. PDI regulates STAT3 signaling and proliferation, which is thought to induce malignancy[22]. PDI is upregulated in CRC cell lines and its upregulation is correlated with cancer cell differentiation[23,24].
GC1q-R is a cell surface glycoprotein, which binds to the globular heads of C1q molecules[25]. C1q molecules bind to a variety of cells such as B cells, monocytes, macrophages, endothelial and smooth muscle cells[26]. C1q elicits responses such as phagocytosis in monocytes and activation of tumor cytotoxicity of macrophages[27,28]. GC1q-R is overexpressed in colon cancer cells and may be involved in tumor metastasis. However, PDI and GC1q-R were downregulated when using average fold change to determine their expression levels.
Proteins are the expression components that regulate cell activity. Differential expression of proteins is expected upon transformation of normal cells to cancerous cells. These differentially expressed proteins are useful in diagnosis and prognosis of the disease. In the present study, the specimens used in the analysis comprised tissues from female and male patients who were diagnosed with various stages, grades and locations of CRC. Regardless of the sex of the patients and pathological specification of the tissues, we showed that the differentially expressed protein identified from 2D protein profiles of cancerous and normal tissues could be used to separate and classify normal and cancerous tissues by combining PCA and LDA. The data reduction technique of PCA was sufficient to provide a classification of tissues according to CRC disease state. These statistical models simplify the data management through the reduced dimensionality of protein spots from the 2D gel images. Therefore, multivariate analysis of differentially expressed proteins identified from cancerous and normal tissues may be used as a tool for diagnosis and prognosis of CRC disease state.
Colorectal cancer (CRC) is one of the leading causes of death worldwide. Differentially expressed proteins between cancerous and normal colonic tissues were identified using 2D gel separation followed by LC/MS/MS analysis. The protein spot intensities of the 2D gel images were analyzed using principal component analysis (PCA) and linear discriminant analysis (LDA) for their possible use in classification of disease state.
Multivariate analyses, including the dimension reduction method known as PCA and classification methods such as LDA, are used in cancer proteomic studies to identify the protein variables that provide the best discrimination between the cancerous and normal tissues.
The authors used sequential protein extraction to extract aqueous soluble and membrane-associated proteins from colorectal tissues. Differentially expressed proteins were analyzed using a combination of PCA and LDA to determine their usability in differentiating normal and cancerous colonic tissues. Using this method, the authors successfully classified the tissues according to their respective types. DJ-1 protein and glutathione S transferase P1 of the aqueous soluble proteins, protein disulfide isomerase and complement component 1 Q subcomponent-binding protein of the membrane-associated proteins gave the best classification of the tissues.
The identified biomarkers may be used for the diagnosis and prognosis of CRC.
Chemometrics is defined as the information aspects of complex biological and chemical systems. Chemometrics utilize mathematical, statistical or formal logic-based methods to extract chemical information, which in this case, is for biomarker discovery.
This study investigated the use of PCA and LDA of differential protein expression between normal and cancerous tissues for classification of disease state. The method gave good classification of cancerous and normal colonic tissues.
| 1. | Cowan ML, Vera J. Proteomics: advances in biomarker discovery. Expert Rev Proteomics. 2008;5:21-23. |
| 2. | Rodríguez-Piñeiro AM, Rodríguez-Berrocal FJ, Páez de la Cadena M. Improvements in the search for potential biomarkers by proteomics: application of principal component and discriminant analyses for two-dimensional maps evaluation. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849:251-260. |
| 3. | Hilario M, Kalousis A. Approaches to dimensionality reduction in proteomic biomarker studies. Brief Bioinform. 2008;9:102-118. |
| 4. | Karson MJ. Multivariate statistical methods: An introduction. Iowa: Iowa State University Press 1982; 159, 191. |
| 5. | Giri NC. Multivariate statistical analysis. New York: Marcel Dekker 1996; 293-294. |
| 6. | Djidja MC, Claude E, Snel MF, Francese S, Scriven P, Carolan V, Clench MR. Novel molecular tumour classification using MALDI-mass spectrometry imaging of tissue micro-array. Anal Bioanal Chem. 2010;397:587-601. |
| 7. | Kamath SD, Mahato KK. Principal component analysis (PCA)-based k-nearest neighbor (k-NN) analysis of colonic mucosal tissue fluorescence spectra. Photomed Laser Surg. 2009;27:659-668. |
| 8. | Zwielly A, Mordechai S, Sinielnikov I, Salman A, Bogomolny E, Argov S. Advanced statistical techniques applied to comprehensive FTIR spectra on human colonic tissues. Med Phys. 2010;37:1047-1055. |
| 9. | Ragazzi E, Pucciarelli S, Seraglia R, Molin L, Agostini M, Lise M, Traldi P, Nitti D. Multivariate analysis approach to the plasma protein profile of patients with advanced colorectal cancer. J Mass Spectrom. 2006;41:1546-1553. |
| 10. | Yeoh LC, Loh CK, Gooi BH, Singh M, Gam LH. Hydrophobic protein in colorectal cancer in relation to tumor stages and grades. World J Gastroenterol. 2010;16:2754-2763. |
| 11. | McGarigal K, Cushman S, Stafford S. Ordination: Principal component analysis. Multivariate statistics for wildlife and ecology research. New York: Springer-Verlag 2000; 41-42. |
| 12. | Dziuda DM. Biomarker discovery and classification. Data mining for genomics and proteomics: Analysis of gene and protein expression data. Hoboken: John Wiley and Sons 2010; 110-112. |
| 13. | Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, Iguchi-Ariga SM, Ariga H. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509-513. |
| 14. | Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci USA. 2006;103:15091-15096. |
| 15. | Yuen HF, Chan YP, Law S, Srivastava G, El-Tanani M, Mak TW, Chan KW. DJ-1 could predict worse prognosis in esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:3593-3602. |
| 16. | Mannervik B, Danielson UH. Glutathione transferases--structure and catalytic activity. CRC Crit Rev Biochem. 1988;23:283-337. |
| 17. | Park HJ, Lee KS, Choo SH, Kong KH. Functional studies of cysteine residues in human glutathione S-transferase P1-1 by site-directed mutagenesis. Bull Korean Chem Soc. 2001;22:77-83. |
| 18. | Lo HW, Antoun GR, Ali-Osman F. The human glutathione S-transferase P1 protein is phosphorylated and its metabolic function enhanced by the Ser/Thr protein kinases, cAMP-dependent protein kinase and protein kinase C, in glioblastoma cells. Cancer Res. 2004;64:9131-9138. |
| 19. | Tsuchida S, Sato K. Glutathione transferases and cancer. Crit Rev Biochem Mol Biol. 1992;27:337-384. |
| 20. | Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35-44. |
| 21. | Hirano N, Shibasaki F, Sakai R, Tanaka T, Nishida J, Yazaki Y, Takenawa T, Hirai H. Molecular cloning of the human glucose-regulated protein ERp57/GRP58, a thiol-dependent reductase. Identification of its secretory form and inducible expression by the oncogenic transformation. Eur J Biochem. 1995;234:336-342. |
| 22. | Coe H, Jung J, Groenendyk J, Prins D, Michalak M. ERp57 modulates STAT3 signaling from the lumen of the endoplasmic reticulum. J Biol Chem. 2010;285:6725-6738. |
| 23. | Katayama M, Nakano H, Ishiuchi A, Wu W, Oshima R, Sakurai J, Nishikawa H, Yamaguchi S, Otsubo T. Protein pattern difference in the colon cancer cell lines examined by two-dimensional differential in-gel electrophoresis and mass spectrometry. Surg Today. 2006;36:1085-1093. |
| 24. | Stierum R, Gaspari M, Dommels Y, Ouatas T, Pluk H, Jespersen S, Vogels J, Verhoeckx K, Groten J, van Ommen B. Proteome analysis reveals novel proteins associated with proliferation and differentiation of the colorectal cancer cell line Caco-2. Biochim Biophys Acta. 2003;1650:73-91. |
| 25. | Ghebrehiwet B, Lim BL, Peerschke EI, Willis AC, Reid KB. Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular "heads" of C1q. J Exp Med. 1994;179:1809-1821. |
| 26. | Ghebrehiwet B. Functions associated with the C1q receptor. Behring Inst Mitt. 1989;204-215. |
| 27. | Bobak DA, Frank MM, Tenner AJ. C1q acts synergistically with phorbol dibutyrate to activate CR1-mediated phagocytosis by human mononuclear phagocytes. Eur J Immunol. 1988;18:2001-2007. |
Peer reviewer: Ki-Baik Hahm, MD, PhD, Professor, Gachon Graduate School of Medicine, Department of Gastroenterology, Lee Gil Ya Cancer and Diabetes Institute, Lab of Translational Medicine, 7-45 Songdo-dong, Yeonsu-gu, Incheon, 406-840, South Korea
S- Editor Sun H L- Editor Kerr C E- Editor Zheng XM