INTRODUCTION
Meta-analysis is a quantitative epidemiological study design that is used to systemically assess previous research studies and allows examination of variable and often heterogeneous studies to provide a consolidated review of a body of literature, which is often complex and may contain elements that are contradictory. A meta-analytical approach involves the aggregation of information to yield a summary effect estimate that often has greater statistical precision than is possible to obtain in any individual study. This will typically involve the calculation of a weighted average of the effect estimates from the individual studies, which is then used to draw conclusions and address the hypothesis being investigated[1]. Meta-analysis, along with systematic review, provides the highest quality evidence and is considered integral to evidence-based medicine[2]. Whilst a good meta-analysis relies on robust and appropriate study design, data preparation, and statistical comparison, if the data that are being included in the study are weak, then inaccurate conclusions will be drawn, and treatment effects can often be overstated[3].
Rare diseases, defined as those that affect less than 1 in 2000 individuals[4], pose a challenge when conducting meta-analysis as their low prevalence results in both a smaller number of available studies and small sample sizes within these studies. This can increase the risk of type I and type II errors and reduce the statistical power[5], making it difficult to detect true associations and effect estimates. There are also challenges posed by data sparsity, particularly in sub-groups, a reduced quality of studies that can be included in the analysis, and the introduction of bias. There are also some populations, such as transgender individuals with inflammatory bowel disease, where data are only available from observational studies, which makes it impossible to perform a traditional meta-analysis[6]. These issues can reduce the robustness of meta-analytic conclusions, resulting in a lack of high-quality evidence to guide practice in rare pathologies.
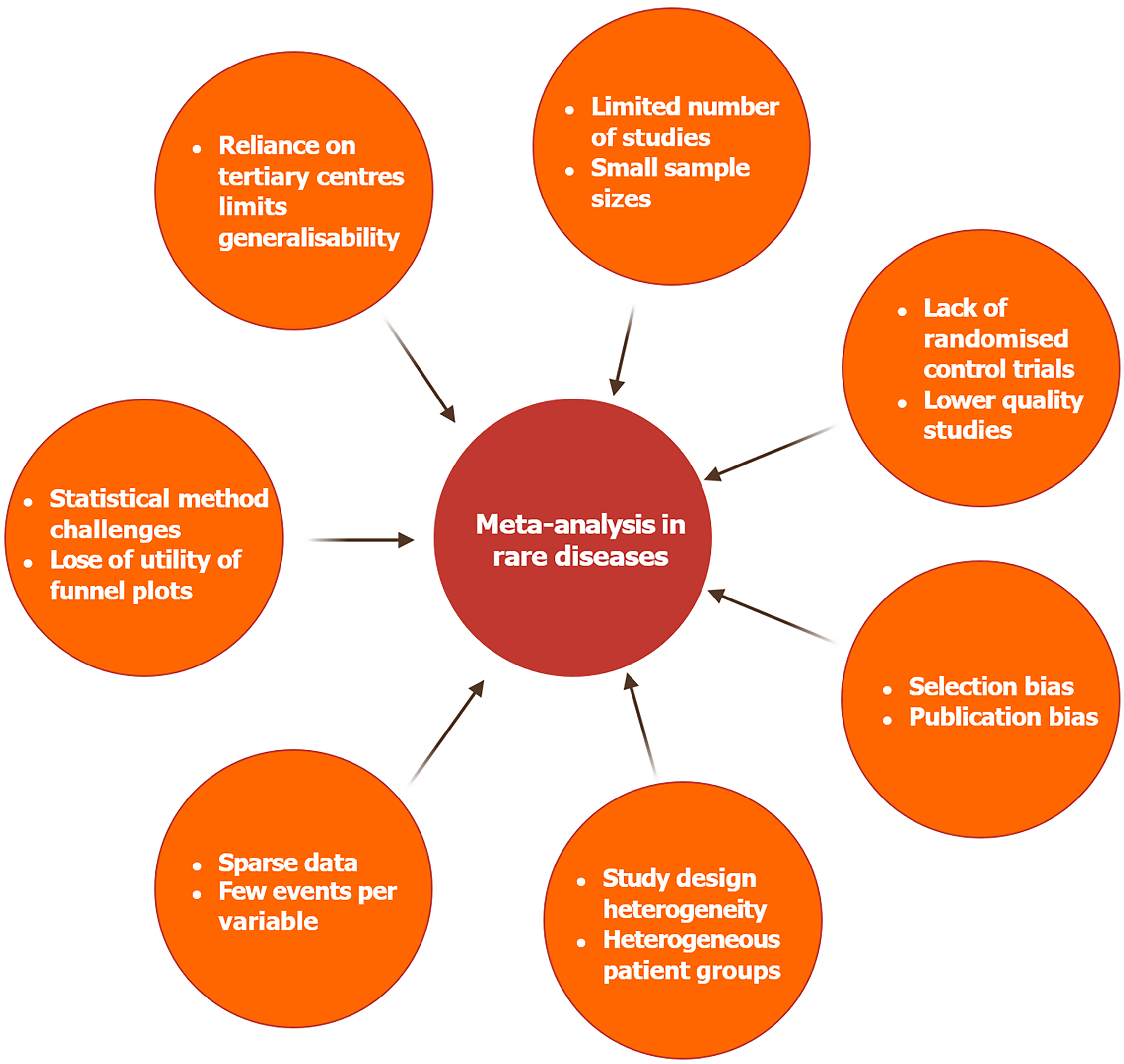
This narrative aims to review the challenges of performing meta-analyses in rare conditions (summarised in Figure 1). To identify relevant articles, a PubMed and Google Scholar search was conducted in May 2025, in keeping with EQUATOR network principles[7], using the MeSH terms “meta-analysis as topic” and “rare disease OR orphan disease” and “challenges OR limitations”. Filters for publication year and language were applied, with studies not in English or published prior to 2000 excluded. 146 studies were reviewed, and reference lists of relevant articles were also screened by the authors to identify additional sources. A review protocol was not utilised.
CHALLENGES OF META-ANALYSIS
Limited studies and small sample sizes
The low prevalence of rare diseases creates a challenge when recruiting into clinical trials[8], and data have shown that up to 25% of trials investigating rare diseases are terminated early, with low recruitment being the commonest reason[9]. This challenge in recruitment, as well as financial and logistical pressures, means that for many rare diseases, there is a distinct lack of randomised control trials (RCTs) which can inform practice. An example is neuronal ceroid lipofuscinoses, a group of degenerative lysosomal storage diseases characterised by intracellular accumulation of autofluorescent lipofuscin and which affects approximately 1 in 100000 births globally[10], but for which there have been only five completed prospective parallel group clinical trials since 1977[11]. The challenges in recruitment mean that not only is there a limited number of studies performed, but the sample sizes within these studies are often smaller. An example is Gaucher's disease, where, whilst a systematic review identified 63 studies, the average number of participants in the two included RCTs was 29.5, and similarly small numbers for other study designs[12]. Small sample sizes, combined with heterogeneity between patients, which is discussed in greater detail below, negatively impact the ability to design and perform trials that are adequately blinded and randomised, resulting in lower quality studies[13].
The limited number of studies and small sample sizes reduce the ability of meta-analysis to draw meaningful conclusions due to insufficient sample sizes and wider confidence intervals when describing effect estimates. As a result, the pooled findings may lack precision and reliability. Furthermore, when only a handful of studies are available, each study disproportionately influences the overall result, increasing the risk of bias and reducing the robustness of the meta-analysis[14]. Efforts to improve the number and size of trials, such as the establishment and utilisation of disease registries, are discussed below.
Quality of included studies: Observational studies vs RCTs
RCTs, considered the gold standard of trial design, minimise selection bias through blinding and randomisation; however this is time-consuming, expensive, and generally requires large sample sizes in order to demonstrate a significant effect. In more common diseases such as depression, meta-analysis can be performed using data predominantly from large RCTs, which increases statistical power and adds precision in estimating treatment effects[15]. However, in rare diseases, this is less feasible, and clinical trials are therefore more likely to be uncontrolled or unblinded in their design[16,17]. Guidelines, treatment protocols, and meta-analysis for rarer diseases often also rely on observational studies, which, whilst providing valuable insights, are inherently more susceptible to bias, confounding, and measurement errors[18].
This lower-quality evidence creates a challenge in performing a meta-analysis. The Grading of Recommendations Assessment, Development, and Evaluation Framework[19] is well-established guidance for rating the quality of evidence and emphasises the importance of study design when performing a meta-analysis. The framework highlights that poorly designed studies often report exaggerated treatment effects due to methodological flaws, which have been shown to distort pooled estimates[20]. Lower-quality studies are also more likely to introduce various forms of bias, such as selection or publication bias, which further compounds challenges when performing a robust and accurate meta-analysis.
Bias
An important purpose of performing meta-analyses is to reduce bias in the conclusions of systematic reviews. However, as discussed for rare diseases, there are often few studies available, and those that are available contain small sample sizes and are often of low quality. This is exaggerated by publication bias, where preferential publication is given to studies with statistically significant or positive results. The inherent difficulties of examining rare pathologies mean that it is often not possible to achieve sufficient power to provide significant positive results, resulting in negative publication bias. This skews the evidence base, and a meta-analysis may therefore rely disproportionately on positive studies, which could inflate the estimates of treatment effects or disease associations[21]. This publication bias can also increase the between-study variance, which subsequently reduces the precision of results[22]. The small sample sizes of included studies further increase the risk of bias. Studies with small sample sizes, as commonly seen in rare pathologies, are more likely to be unblinded and therefore more prone to selection bias[23]. Selection bias is also inherent to disease registries, often used to overcome the epidemiological challenges posed by rare pathologies, given the process by which researchers enrol participants into these entities. Methods to address this bias are discussed in greater detail below.
Heterogeneity
Heterogeneity in meta-analysis poses a challenge, particularly in rarer diseases, where it is more common[24], and can arise from several sources. Patients who suffer from rare diseases often exhibit a wide range of symptoms, disease progression, and responses to treatment, and this heterogeneity can make it more difficult to draw meaningful conclusions from pooled data, exacerbated by the low numbers usually involved in studies[25]. The paucity of data available for many rare diseases also results in a lack of standardised protocols for diagnosis and treatment, resulting in patients with diverse phenotypes and treatment requirements being classified under the same rare disease. Treatment approaches may also vary significantly between different centers or countries, contributing to inter-study differences and compounding the heterogeneity[26]. Heterogeneity within the study design, specifically with regard to trial protocols, outcomes, and the methods used to measure outcome results, also hinders data pooling and meta-analysis comparability. For example, a meta-analysis investigating treatment strategies in patients with mucopolysaccharidosis identified 23 studies with a wide variety of efficacy outcomes, such as measuring glycosaminoglycan levels in cerebrospinal fluid in 3 studies or respiratory function in 6 studies[13]. This resulted in low or extremely low confidence in the evidence and an inability to establish definite conclusions.
Furthermore, whilst it is well established that differences in patient demographics such as age, ethnicity, or co-existing conditions can influence treatment outcomes[27], accounting for these differences in relatively small sample sizes and limited studies can be challenging[28]. Using Bayesian methodology in this scenario has been proposed as a superior method to account for heterogeneity when conducting a meta-analysis[29]. Additionally, established statistical measures such as I2 and Q tests, which attempt to quantify heterogeneity, can be imprecise when a small number of studies are included in the meta-analysis and potentially lead to incorrect inferences about heterogeneity[30].
Sparsity of data
As discussed, the low prevalence of rare diseases leads to both a limited number of available studies and small sample sizes within these studies. This results in few events per variable and therefore outcome data can be sparse, which, in meta-analyses, results in wide confidence intervals and unstable estimates[31]. Undertaking a meta-analytical summary of logistic regression estimates using small sample sizes and sparse data points can also introduce or propagate finite sample bias. This has been demonstrated by Richardson et al[32], who identified, using simulated data, that results were biased away from the null in smaller sample sizes and when the number of covariates was large, which is common in healthcare research.
Inappropriate statistical methods
Sparse data is common when investigating rare diseases, which means that zero-event studies are more common, which introduces difficulties when estimating effect size and variance. Whilst the continuity corrections method can be utilised to adjust for this, this itself can introduce bias and may underestimate the size of treatment effects[33]. Methods such as Peto's odds ratio offer reliable estimates when events are rare and effect sizes are small, though they require balanced group sizes[34]. Alternative approaches, such as generalised linear mixed models and beta-binomial models, can also better account for between-study variability and sparse data[35]. These approaches avoid the bias introduced by continuity correction methods and provide more accurate and interpretable estimates, especially when handling zero-event or small-sample studies. As discussed, performing research into rare diseases often involves both insufficient and lower-quality data and conventional meta-analytic techniques, which assume the opposite, are therefore inappropriate. For example, the DerSimonian-Laird method, a commonly used random-effects estimator, has been shown to underestimate between-study variance when data are sparse, leading to misleadingly narrow confidence intervals[36].
Fixed-effect models, which assume homogeneity across studies, are also not useful in rare diseases due to the high inter-study and inter-individual variability[37]. Similarly, studies have demonstrated that random-effects models can also perform poorly with few studies and high heterogeneity[38]. Methods used to detect bias, such as the funnel plot for publication bias, lose diagnostic utility in the context of rare diseases due to the limited number of studies and high inter-study variability[39]. Sterne et al[40] performed a simulation using meta-analyses containing 5, 10, 20, and 30 trials and found that the ability of statistical methodology, such as the rank correlation test or a weighted regression method, to detect even moderate bias significantly decreased as the number of trials became smaller.
Generalisability
The prevalence of rare diseases often necessitates studies to be conducted in specialised tertiary centers with unique expertise, experience, and resources. Subsequently, patients enrolled in rare disease studies may not represent the broader patient population due to referral bias, geographic differences, or inclusion criteria that favours more severe cases[41]. Examples of this were studies into cystic fibrosis or Wilson's disease, where academic output, including the publication of clinical trials, is significantly biased towards tertiary centers[42]. When these studies are used to perform a meta-analysis, the bias created by this disparity in academic output raises questions about the generalisability to wider populations.
OVERCOMING THE CHALLENGES OF META-ANALYSIS IN RARE DISEASES
As discussed, a key challenge when performing a meta-analysis in rare pathologies is related to the low prevalence of these diseases, resulting in low numbers of studies, small sample sizes, lower quality studies, and challenges of generalisability. Collaborative research networks, such as the International Rare Diseases Research Consortium[43] or national rare disease collaborative networks run by the National Health Service in the United Kingdom[44], and which are part of the European Reference Networks[45,46], were established to address this. These networks have provided opportunities for researchers to study rare diseases in more detail and perform high-quality RCTs, which were not previously possible for some diseases. Despite their promise, challenges in establishing such collaborative networks include data sharing limitations, regulatory inconsistencies, particularly when establishing international networks, funding disparities, and infrastructural gaps. The European Reference Networks is a positive example of overcoming these challenges; the leadership of the European Union Council initially ensured that this was placed on the agenda with full political support, a clearly defined goal and timeframe was set, full funding was in place with pan-European backing and a structured plan for creation and implementation was developed with a framework for regular re-assessment and modification of the plan as required. Processes were developed for data sharing, guideline sharing, and virtual consultations in order to overcome the challenges faced by collaborative networks[45,46]. It is hoped that as these and other registries and networks mature, their impact will increase, which will improve the quality and quantity of data available for meta-analysis[47]. This will also expand the data pool used to include more diverse populations, which will improve the generalisability of meta-analysis findings[48].
Modifying the methodology employed when performing a meta-analysis also offers opportunities to overcome challenges described earlier. Selection bias is a persistent problem when investigating rare diseases, particularly with case-control trials and, therefore, the meta-analyses that include this data. To correct this, Cole et al[49] demonstrated that, in the case of a disease registry for Gaucher disease, a novel case-control matching analysis using a risk-set method was able to remove bias and has potential utility in future studies.
Using different methods to assess for random effects in meta-analysis may also be beneficial to provide a more rigorous statistical analysis. Commonly employed techniques, such as the DerSimionian-Laird approach, can over-report statistically significant results in rare diseases[50]. The Hartung-Knapp-Sidik-Jonkman (HKSJ) method is an alternative to traditional random-effects meta-analysis and is especially effective when the number of studies is small or heterogeneity is high, both of which are common in rare disease research, as discussed. Unlike the DerSimonian-Laird method, HKSJ adjusts the confidence intervals using a more accurate estimate of between-study variance, providing better error control. A case study by IntHout et al[51] demonstrated that HKSJ yields more conservative and reliable results in small-sample meta-analyses of clinical trials. However, caution is still required as a critique by Jackson et al[52] found that whilst the HKSJ method can produce shorter confidence intervals with smaller P values, it can produce conservative results for effect estimation and overestimate heterogeneity, which can reduce the accuracy of the analysis.
Bayesian methodology combines prior knowledge with observed data using Bayes' Theorem to produce a posterior distribution of the parameter of interest[53]. It uses likelihood functions and priors, often computed via Markov chain Monte Carlo methods, allowing flexible, probabilistic inference; ideal for rare diseases with sparse data and small sample sizes. This approach can also be employed to address issues with regard to bias and sparse data, although this can introduce subjectivity, which requires cautious interpretation[54]. A recently published example is a systemic review and network meta-analysis by Huang et al[13] investigating mucopolysaccharidosis using Bayesian methodology to overcome the challenges described and provide a robust interpretation of the available evidence. Furthermore, Sweeting et al[55] highlighted that both fixed and random effects models have limitations under the conditions of sparse data, such as the introduction of bias when the ratio of study sizes changes, and recommended routinely using Bayesian shrinkage approaches for improved estimation.
Despite these modifications, it may not always be possible to overcome the aforementioned challenges and produce robust and meaningful analysis. Therefore, a scoping review may be a more appropriate methodology[56]. Scoping reviews - defined as “a means to identify the types, characteristics, concepts, or definitions of the evidence available for a topic”[56] - have been used on many occasions when only low-quality data is available for analysis. A recent example was when studying inflammatory bowel disease in the transgender community, a scoping review identified unique and poorly known health needs for this small population[6]. A further example was a review that aimed to assess clinical decision support systems for rare diseases. These cover a wide range of processes in different populations with only low-quality heterogeneous data available. The scoping review was able to clarify the strengths and limitations of these systems and provide useful guidance to clinicians[57].
Type I and II error
In hypothesis testing, a type I error occurs when a true null hypothesis is incorrectly rejected, meaning the test suggests there is an effect or difference when in fact none exists. This is also known as a false positive, and the probability of making this error is denoted by a (alpha), typically set at 0.05. In contrast, a type II error happens when a false null hypothesis is not rejected, leading to a false negative result. This means the test fails to detect a real effect or difference. The probability of a type II error is denoted by β (beta), and the power of a test - its ability to detect an actual effect - is calculated as 1 - β.
Zero-event studies
Zero-event studies are clinical trials or observational studies in which no events of interest occur in one or both comparison groups (e.g., no adverse events, deaths, or disease recurrences). These studies often arise in meta-analyses of rare outcomes or rare diseases, where the event rate is very low. Although they provide valuable information, zero-event studies present statistical challenges, particularly in estimating effect sizes and variances, and may require specialized methods or continuity corrections to be included in meta-analyses.