©The Author(s) 2025.
World J Gastroenterol. Nov 14, 2025; 31(42): 112478
Published online Nov 14, 2025. doi: 10.3748/wjg.v31.i42.112478
Published online Nov 14, 2025. doi: 10.3748/wjg.v31.i42.112478
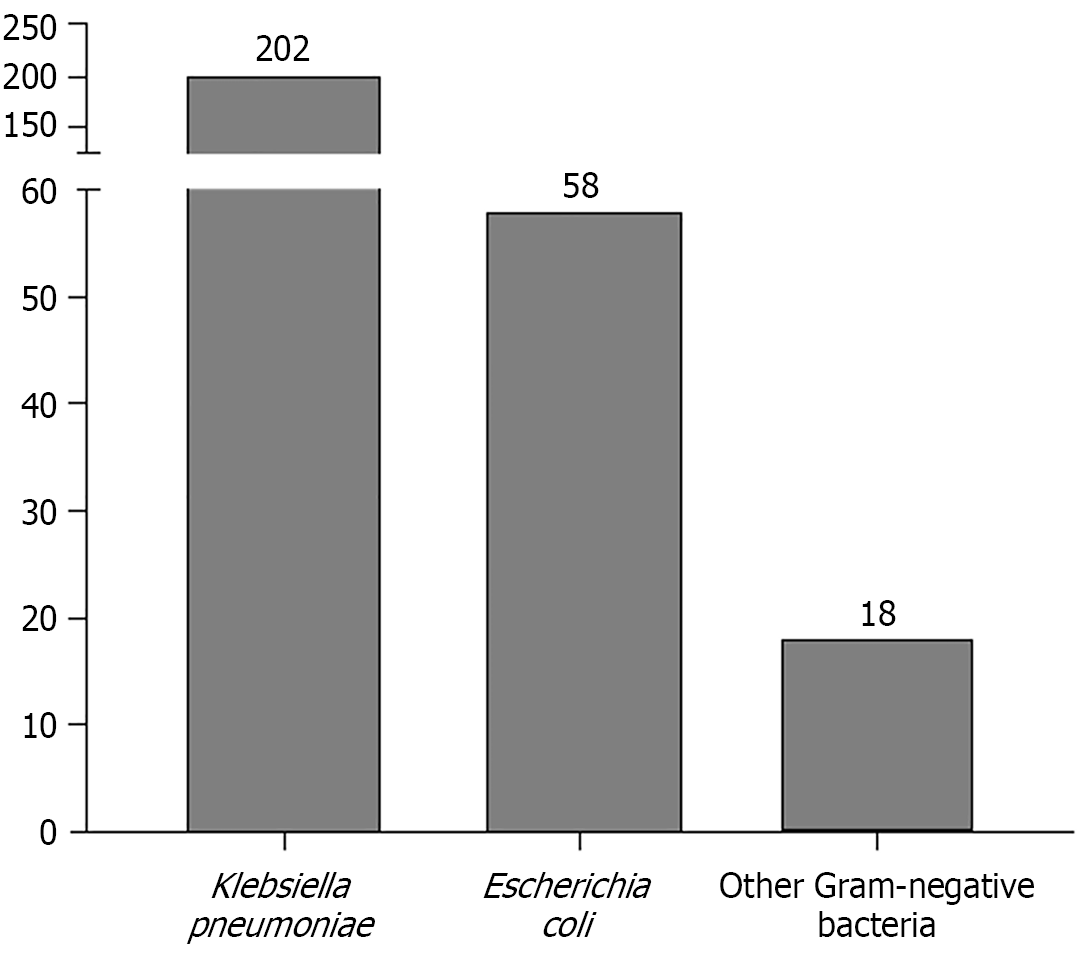
Figure 1 Pathogen distribution of Gram-negative bacteria in bacterial liver abscesses.
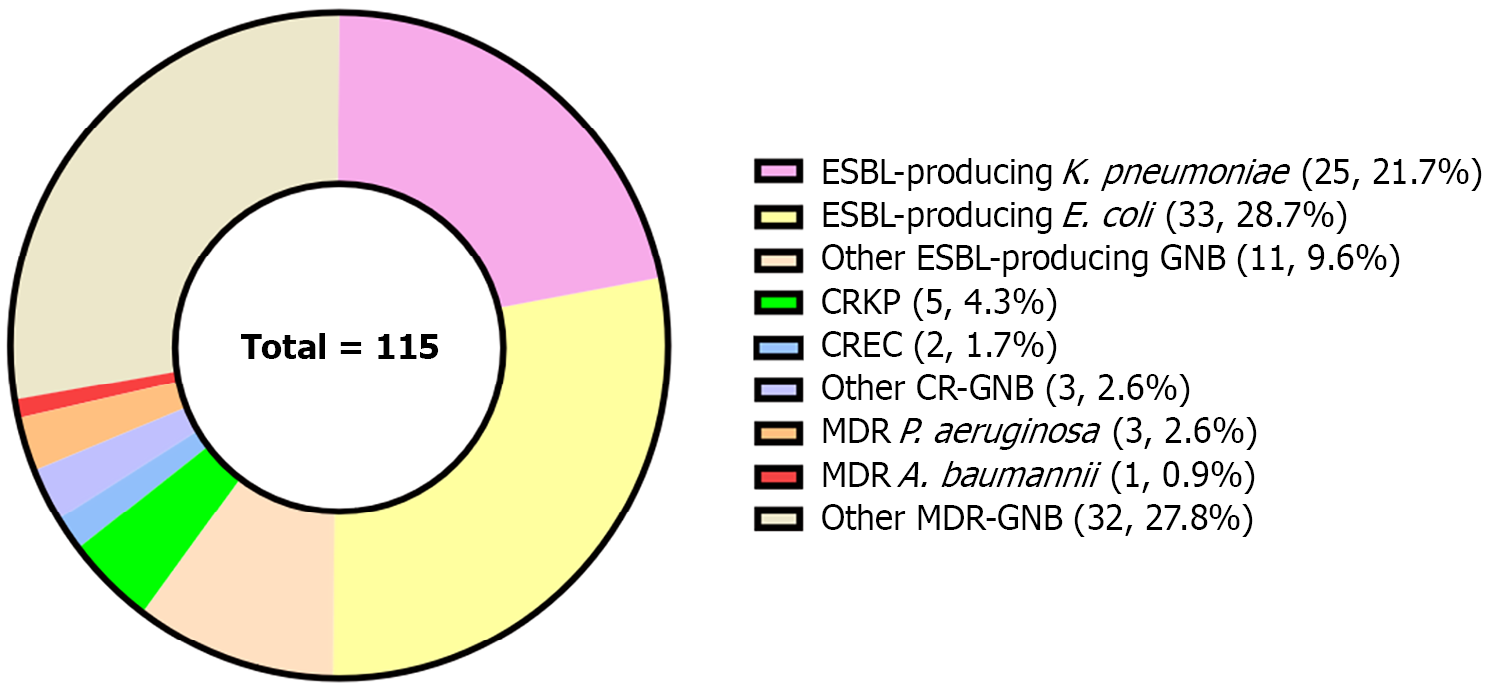
Figure 2 Distribution of multidrug-resistant Gram-negative bacteria in bacterial liver abscesses.
ESBL: Extended-spectrum β-lactamase; K. pneumoniae: Klebsiella pneumoniae; E. coli: Escherichia coli; GNB: Gram-negative bacteria; CRKP: Carbapenem-resistant Klebsiella pneumoniae; CREC: Carbapenem-resistant Enterobacter cloacae; CR-GNB: Carbapenem-resistant Gram-negative bacteria; P. aeruginosa: Pseudomonas aeruginosa; A. baumannii: Acinetobacter baumannii; MDR: Multidrug-resistant.
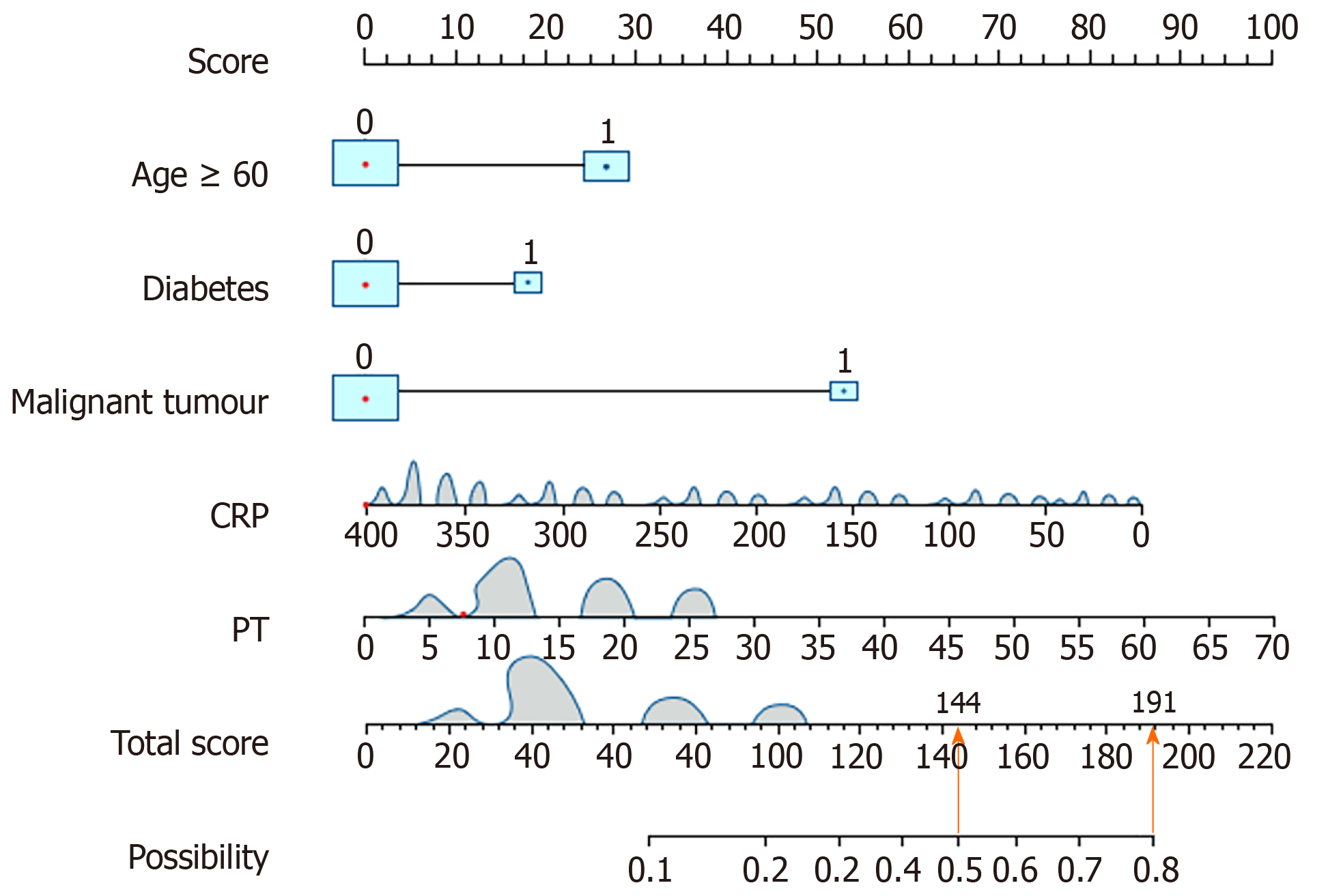
Figure 3 Personalized nomogram model for predicting the risk of multidrug-resistant-Gram-negative bacteria induced bacterial liver abscesses.
CRP: C-reactive protein; PT: Prothrombin time.
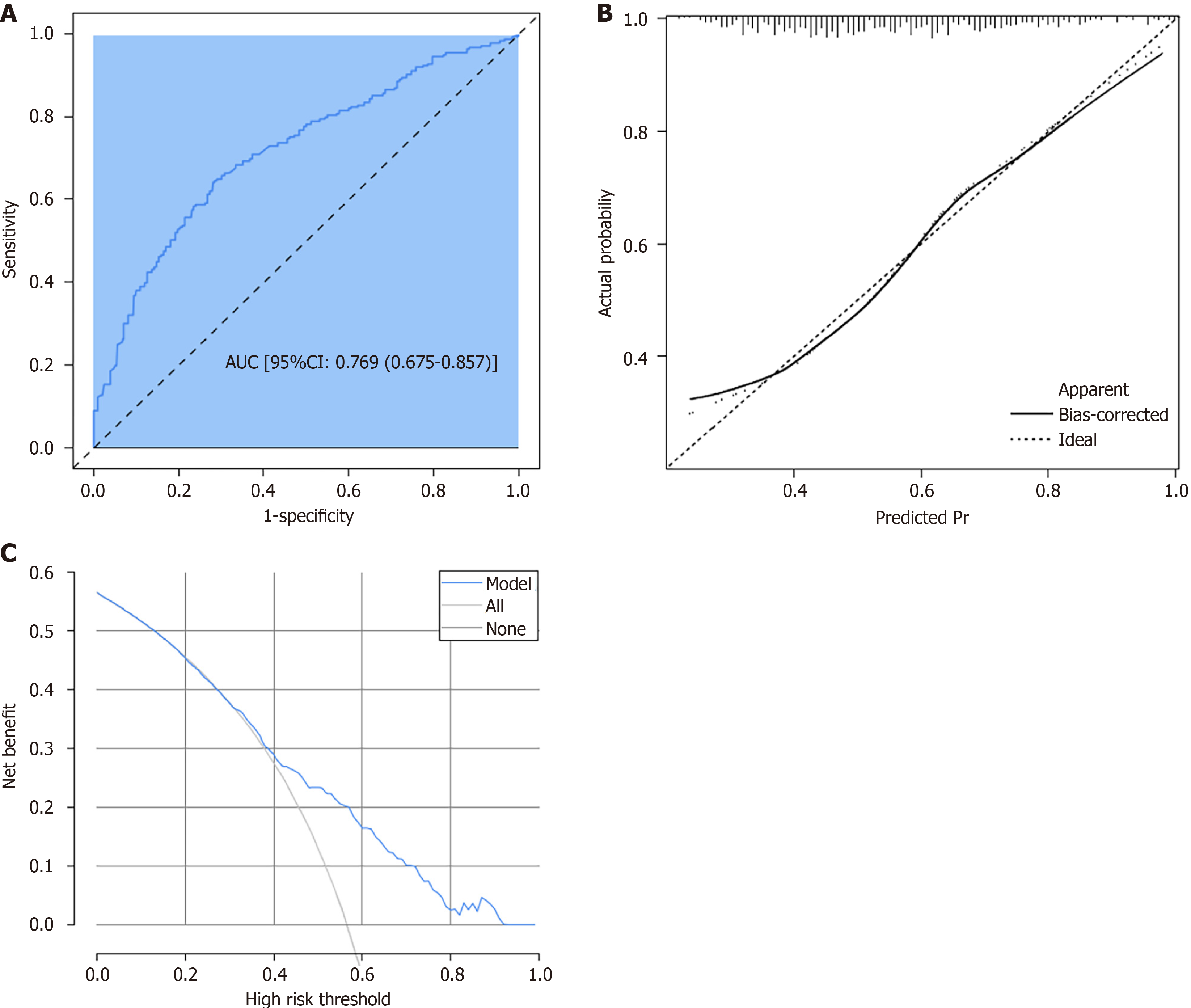
Figure 4 Curves for predicting the risk of multidrug-resistant-Gram-negative bacteria induced bacterial liver abscesses.
A: Receiver operating characteristic curve; B: Calibration curve; C: Decision curve analysis. AUC: Area under the curve; CI: Confidence interval; Pr: Probability.
- Citation: Xu K, Wu DH, Zeng CJ, Guo JY, Xi DY, Wang MJ, Yao ZY, Feng AQ, Ji F, Yan XB, Ye LL, Li CY. Clinical predictors of multidrug-resistant Gram-negative pyogenic liver abscess and nomogram construction: A retrospective analysis. World J Gastroenterol 2025; 31(42): 112478
- URL: https://www.wjgnet.com/1007-9327/full/v31/i42/112478.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i42.112478