©The Author(s) 2025.
World J Gastroenterol. Oct 28, 2025; 31(40): 111307
Published online Oct 28, 2025. doi: 10.3748/wjg.v31.i40.111307
Published online Oct 28, 2025. doi: 10.3748/wjg.v31.i40.111307
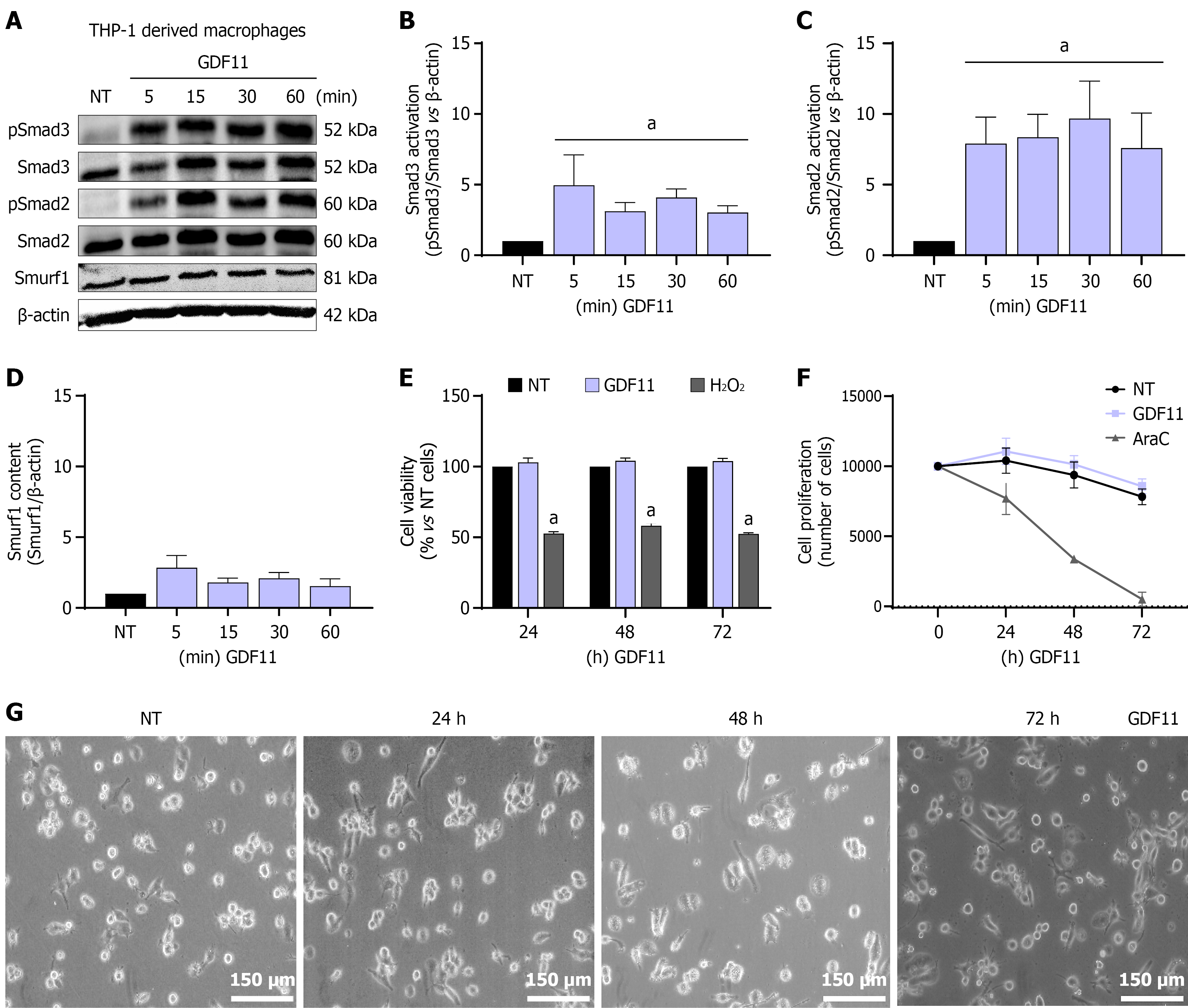
Figure 1 Growth differentiation factor 11 induces Smad signaling pathway activation in THP-1-derived M0 macrophages.
A: Representative images of western blots; B-D: Densitometry analysis (β-actin was used as a loading control); E: Cell viability (H2O2 was used as a negative control); F: Cell proliferation (cytarabine was used as a negative control); G: Cellular morphology, each image is representative of at least three independent experiments. Scale bars: 150 μm (200 ×, original magnification). Each column or point represents the mean ± SEM of at least three independent experiments carried out in triplicate. aP ≤ 0.05 vs not treated cells. GDF11: Growth differentiation factor 11; NT: Not treated cells; AraC: Cytarabine.
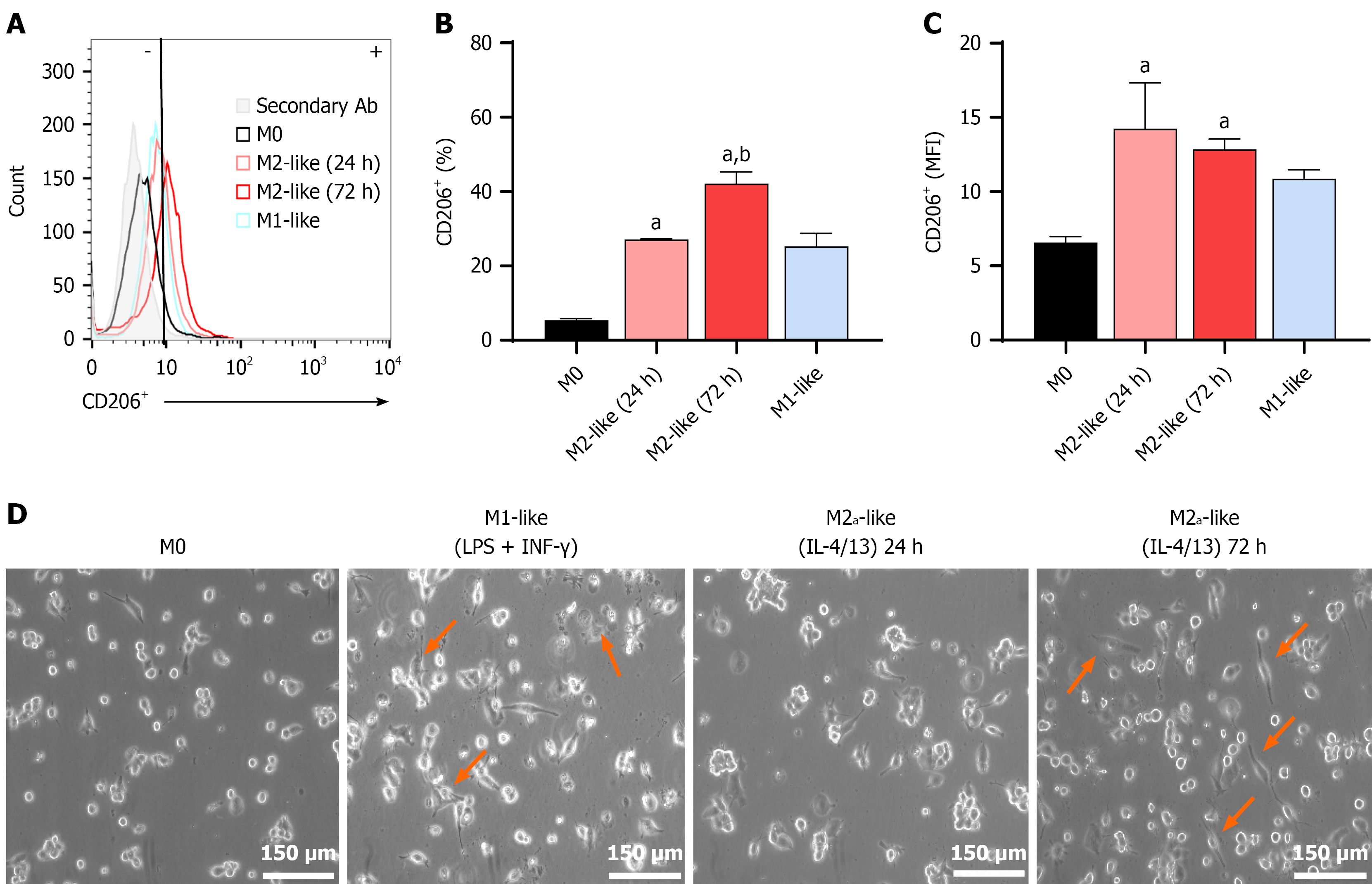
Figure 2 The anti-inflammatory stimulus acquires M2-like macrophage polarization.
A: Fluorescence histogram of the expression of the cluster of differentiation 206 (CD206) marker obtained by flow cytometry; B: Percentage of CD206+ cells in IL-4/IL-13-treated macrophages (M2-like); C: Represents the mean fluorescent intensity of the CD206+ subset; D: Macrophage morphology under the different treatments, orange arrows show specific morphology. Images are representative of at least three independent experiments. Scale bars: 150 μm (200 ×, original magnification). Each column represents the mean ± SEM of at least three independent experiments. aP ≤ 0.05 vs M0; bP ≤ 0.05 vs M2-like (24 hours). CD206: Cluster of differentiation 206; MFI: Mean fluorescent intensity; LPS: Lipopolysaccharide; IFN: Interferon; IL: Interleukin.
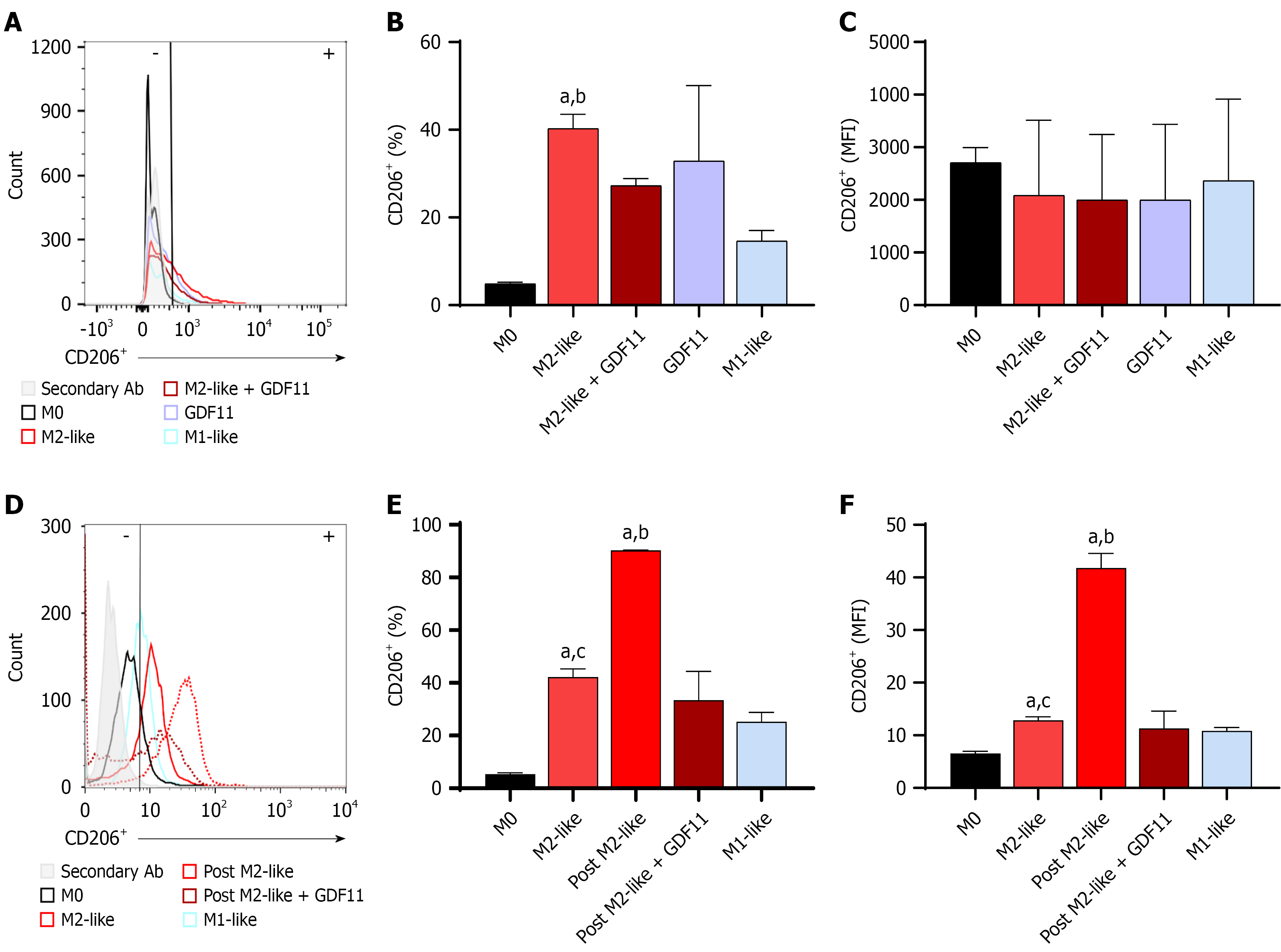
Figure 3 Growth differentiation factor 11 decreases cluster of differentiation 206 in M2-like macrophages.
FCM analysis was performed to evaluate cluster of differentiation 206 (CD206) expression. Cells were treated with growth differentiation factor 11 for 72 hours, as stated in the materials and methods section. A: Fluorescence histogram; B: Percentage of CD206+ cells; C: Represents the median fluorescence intensity of the subset. Cells were treated with growth differentiation factor 11 for 144 hours, as stated in the materials and methods section; D: Fluorescence histogram; E: Percentage of CD206+ events; F: Mean fluorescent intensity in subset. Each column represents the mean ± SEM of at least three independent experiments. aP ≤ 0.01 vs M0; bP ≤ 0.05 vs post M2-like + growth differentiation factor 11; cP ≤ 0.001 vs post M2-like. CD206: Cluster of differentiation 206; GDF11: Growth differentiation factor 11; MFI: Mean fluorescent intensity.
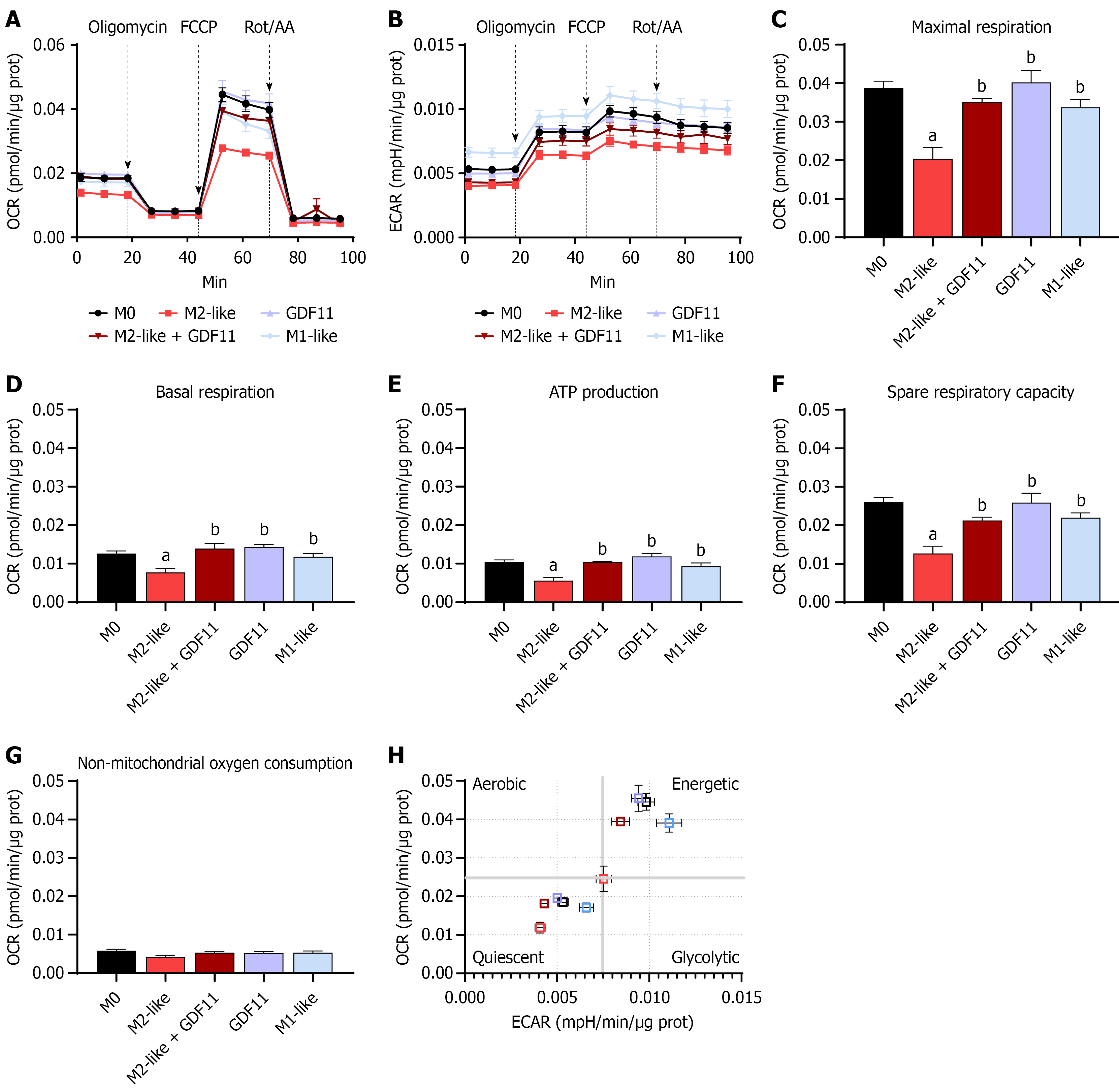
Figure 4 Growth differentiation factor 11 promotes bioenergetics rewiring in M2-like cells.
Mitochondrial stress test was performed to determine. A: Oxygen consumption rate; B: Extracellular acidification rates using Seahorse XF24e flux analyzer; C: Maximal respiration; D: Basal respiration; E: Adenosine triphosphate production; F: Spare respiratory capacity; G: Non-mitochondrial oxygen consumption; H: Defective metabolic profile based on relative values of oxygen consumption rate and extracellular acidification rate before and after metabolic inducing stress. Each column represents the mean ± SEM of at least three independent experiments. aP ≤ 0.05 vs M0; bP ≤ 0.05 vs M2-like. OCR: Oxygen consumption rate; ECAR: Extracellular acidification rate; GDF11: Growth differentiation factor 11; ATP: Adenosine triphosphate.
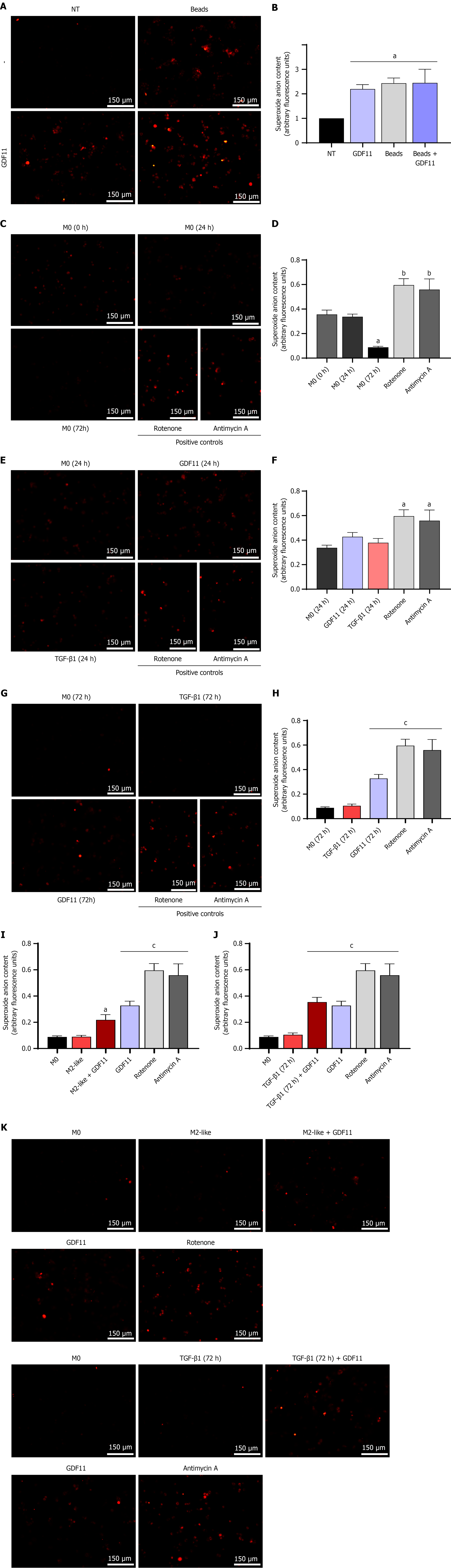
Figure 5 Growth differentiation factor 11 increases reactive oxygen species production in macrophages.
Superoxide anion detection by dihydroethidium (DHE)-derived fluorescence. A: Representative images of DHE-derived fluorescence in the phagocytosis beads assay. Scale bars: 150 μm (200 ×, original magnification); B: Representative densitometry analysis of phagocytosis beads assay; C: Representative images of DHE-derived fluorescence in M0 macrophages. Scale bars: 150 μm (200 ×, original magnification); D: Representative densitometry analysis in M0 macrophages; E: Representative images of DHE-derived fluorescence at 24 hours. Scale bars: 150 µm (200×, original magnification); F: Representative densitometry analysis at 24 hours; G: Representative images of DHE-derived fluorescence at 72 hours. Scale bars: 150 μm (200 ×, original magnification); H-J: Represent the densitometry analysis at 72 hours; K: Representative images of DHE-derived fluorescence at 72 hours. Scale bars: 150 μm (200 ×, original magnification). Rotenone (2 μmol/L) and antimycin A (5 μmol/L) were positive controls. Each column represents the mean ± SEM of at least three independent experiments in triplicate. aP ≤ 0.05 vs not treated cells or M0; bP ≤ 0.05 vs M0; cP ≤ 0.01 vs M0. NT: Not treated cells; GDF11: Growth differentiation factor 11; TGF: Transforming growth factor.
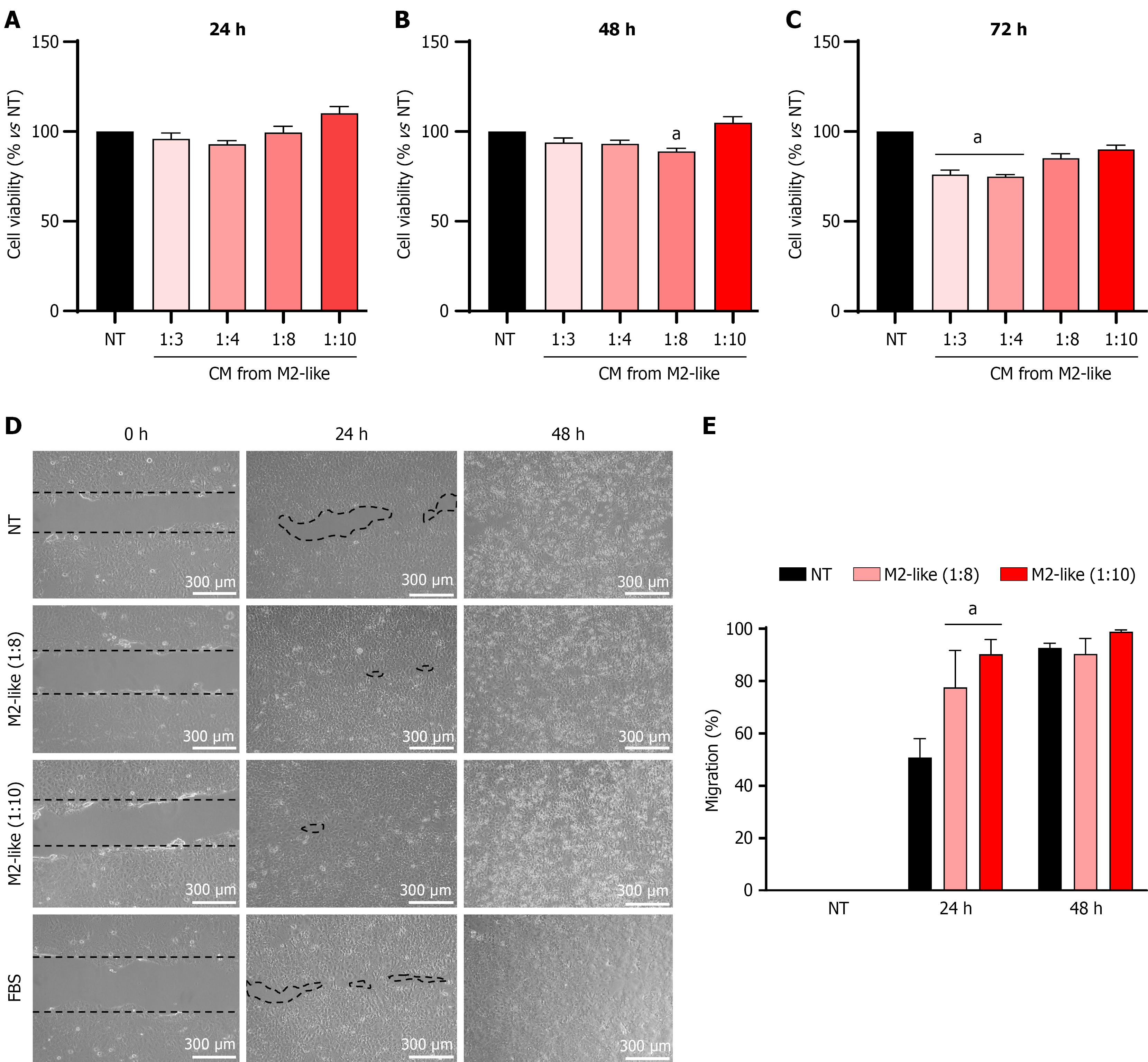
Figure 6 Effects of the conditioned media from M2-like derived macrophages on the viability and migration of Huh7 cells.
A-C: Cell viability was assessed using the Cell Counting Kit-8 kit (24 hours, 48 hours, and 72 hours, respectively), conditioned media from macrophages at different dilutions were used; D: Cell migration assay determined by a wound healing assay, representative images of at least three independent experiments. Scale bars: 300 μm (100 ×, original magnification); E: Plot of the percentage of migration determined by Image J software. Each column represents the mean ± SEM of at least three independent experiments in triplicate. aP ≤ 0.05 vs not treated cells. NT: Not treated cells; CM: Conditioned media; FBS: Fetal bovine serum.
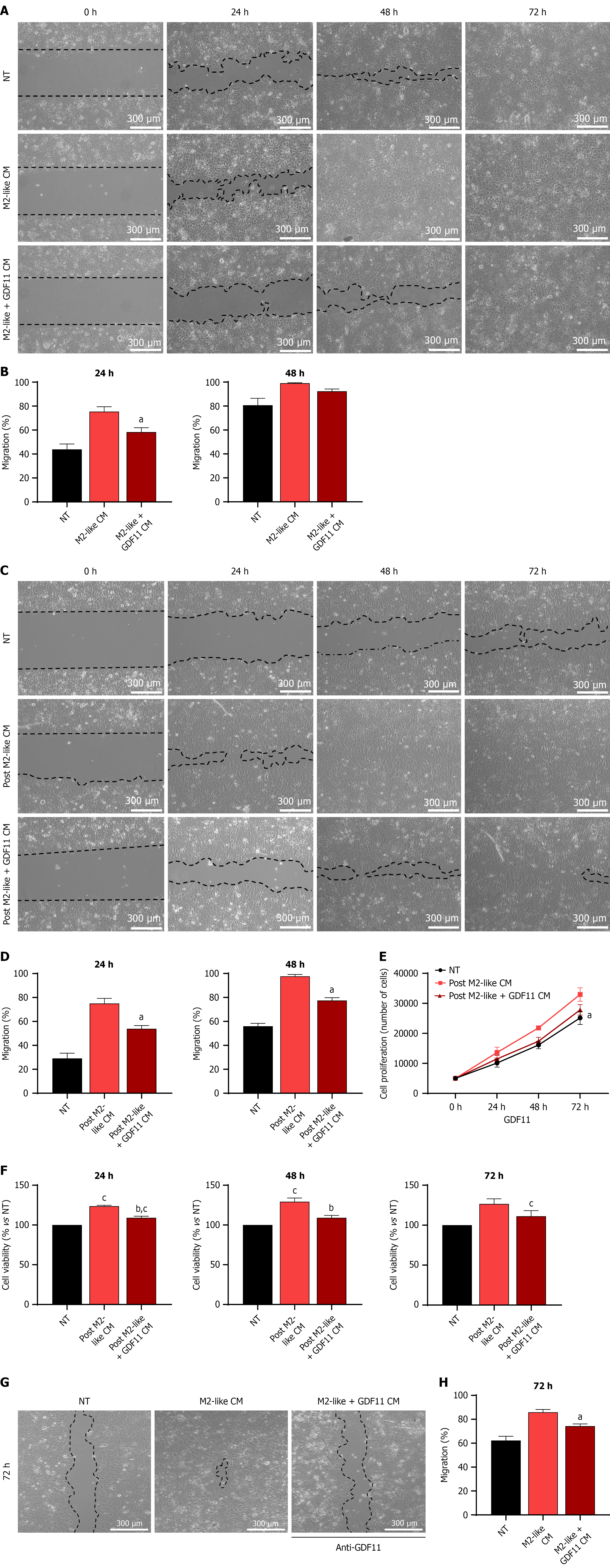
Figure 7 Conditioned media from growth differentiation factor 11-treated macrophages mitigate migration capacity and proliferation in Huh7 cells.
A: Cell migration assay determined by a wound healing assay, representative images of at least three independent experiments. Scale bars: 300 μm (100 ×, original magnification); B: Percentage of migration determined by Image J software; C: Cell migration assay determined by a wound healing assay, representative images of at least three independent experiments. Scale bars: 300 μm (100 ×, original magnification); D: Percentage of migration determined by Image J software; E: Cell proliferation; F: Cell viability at 24 hours, 48 hours, and 72 hours under different treatments; G: Cell migration assay determined by a wound healing assay using anti-GDF11. Scale bars: 300 μm (100 ×, original magnification); H: Percentage of migration determined by Image J software. Each column or point represents the mean ± SEM of at least three independent experiments carried out in triplicate. aP ≤ 0.05 vs M2-like conditioned media or post M2-like conditioned media; bP ≤ 0.05 vs post M2-like conditioned media; cP ≤ 0.05 vs not treated cells. Each column or point represents the mean ± SEM of at least three independent experiments carried out in triplicate. Anti-growth differentiation factor 11 was used to inhibit the remnants of the recombinant growth differentiation factor 11. NT: Not treated cells; CM: Conditioned media; GDF11: Growth differentiation factor 11.
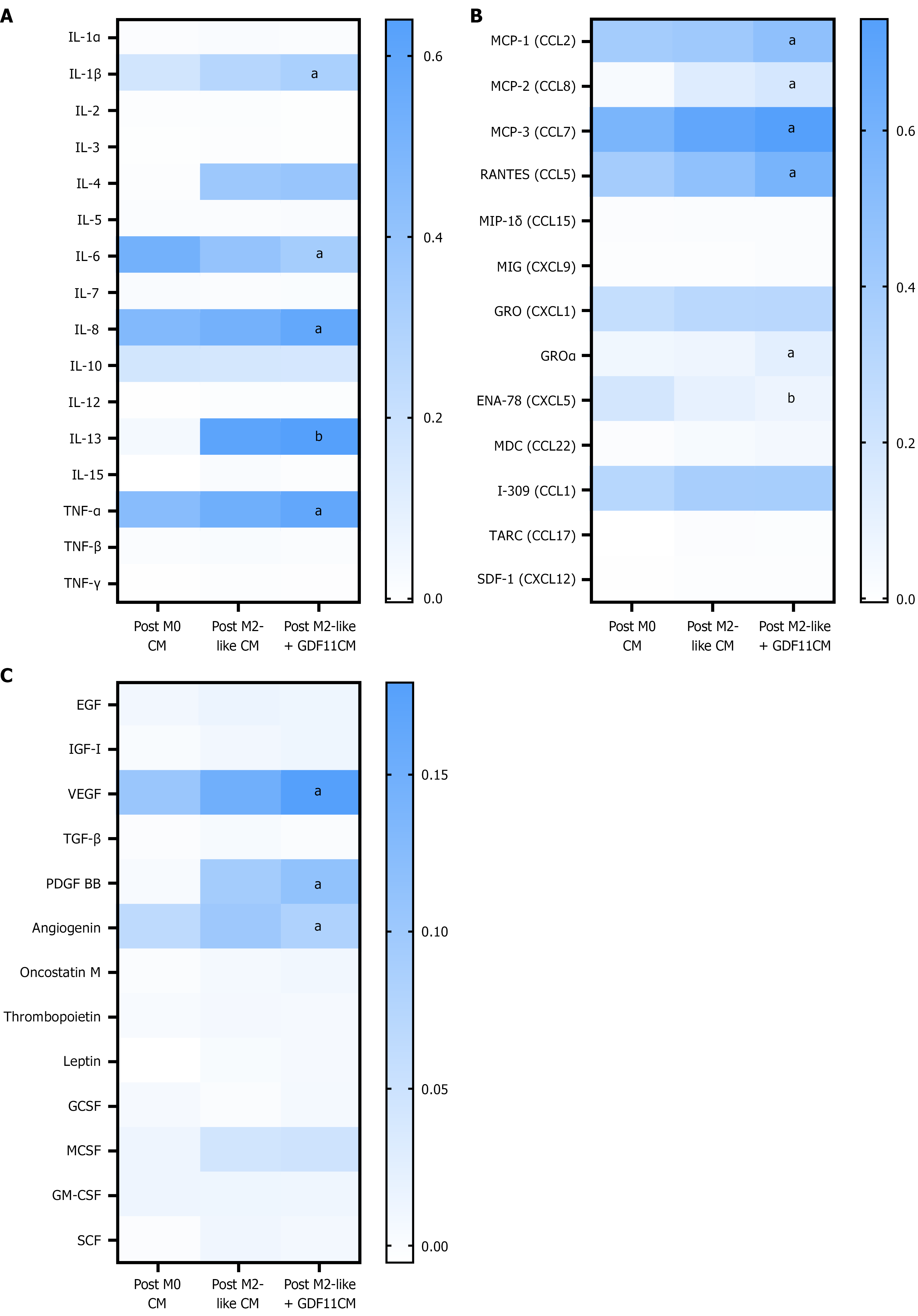
Figure 8 Cytokine profile changes induced by growth differentiation factor 11 on post M2-like macrophages.
A: Interleukin and interferon family alteration by growth differentiation factor 11 (GDF11) on post M2-like macrophages, aP < 0.0001 vs post M2-like conditioned media (CM), bP = 0.0012 vs post M2-like CM; B: Chemokine family alteration by GDF11 on post M2-like macrophages, aP < 0.0001 vs post M2-like CM, bP = 0.0050 vs post M2-like CM; C: Growth factors alteration by GDF11 on post M2-like macrophages, aP ≤ 0.0001 vs post M2-like CM. The mean pixel density in each column on the heat map represents the mean ± SEM of at least three independent experiments. CM: Conditioned media; GDF11: Growth differentiation factor 11.
- Citation: Escobedo-Calvario A, Chávez-Rodríguez L, Souza-Arroyo V, Bucio-Ortiz L, Miranda-Labra RU, Masso F, Páez-Arenas A, Hernández-Pando R, Marquardt J, Gutiérrez-Ruiz MC, Gomez-Quiroz LE. Growth differentiation factor 11 modulates metabolism, mitigating the pro-tumoral behavior provided by M2-like macrophages in hepatocellular carcinoma-derived cells. World J Gastroenterol 2025; 31(40): 111307
- URL: https://www.wjgnet.com/1007-9327/full/v31/i40/111307.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i40.111307